 |
 |
| Plant Pathol J > Volume 38(5); 2022 > Article |
|
Abstract
Brown blotch disease, caused by Pseudomonas tolaasii, is one of the most serious diseases in mushroom cultivation, and its control remains an important issue. This study isolated and evaluated pathogen-specific bacteriophages for the biological control of the disease. In previous studies, 23 varieties of P. tolaasii were isolated from infected mushrooms with disease symptoms and classified into three subtypes, Ptα, Ptβ, and Ptγ, based on their 16S rRNA gene sequences analysis and pathogenic characters. In this study, 42 virulent bacteriophages were isolated against these pathogens and tested for their host range. Some phages could lyse more than two pathogens only within the corresponding subtype, and no phage exhibited a wide host range across different pathogen subtypes. To eliminate all pathogens of the Ptα, Ptβ, and Ptγ subtype, corresponding phages of one, six, and one strains were required, respectively. These phages were able to suppress the disease completely, as confirmed by the field-scale on-farm cultivation experiments. These results suggested that a cocktail of these eight phages is sufficient to control the disease induced by all 23 P. tolaasii pathogens. Additionally, the antibacterial effect of this phage cocktail persisted in the second cycle of mushroom growth on the cultivation bed.
Oyster mushrooms (Pleurotus ostreatus) rank first in terms of cultivation area, production, and consumption in the Korean mushroom industry. Although they show high profitability, the number of farms and cultivation areas have decreased due to the frequent occurrence of the bacterial brown blotch disease (Iacobellis and Lo Cantore, 1998; Ministry of Agriculture, Food and Rural Affairs, 2021). Brown blotch disease is caused by the bacterial peptide toxin, tolaasin, and its structural analogs, secreted by Pseudomonas tolaasii (Tolaas, 1915). These peptides form pores in cellular membranes, disrupt mushroom tissue, and form blotches on the surface of mushroom caps. The occurrence of brown blotch disease is typically prevented by sterilizing underground water used for mushroom cultivation, cleaning the cultivation house, and fumigating it with hot air (Geels et al., 1991). Mushroom cultivation under plastic mulching or in plastic bags and bottles has been proved to provide greater protection against the disease; however, these physical methods are frequently unsuccessful in consecutive cultivation.
Biological control of the brown blotch disease using antagonistic bacteria reduced the loss of mushroom production (Lee et al., 2014). Antibiotics are powerful for controlling diseases (Geels, 1995), but their field application for edible mushrooms is limited. Significant efforts have been made to develop methods to suppress these pathogenic bacteria. Bacteriophages have been recognized as an important alternative tool in controlling antibiotic-resistant pathogens. Phage therapy employs a specific virulent bacteriophage to kill pathogenic bacteria, and it had been extensively studied and improved until the 1940s when its popularity declined with the advent of antibiotics. Recently, the rise of multidrug-resistant bacteria, such as methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus, has become a threat to human health. Therefore, phage therapy is actively being researched as a substitute for antibiotics. Notably, the U.S. Food and Drug Administration approved the use of phages to target Listeria species and Escherichia coli strains for which no appropriate antibiotics are available (Food and Drug Administration, 2006). In addition, phage therapy has been studied for the control of Vibrio cholerae (Yen et al., 2017), Pseudomonas aeruginosa (Oechslin et al., 2017), Escherichia coli (Pereira et al., 2017), Enterococcus faecalis (Khalifa et al., 2016), and Staphylococcus species (Abedon, 2016).
The application of bacteriophages in agricultural practices and food product industries has been legally approved in several countries (Fernández et al., 2018). Bacteriophages have been found to be effective for the control of several pathogenic phytobacterial strains, such as Erwinia sp., causing bacterial soft rot and fire blight on apple and pear (Park et al., 2018), Xanthomonas sp., pathogenic strains of bacterial spot of tomato, peach, geranium, citrus, walnut blight, leaf blight of onion, and citrus canker (Dong et al., 2018), Ralstonia solanacearum, provoking bacterial wilt of tomato (Wang et al., 2019), and Streptomyces scabies, producing potato scab (Abdelrhim et al., 2021).
In previous studies, 23 strains of P. tolaasii were isolated and divided into three subtypes, Ptα, Ptβ, and Ptγ, based on the phylogenetic analysis (Yun et al., 2013). Their pathogenic toxicities were evaluated by the pitting test, white line formation, and hemolytic activity. Although all three subtypes caused the blotch disease, only the Ptα subtype strains produced tolaasin and its analog peptides, indicating that pathogenic properties of both Ptβ and Ptγ subtype strains differ from those of the Ptα subtype (Yun and Kim, 2020). In this study, bacteriophages were isolated and bactericidal activities were tested against these host P. tolaasii strains. They were successfully able to suppress the disease development on the mushrooms infected with the pathogens. Therefore, the present study aimed to develop an optimal protocol for the biological control of brown blotch disease using a cocktail of the selected bacteriophages.
Various P. tolaasii strains isolated in previous studies were used as host strains of bacteriophages. P. tolaasii strains were isolated from oyster mushrooms severely damaged by brown blotch disease, as described by Lee et al. (1997). The mushroom tissues were homogenized, and the homogenates were subjected to centrifugation. The supernatants were spread onto a Pseudomonas Agar F media (PAF; bacto-peptone 10 g, bacto-tryptone 10 g, K2HPO4 1.5 g, MgSO4·7H2O 1.5 g, glycerol 10 ml, agar 15 g/l), and P. tolaasii strains were isolated and identified by 16S rRNA gene sequencing, a white line test (Munsch et al., 1991), and a pitting test on the mushroom tissue (Yun et al., 2013). After that, the isolated bacteria were stored at −70°C in a PAF broth containing 20% (v/v) glycerol until use.
The collected P. tolaasii strains were divided into three subtypes, Ptα, Ptβ, and Ptγ subtype. The isolated 23 strains include six of Ptα subtype: P. tolaasii 6264 (GenBank accession no. JN187439), HK1 (MK355659), HK2 (MK355660), HK3 (MK355661), HK4 (JX417441), and HK5 (JX417440), 16 of Ptβ: HK7 (JQ684100), HK8 (MK355662), HK9 (JX417437), HK10 (JX417439), HK11 (MK355663), HK12 (MK355664), HK13 (MK355665), HK14 (MK355666), HK15 (MK355667), HK16 (MK355668), HK17 (JX417438), HK18 (MK355669), HK19 (MK355670), HK20 (MK355671), HK21 (MK355672), and HK22 (JQ684101), and one of Ptγ, P. tolaasii: HK23 (JQ684099) (Yun et al., 2013).
Bacteriophages were isolated by using P. tolaasii strains as the host strains. Various sewage samples obtained from a rural area of Cheongju, Korea, were mixed with the host bacterial culture for primary isolation of the phages. The mixture was added to a 0.75% semisolid agar medium at a 1:2 ratio (v/v) and poured onto a 1.5% solid agar plate. The double-layered plate was incubated for 15 h at 25°C. One of the phage plaques in the incubated plate was chosen and added to the overnight culture of the corresponding P. tolaasii host strain.
Following the method described by Chibani-Chennoufi et al. (2004), phage lysate was a supernatant obtained by centrifuging the culture medium inoculated with the phage and host strain. NaCl was added to the phage lysate at a concentration of 10%, incubated for 1 h at 0°C, and centrifuged at 6,000 ×g for 10 min. Polyethylene glycol (PEG-6000) was added to the supernatant at a concentration of 10% and incubated for 1 h at 0°C. Phages were collected by centrifugation at 19,000 ×g for 10 min (XL-90; Beckman Coulter, Pasadena, CA, USA). The precipitated phages were resuspended in a phage buffer (50 mM tris(hydroxymethyl)aminomethane-HCl, 150 mM NaCl, 20 mM NH4Cl, 10 mM MgCl2, 1 mM CaCl2, 0.2% gelatin, pH 7.4). The resuspended phages were filtered using a 0.2 μm microfilter and stored at −70°C.
Phage titer was determined using the double-layer agar technique (Chibani-Chennoufi et al., 2004). Phage suspensions were diluted 104-109-fold and mixed with the culture media of the host bacteria that was previously added to 3 ml of soft agar and poured onto a PAF hard agar plate. The plate was incubated for 15 h at 25°C, and plaques formed in the plate were counted. To measure the bactericidal activity, the phage lysate at a final concentration of 1% (v/v) was added to the PAF broth containing 5% of host cell culture and incubated at 25°C for 15 h. The final concentrations of bacterial strains and phage cocktail were 2 × 106 cfu/ml and 1 × 105 pfu/ml, respectively. Bacterial growth was measured by using a UV/Vis spectrophotometer (U-2000, Hitachi, Tokyo, Japan) at 600 nm.
Seedlings of oyster mushrooms were cultivated in three replicated beds 1 m wide and 2 m long filled with a sterilized sawdust medium. After 3 weeks, when the bed was fully covered with mycelia, the temperature in the cultivation house was lowered to below 18°C to induce the development of the fruiting body. The mixture of pathogen culture media (5 × 106 cfu/ml) and the mixture of eight phages (2 × 108 pfu/ml) were sprayed onto the beds. Acrylic plate walls 20 cm high were installed to minimize the disturbance among the control and the experimental plots. The bacterial mixture (Ptαβγ) and phage mixture (ɸPtα, ɸPtβ, ɸPtγ, and ɸPtαβγ) were made independently and the mixtures applied sequentially. There were three replicated mushroom-cultivating beds and each mushroom bed was divided into six sections (Control, Ptαβγ + ɸPtαβγ, Ptαβγ, Ptαβγ + ɸPtα, Ptαβγ + ɸPtβ, and Ptαβγ + ɸPtγ) and each section was 0.3 m wide and 1 m long. Five milliliters of each mixture were applied. The ratio of each one of 23 strains in the mixture was about 4% and the 0.5 ml of each bacterial culture was added to the mixture. Each bacterial strain had a level of 5 × 106 cfu/ml in the mixture. Furthermore, the ratio of each phage in the mixture was 12.5% and the 0.5 ml of each phage lysate was added to the mixture and each phage had a level of 2 × 108 pfu/ml. Protective effects of the phage cocktail were measured by the simultaneous treatment of phage cocktail right after pathogen treatment and by the treatment of phage cocktail at 12 h after the pathogen treatment, before blotch disease development.
Disease symptoms appeared within 1-3 days after the treatment of pathogens and phages. When the seedlings of oyster mushrooms turned brown, pictures of the control, pathogen-treated, and pathogen plus phage-treated mushrooms were taken. The effects of phage therapy were evaluated by color analysis using the Adobe Photoshop CS program (Adobe Inc., San Jose, CA, USA). On the mushroom surface, more than 10 pixels were chosen and the values of three colors, red, green, and blue (RGB), were measured and averaged. The RGB ratio is the value obtained by dividing the number of pixels in the area occupied by each color by the total number of all pixels. Then, the RGB ratios taken from each experimental plot were drawn.
For the biological control of brown blotch disease, virulent bacteriophages against P. tolaasii strains were isolated from various sewage samples, streams, and ponds in Cheongju City. A total of 31, 9, and 2 isolated phages were virulent to Ptα, Ptβ, and Ptγ subtype pathogens, respectively (Supplementary Table 1). To prepare a phage cocktail containing the least number of phages, phages were selected based on the range of their host specificity. Since the isolated phages showed different host ranges, 3, 6, and 1 phage types were identified against Ptα, Ptβ, and Ptγ subtype pathogens, respectively (Fig. 1).
Phages were often virulent to other host strains only within the same subtype of pathogens, but none of the phages showed bacteriolytic activity against the strains of another subtypes. Three phage types were isolated against the host strains of the Ptα pathogen. All phages of the Ptα subtype were virulent to host strains, P. tolaasii 6264 and HK2. Phages ɸ6264 and ɸHK1 killed host strains, P. tolaasii HK1 and HK3, and phage ɸ6264 killed P. tolaasii HK4 and HK5. Six types of phages were isolated against the Ptβ subtype pathogens, while Ptβ pathogens were divided into 10 host groups depending on their phage sensitivity. P. tolaasii HK7 was susceptible to four different phage types, and six out of the 10 host groups were susceptible to only one type of phage. These results show that the host strains of Ptβ subtype are diverse in phage sensitivity. Therefore, a minimal combination of phages should include eight selected phages: one, six, and one against the host strains of Ptα, Ptβ, and Ptγ subtype, respectively (highlighted in green in Fig. 1). There were phages with a very narrow host range, such as ɸHK3, ɸHK8, and ɸHK15, killing only their corresponding host strains. Since these host strains were also killed by other phages, these phages were excluded from Fig. 1. Host ranges of 42 bacteriophages isolated in this study were investigated. Most of the phages killed only their corresponding host strains showing a specific host range. However, seven out of 42 phages had a wide host range killing other strains as well as their host strains. The aim of phage therapy was to kill the maximum number of pathogenic bacterial strains using minimum number of phages. Therefore, the host ranges of the isolated phages were investigated and the results were showed in Supplementary Tables 2-4.
For successful phage therapy, we examined the effect of phage combinations of all phage subtypes. Pathogen mixtures of the 23 strains, including six Ptα, 16 Ptβ, and one Ptγ subtype strains, were prepared. Phages were selected based on their host specificity to obtain the minimum number of phages required for the cocktail: phage ɸ6264 against the Ptα subtype pathogens; phages ɸHK7, ɸHK11, ɸHK14, ɸHK16, ɸHK19, and ɸHK22 against the Ptβ subtype; and phage ɸHK23 against the Ptγ subtype. Growth curves of the three subtype pathogen mixtures are shown in Fig. 2. All pathogen mixtures showed similar growth rates, reaching the stationary phase after 10 h of incubation. However, when the pathogen mixtures were incubated with the corresponding selected phages, complete bacterial lysis was observed for the Ptα and Ptγ subtype pathogens. In contrast, the absorbance of the culture medium inoculated with Ptβ subtype strains increased until about 8 h of incubation, and then gradually decreased as bacterial lysis proceeded.
Protective effects of the phage cocktail were evaluated on oyster mushroom cultivation. P. tolaasii strains were inoculated on the surface of young fruiting bodies, and the corresponding phages were applied around the inoculation area (Fig. 3). The effect of phages was evaluated for 5 days after the treatment. The pathogen mixture of the 23 strains (Ptαβγ) produced disease symptoms, such as brown coloration, blotches, and shrinkage of small fruiting bodies after 3 days of treatment. These symptoms were followed by tissue collapse of the small fruiting bodies, severe blotching, and the decay of mushroom caps 2 days later. However, no noticeable symptoms of brown blotch disease were observed in mushrooms treated with the phage cocktail of eight phages (Ptαβγ + ɸPtαβγ), and the shape and color of fruiting bodies remained healthy. Moreover, severe brown blotches were observed in all three treatments with the 23 pathogens (Ptαβγ) and one of the three phage subtypes (Ptαβγ + ɸPtα, Ptαβγ + ɸPtβ, and Ptαβγ + ɸPtγ). The subtype phages in each treatment lysed only their corresponding pathogens. These results demonstrate that the phage cocktail of the selected eight phages successfully controlled the disease by killing all P. tolaasii pathogens or by suppressing the increase in pathogen population.
The effects of the phage cocktails were evaluated by comparing color changes of fruiting bodies obtained by various pathogen and phage treatments (Fig. 4). From the color pictures of fruiting bodies, several spots of mushroom surfaces were analyzed by measuring components of three colors, RGB. In both the control and all pathogens plus phage cocktail-treated (Ptαβγ + ɸPtαβγ) mushrooms, the proportions of the three colors evaluated were very similar at 30-35%. However, in the pathogen-treated (Ptαβγ) and pathogen plus one type phage-treated (Ptαβγ + ɸPtα, Ptαβγ + ɸPtβ, and Ptαβγ + ɸPtγ) mushrooms, the proportion of red appeared over 50%, while that of blue was only near 15%. Thus, the results of the color proportion analysis indicate that the phage cocktail-treated mushrooms were very similar to the normal mushrooms in color, indicating that the cocktail is highly successful in suppressing the blotch disease.
To investigate the persistence of the phage cocktail effects, both healthy and infected mushrooms were harvested from the experimental beds, and the surface of the beds was briefly washed with sterilized distilled water. To stimulate the mushroom growth for the second cultivation cycle, the temperature was maintained at 18°C. Compared with the control, brown spots and blotches were observed on the surface of the newly produced fruiting bodies only in beds pretreated with the pathogen mixture and the mixture plus one phage subtype (Ptαβγ, Ptαβγ + ɸPtα, Ptαβγ + ɸPtβ, and Ptαβγ + ɸPtγ) (Fig. 5). These results showed that the pathogenic bacteria remained in the beds and caused the blotch disease during the second cultivation cycle. In the second growth cycle, the number of young fruiting bodies decreased, and their discoloration became severe with the growth. However, in the bed treated with the mixture of pathogen strains and the phage cocktail (Ptαβγ + ɸPtαβγ), healthy mushrooms developed, and the disease was completely suppressed. The bactericidal effect of the phage cocktail may persist throughout the second cycle of mushroom growth and suppressed the development of brown blotch disease.
In another set of experiments, inoculation with the mixture of all pathogens (Ptαβγ) resulted in the development of brown spots across the entire fruiting bodies of the mushrooms. When the phage cocktail was sprayed 12 h after pathogen inoculation, complete suppression of the disease was observed (Fig. 6). These results suggested that the phage cocktail successfully attacked and killed the pathogens when applied to the infected mushrooms before the disease symptoms developed. Therefore, the control effect of phage therapy on the pathogens lasted up to the second cycle of the mushrooms production and it had an excellent control effect even though the mushrooms were already contaminated by pathogens.
The analysis of 16S rRNA gene sequence has been used previously for the identification of pseudomonads (Godfrey et al., 2001). However, identifying P. tolaasii strains based on the 16S rRNA gene sequences may not reveal the full diversity of the genus (Ranjan et al., 2016). Therefore, in other to classify various strains of pathogenic P. tolaasii, pathogenic characters, such as tolaasin secretion, white line formation with P. reactans culture, and blotch formation through the pitting test, were measured along with the analysis of 16S rRNA gene. Significant differences in pathogenic properties among the P. tolaasii subtypes were observed in previous studies. The Ptα subtype exhibited typical pathogenic traits, including secretion of tolaasin and its analog peptides, hemolytic activity, blotch-forming ability, and white line formation in the presence of white line-inducing principle, secreted by P. reactans (Munsch et al., 1991). The strains of the Ptβ subtype did not secrete peptide molecules of the tolaasin type and did not exhibit hemolytic activity; however, they still caused the brown blotch disease. The Ptγ subtype also did not secrete tolaasin peptides, but it showed hemolytic activity and blotch-forming ability (Yun and Kim, 2020). Therefore, pathogenic strains of three subtypes causing the brown blotch disease were used as host strains to expand the therapeutic applications of the phage cocktail.
The phages used in the present study virulent to the pathogens of brown blotch disease were isolated from a wide array of agricultural and urban areas, indicating the presence of the host pathogens outside the mushroom farming area. Phages lethal to host strains of the Ptα subtype were obtained from most sewage samples. The pathogenic strains of the Ptα and Ptγ subtypes were very sensitive to their corresponding phages and were killed by a short incubation period with phages. However, the strains of the Ptβ subtype were less sensitive to their corresponding phages, and bacterial lysis was retarded and occurred after 8 h of incubation (Fig. 2). Phages were classified based on their host specificity. The host range of the phage is defined as the span of hosts sensitive to the phage, and host specificity is dependent on many bacterial characters, such as the cell wall structure and immunity determined by the presence of restriction enzymes (Hyman and Abedon, 2010). In this study, the host range of the isolated phages was limited within the host strains of the same subtype. Therefore, phage collection for the biological control of brown blotch disease should be performed separately for each host subtype. To prepare a phage cocktail, the number of selected phages in each host subtype should be minimized such that the number of phages is smaller than that of the host strains. We predicted that a mixture of eight phages (one in ɸPtα, six in ɸPtβ, and one in ɸPtγ) could control the 23 strains of P. tolaasii pathogens: six, 16, and one of the Ptα, Ptβ, and Ptγ subtypes, respectively. This mixture successfully prevented blotch formation, suggesting that the selected phage cocktail effectively suppressed brown blotch disease by destroying all 23 P. tolaasii strains.
When various P. tolaasii strains were co-cultured, it could be dominated by one or few strains. In Fig. 3, while the treatment of each subtype phage killed their corresponding bacterial strains, the pathogens of other subtypes grew and caused brown blotch disease on cultivated mushrooms. These results show that pathogenicity of each subtype pathogen is not limited by the co-culture of other subtype pathogens. Since the cocktail of eight phages (ɸPtαβγ) was able to control the mixture of all 23 pathogens (Ptαβγ) and the disease symptom was not observed, the possible dominancy of one or a few pathogens may not influence the positive effect of phage therapy. In the co-culture of interspecies or intraspecies strains, competition, cooperation, or co-evolution between the strains may occur (Hibbing et al., 2010). Also, they are able to induce quorum sensing of the strains by secreting signaling molecules (Smith and Schuster, 2019).
The phage cocktail was confirmed to prevent the brown blotch disease in the shelf cultivation of oyster mushrooms (Figs. 3 and 6), and its effectiveness extended over two successive cultivation cycles (Fig. 5). Although positive effects of the phage cocktail were observed, monitoring the populations of pathogenic bacterial strains that decreased by phages in the mushroom tissues may be practically difficult since the presence of other natural bacterial strains and phages interferes with the identifications of pathogenic strains and phages used in this experiment. However, in the treatments of all 23 pathogens plus eight phages of the cocktail, no symptoms of blotch disease were observed. These results indicate that the phage cocktail was successful in controlling all pathogenic bacterial strains and each phage in the cocktail killed the corresponding pathogens independent of the presence of other phages. Furthermore, the effect of phage treatment was clearly obtained in the experiments with both simultaneously and consecutively treatments of pathogen and phage. Since phages were able to survive longer than a month in the mushroom-growing media, the bactericidal activity was always obtained with phages and it was independent of the order of pathogen and phage treatments.
To evaluate the effects of the phage cocktail, color analysis of the mushroom cap picture was performed. The control and pathogen (Ptαβγ) plus phage cocktail (ɸPtαβγ)-treated mushrooms had a normal greyish color. However, disease symptoms were very clear in the mushrooms treated with pathogen and pathogen plus one component of subtype phages. Color compositions of the disease-symptom mushrooms were very similar among the mushrooms treated with different subtypes of pathogens, suggesting that the mechanism and phenotype of blotch disease may be identical among different subtypes of pathogenic bacteria. Surface color analysis has been used in quality assessment studies of foods and is considered as one of the primary characteristics of evaluating food quality (Cubero et al., 2011; Pedreschi et al., 2010). In our previous study, we also used a color analysis for measuring the effect of inhibiting the green mold disease that occurred by Trichoderma harzianum in oak mushrooms (Lentinus edodes) (Lee et al., 2017; Yun et al., 2018b).
In conclusion, a combination of eight phages was used to prepare a phage cocktail that successfully suppressed the brown blotch disease by killing all pathogens. These results further imply that the bactericidal activity of individual phages in the phage cocktail was not interrupted by the presence of other bacterial strains and phages. Therefore, when a new pathogen is detected, the cocktail can be optimized by adding a corresponding phage. However, one major weakness of phage therapy is the development of phage resistant mutant (PRM) strains, which is frequently induced by the presence of phages. Our previous study demonstrated that the PRM strains showed a relatively weaker pathogenic activity than the original pathogen (non-resistant strains) (Yun et al., 2018a). Nevertheless, the effectiveness of the phage cocktail in the presence of various PRM strains is currently under investigation. Phage cocktails were found to be effective against many pathogens including Escherichia coli (Gundogdu et al., 2016). Phage therapy with a relatively low production cost will be a great advantage in commercial use after completion of the related research on the composition and storage conditions of phage strains to maintain their bactericidal activities.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2017R1D1A3B03032718).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Phage typing according to host specificity. Green shades indicate phages selected for phage cocktail preparation. aO, susceptible to phage. bX, resistant to phage.
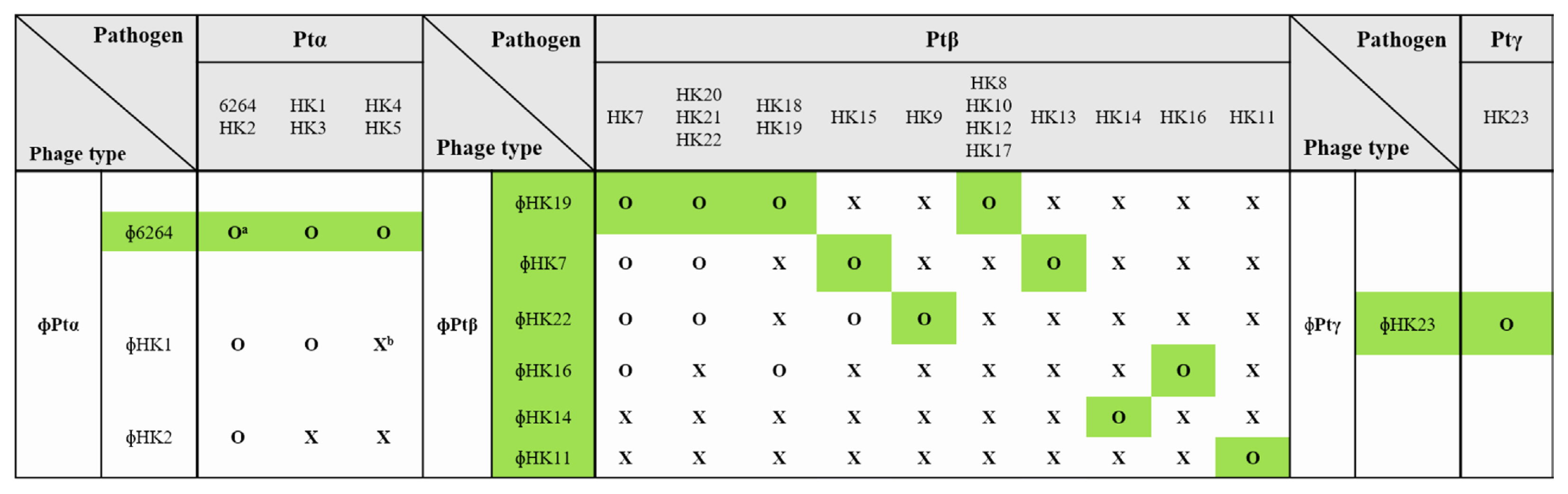
Fig. 2
Lysis curves of Pseudomonas tolaasii strains by the corresponding bacteriophages. Growth curves of Ptα, Ptβ, and Ptγ were obtained with the mixture of six Ptα, 16 Ptβ, and one Ptγ subtype strains, respectively. Ptα + ɸPtα: a mixture of six Ptα pathogen strains plus phage ɸ6264. Ptβ + ɸPtβ: a mixture of 16 Ptβ subtype strains plus six Ptβ phages selected in Fig. 1. Ptγ + ɸPtγ: P. tolaasii HK23 strain plus phage ɸHK23.

Fig. 3
Effects of the phage cocktail on the development of blotch disease. Control: treated with sterilized culture medium. Ptαβγ: all 23 strains of P. tolaasii pathogen. Ptαβγ + ɸPtαβγ: all P. tolaasii strains plus eight phages of the cocktail. ɸPtα: Ptα phage, ɸ6264. ɸPtβ: six Ptβ phages, ɸHK7, ɸHK11, ɸHK14, ɸHK16, ɸHK19, and ɸHK22. ɸPtγ: Ptγ phage ɸHK23. In each experiment phage mixture was sprayed on the mushroom immediately after pathogen inoculation.

Fig. 4
Color component evaluation of the brown blotch area obtained from shelf-cultivated mushrooms. Red-green-blue (RGB) ratios were calculated from photos of mushrooms obtained by the control, Ptαβγ + phage cocktail (ɸPtαβγ), and Ptαβγ treatments. Those of all pathogens plus one type of phage cocktail treatments were shown in Ptαβγ + ɸPtα, Ptαβγ + ɸPtβ, and Ptαβγ + ɸPtγ. Data are mean ± standard error percentages of the results from 7-8 replicates of more than three mushrooms. The same letters are not statistically significances at P < 0.05 (Tukey’s test).
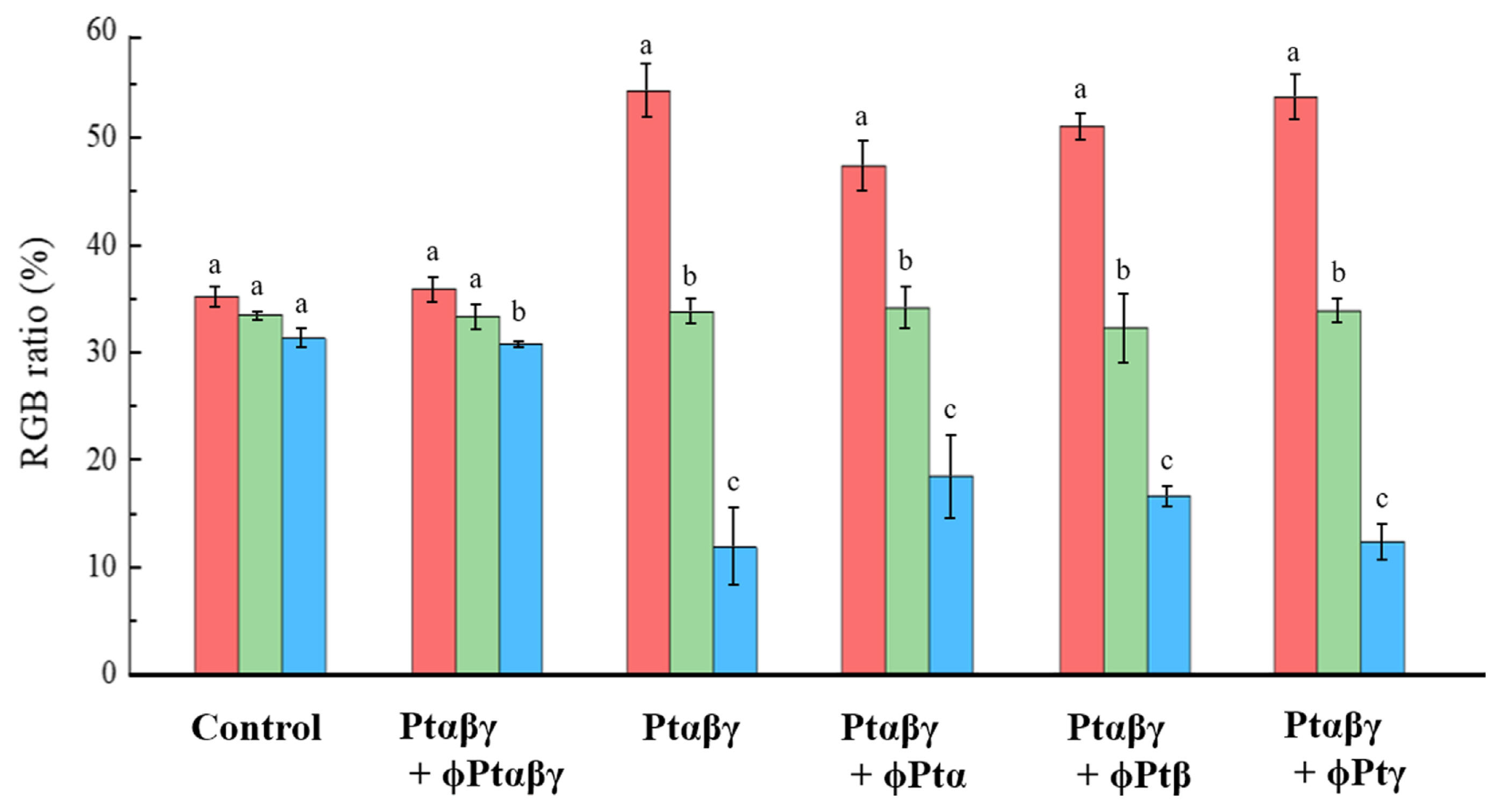
Fig. 5
Persistent effect of the phage cocktail during the second cultivation cycle. After the removal of mushrooms from experimental beds shown in Fig. 2, the second growth cycle was monitored without any treatment. Disease development was evaluated by brown coloration and shrinkage of the fruiting bodies. Ptαβγ: mushrooms grown after 1st generation in the shelf treated with all 23 pathogens, Ptαβγ + ɸPtαβγ: mushrooms grown in the shelf treated with Ptαβγ + phage cocktail.

Fig. 6
Protective effect of the phage cocktail ensured by the treatment before blotch disease development. Control: treated with sterilized culture medium. Ptαβγ: treated with all pathogenic strains of Pseudomonas tolaasii. Ptαβγ + ɸPtαβγ: the phage cocktail was sprayed on the mushrooms before the development of disease at 12 h after pathogen inoculation.

References
Abdelrhim, AS, Ahmad, AA, Omar, MOA, Hammad, AMM and Huang, Q 2021. A new Streptomyces scabies-infecting bacteriophage from Egypt with promising biocontrol traits. Arch. Microbiol 203:4233-4242.



Abedon, ST 2016. Commentary: phage therapy of Staphylococcal chronic osteomyelitis in experimental animal model. Front. Microbiol 7:1251.



Chibani-Chennoufi, S, Sidoti, J, Bruttin, A, Kutter, E, Sarker, S and Brüssow, H 2004.
In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy. Antimicrob. Agents Chemother 48:2558-2569.




Cubero, S, Aleixos, N, Moltó, E, Gómez-Sanchis, J and Blasco, J 2011. Advances in machine vision applications for automatic inspection and quality evaluation of fruits and vegetables. Food Bioproc. Technol 4:487-504.


Dong, Z, Xing, S, Liu, J, Tang, X, Ruan, L, Sun, M, Tong, Y and Peng, D 2018. Isolation and characterization of a novel phage Xoo-sp2 that infects Xanthomonas oryzae pvoryzae
. J. Gen. Virol 99:1453-1462.


Fernández, L, Gutiérrez, D, Rodríguez, A and García, P 2018. Application of bacteriophages in the agro-food sector: a long way toward approval. Front. Cell Infect. Microbiol 8:296.


Food and Drug Administration 2006. Food additives permitted for direct addition to food for human consumption; bacteriophage preparation. Fed. Regist 71:47729-47732.
Geels, FP 1995.
Pseudomonas tolaasii control by kasugamycin in cultivated mushrooms (Agaricus bisporus). J. Appl. Bacteriol 79:38-42.

Geels, FP, van Griensven, LJLD and Rutjens, AJ 1991. Chlorine dioxide and the control of bacterial blotch on mushrooms, caused by Pseudomonas tolaasii
. In: Science and cultivation of edible fungi, eds. by MJ Maher, pp. 437-442. Balkema, Rotterdam, Netherlands.
Godfrey, SA, Harrow, SA, Marshall, JW and Klena, JD 2001. Characterization by 16S rRNA sequence analysis of pseudomonads causing blotch disease of cultivated Agaricus bisporus
. Appl. Environ. Microbiol 67:4316-4323.




Gundogdu, A, Bolkvadze, D and Kilic, H 2016.
In vitro effectiveness of commercial bacteriophage cocktails on diverse extended-spectrum beta-lactamase producing Escherichia coli strains. Front. Microbiol 7:1761.



Hibbing, ME, Fuqua, C, Parsek, MR and Peterson, SB 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol 8:15-25.




Hyman, P and Abedon, ST 2010. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol 70:217-248.


Iacobellis, NS and Lo Cantore, P 1998. Recenti acquisizioni sul determinismo della batteriosi del cardoncello (Pleurotus eryngii). Agricoltura Ricerca 176:51-54 (in Italian).
Khalifa, L, Shlezinger, M, Beyth, S, Houri-Haddad, Y, Coppenhagen-Glazer, S, Beyth, N and Hazan, R 2016. Phage therapy against Enterococcus faecalis in dental root canals. J. Oral Microbiol 8:32157.



Lee, C-J, Yoo, Y-M, Han, J-Y, Jhune, C-S, Cheong, J-C, Moon, J-W, Suh, J-S, Han, H-S and Cha, J-S 2014.
Pseudomonas azotoformans HC5 effective in antagonistic of mushrooms brown blotch disease caused by Pseudomonas tolaasii
. Korean J. Mycol 42:219-224 (in Korean).

Lee, HI, Lee, SD, Park, KS, Kim, YK and Cha, JS 1997. Pathogenicity of bacterial isolates from brown blotch-diseased oyster mushrooms in Chungcheongbuk-do. J. Agric. Sci. Chungbuk Nat. Univ 14:121-132 (in Korean).
Lee, H-J, Yun, Y-B, Huh, J-H and Kim, Y-K 2017. Suppression of green mold disease on oak mushroom cultivation by antifungal peptides. J. Appl. Biol. Chem 60:149-153 (in Korean).

Ministry of Agriculture, Food and Rural Affairs 2021 Special crop production statistics 2019 URL https://lib.mafra.go.kr/skyblueimage/5729.pdf
. 20 August 2022.
Munsch, P, Oliver, JM and Houdeau, G 1991. Experimental control of bacterial blotch by bacteriophages. In: Science and cultivation of edible fungi, eds. by MJ Maher, pp. 389-396. Balkema, Rotterdam, Netherlands.
Oechslin, F, Piccardi, P, Mancini, S, Gabard, J, Moreillon, P, Entenza, JM, Resch, G and Que, YA 2017. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J. Infect. Dis 215:703-712.



Park, J, Lee, GM, Kim, D, Park, DH and Oh, C-S 2018. Characterization of the lytic bacteriophage phiEaP-8 effective against both Erwinia amylovora and Erwinia pyrifoliae causing severe diseases in apple and pear. Plant Pathol. J 34:445-450.




Pedreschi, R, Hertog, M, Lilley, KS and Nicolaï, B 2010. Proteomics for the food industry: opportunities and challenges. Crit. Rev. Food Sci. Nutr 50:680-692.


Pereira, C, Moreirinha, C, Teles, L, Rocha, R, Calado, R, Romalde, JL, Nunes, ML and Almeida, A 2017. Application of phage therapy during bivalve depuration improves Escherichia coli decontamination. Food Microbiol 61:102-112.


Ranjan, R, Rani, A, Metwally, A, McGee, HS and Perkins, DL 2016. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun 469:967-977.



Tolaas, AG 1915. A bacterial disease of cultivated mushrooms. Phytopathology 5:51-54.
Wang, X, Wei, Z, Yang, K, Wang, J, Jousset, A, Xu, Y, Shen, Q and Friman, V-P 2019. Phage combination therapies for bacterial wilt disease in tomato. Nat. Biotechnol 37:1513-1520.



Yen, M, Cairns, LS and Camilli, A 2017. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat. Commun 8:14187.




Yun, Y-B, Han, J-H and Kim, Y-K 2018a. Characterization of phage-resistant strains derived from Pseudomonas tolaasii 6264, which causes brown blotch disease. J. Microbiol. Biotechnol 28:2064-2070.


Yun, Y-B and Kim, Y-K 2020. Molecular analysis of peptide toxins secreted by various Pseudomonas tolaasii strains. J. Appl. Biol. Chem 63:387-392 (in Korean).

- TOOLS
-
METRICS

- ORCID iDs
-
Young-Kee Kim

https://orcid.org/0000-0001-9279-6832 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



