 |
 |
| Plant Pathol J > Volume 36(5); 2020 > Article |
|
Abstract
Pepper and tomato plants infected with two Clavibacter species, C. capsici and C. michiganensis have shown different patterns of disease development depending on their virulence. Here, we investigated how pepper and tomato plants respond to infection by the high-virulent or low-virulent Clavibacter strains. For this, we chose two strains of each Clavibacter species to show different virulence level in the host plants. Although low-virulent strains showed less disease symptoms, they grew almost the same level as the high-virulent strains in both plants. To further examine the response of host plants to Clavibacter infection, we analyzed the expression patterns of plant defense-related genes in the leaves inoculated with different strains of C. capsici and C. michiganensis. Pepper plants infected with high-virulent C. capsici strain highly induced the expression of CaPR1, CaDEF, CaPR4b, CaPR10, and CaLOX1 at 5 days after inoculation (dai), but their expression was much less in low-virulent Clavibacter infection. Expression of CaSAR8.2 was induced at 2 dai, regardless of virulence level. Expression of GluA, Pin2, and PR2 in tomato plants infected with high-virulent C. michiganensis were much higher at 5 dai, compared with mock or low-virulent strain. Expression of PR1a, Osmotin-like, Chitinase, and Chitinase class 2 was increased, regardless of virulence level. Expression of LoxA gene was not affected by Clavibacter inoculation. These results suggested that Clavibacter infection promotes induction of certain defense-related genes in host plants and that differential expression of those genes by low-virulent Clavibacter infection might be affected by their endophytic lifestyle in plants.
The genus Clavibacter belongs to actinobacteria carrying genomic DNA with high G + C content (Eichenlaub and Gartemann, 2011). Two species, C. michiganensis and C. capsici have been shown to cause bacterial canker in tomato and pepper plants, respectively (Thapa et al., 2019). In particular, C. capsici was first isolated from infected pepper leaves in Korea and reported in 1999 (Lee et al., 1999). Since then, it has been considered as C. michiganensis, but recently it was reclassified as a separate species (Li et al., 2018; Oh et al., 2016). A recent study reported that C. capsici strains with different levels of virulence exist in Korea and that this virulence depends on the plasmid carrying the virulence genes (Hwang et al., 2020). The small plasmid, pCM1Cc of C. capsici carries at least five virulence genes, and these genes play a role in virulence during infection into host plants (Hwang et al., 2018).
C. michiganensis has been used as a representative species as Gram-positive bacterium for research on virulence genes and mechanisms in plants (Eichenlaub and Gartemann, 2011). Although, in Korea, this pathogen resided in xylem vessels was first observed in 1997, it was reported in 2008 because it became severe in tomato production (Myung et al., 2008). C. michiganensis also carries two plasmids like C. capsici, and it was shown that both plasmids play an important role in virulence. The plasmid-borne celA (cellulase) gene in pCM1 and pat-1 (putative serine protease) gene in pCm2 are critical for the virulence of C. michiganensis in tomato (Hwang et al., 2019; Jahr et al., 2000). In addition, a pathogenicity island region located in the chromosome of C. michiganensis also contains several virulence genes for putative serine proteases and enzymes that seem to be involved in disease development (Eichenlaub and Gartemann, 2011; Gartemann et al., 2008). Importantly, in both C. capsici and C. michiganensis, there was no positive correlation between bacterial growth and virulence level (Hwang et al., 2020; Thapa et al., 2017), meaning that both the high-virulent and low-virulent strains of both bacteria could grow well similarly inside host plants.
During the long-term battle between plants and pathogens, plants have developed defense mechanisms to protect themselves from pathogen attack, and these interactions have been intensively studied. For successful colonization inside the plants, molecules secreted by pathogens have to overcome physical barriers such as the cell wall. The plant also responds by the recognition of invading pathogens, and plant defense responses were promoted via activation of defense-related genes and production of defensive compounds (Abramovitch et al., 2006; Dodds and Rathjen, 2010; Jones and Dangl, 2006). Infection of Xanthomonas campestris pv. vesicatoria resulted in induced expression of many defense-related genes, such as CaPR1, CaDEF, CaSAR8.2, and CaPR4b, and these gene transcripts were accumulated at various time point in plants (Choi and Hwang, 2015; Hong et al., 2017; Lee and Hwang, 2006). Infection with a soil-borne pathogen Phytophthora capsici also triggered the expression of defense genes in leaves and roots, and these expressed genes might be involved in defense responses (Jin et al., 2015; Wang et al., 2013). Defense-related genes mentioned above have been shown to be differentially expressed depending on the virulence level of X. campestris pv. vesicatoria and P. capsici in pepper plants. The expression of the pepper defense-related genes by infection with incompatible pathogens were significantly and strongly induced at early time point in plants, compared to compatible pathogens (Choi and Hwang, 2015; Kim et al., 2010; Wang et al., 2013).
Infected tomato plants with Ralstonia solanacearum induced expression of defense-related genes, such as PR1a, GluA, Pin2, LoxA, Osmotin-like, PR2, Chitinase, and Chitinase class 2 (Milling et al., 2011). Expression of PR1 gene was detected in tomato plants infected with Pseudomonas syringae pv. tomato under greenhouse condition (Herman et al., 2008). Despite of extensive researches on pathogen-induced defense responses in plants, including pepper and tomato, how transcriptional changes in plants after inoculation with Clavibacter species can protect plants remains unknown. The only finding that infection with C. michiganensis activates tomato basal defense responses and defense-related hormones by expression of defense-related genes, implying that plants may also activate defense responses against Clavibacter infection (Balaji et al., 2008).
In this study, we focused on the defense responses during infection of Clavibacter species in their host plants. In particular, we used the high-virulent and low-virulent strains of C. capsici and C. michiganensis in pepper and tomato, respectively, which have no positive correlation between virulence level and bacterial growth in host plants. Our study provides a detailed description of transcriptional changes induced by various Clavibacter strains infection in host plants.
Pepper (Capsicum annuum ŌĆśNockwangŌĆÖ) and tomato (Solanum lycopersicum cv. ŌĆśBetatiniŌĆÖ) plants used in this study were grown in a growth chamber at 26┬░C with a 14 h:10 h light:dark photoperiod and 50% relative humidity. For all inoculation experiments, 3-week-old pepper plants and 2-week-old tomato plants were used.
For pepper plant inoculation, the highvirulent strains C. capsici PF008 and 822 and the low-virulent strains PF008ΔpCM1Cc and 1101 (lacking pCM1Cc) (Hwang et al., 2020) and for tomato plant inoculation, the high-virulent strains C. michiganensis LMG7333 (Hwang et al., 2019) and KACC16995 and the low-virulent strain LMG3685 (Yim et al., 2012; Oh et al., 2016) were used in this study.
For virulence assay, bacteria were incubated in KB (KingŌĆÖs B) medium (proteose peptone no. 3; 20 g, K2HPO4; 1.5 g, 1 M MgSO4; 6 ml, and 50% glycerol; 16 ml in total volume 1 L) at 26┬░C, after single colony propagation. Cultured bacteria were then harvested and resuspended in 10 mM MgCl2. Bacterial suspension adjusted to an optical density of 0.1 at 600 nm was infiltrated into leaves of pepper plants with the needleless syringe. OD600 = 0.4 of C. michiganensis bacterial suspension was prepared in 10 mM MgCl2, and inoculated into tomato leaves by leaf clipping method using scis sors. Ten mM MgCl2 was used as a mock inoculation.
After pepper inoculation with C. capsici strains, disease symptoms on inoculated leaves were scored at 7 days after inoculation (dai) based on the following 0 to 5 scales: 0, no visible symptoms; 1, <5% leaf area affected; 2, 6-25% leaf area affected; 3, 26-50% leaf area affected; 4, 51-75% leaf area affected; 5, >75% leaf area affected (Hwang et al., 2018). Tomato plants infected with C. michiganensis were scored at 23 dai. Disease severity was evaluated by scoring of wilting symptoms, using an index ranging from 0 to 5: 0, no symptoms; 1, one or two leaves showed wilting symptoms; 2, less than 25% leaves wilted; 3, more than 25% but less than 50% leaves wilted; 4, 50 to 75% leaves showed wilting symptoms; 5, more than 75% leaves wilted or plants dead (Hwang et al., 2019). All experiments were repeated at least 3 times independently.
In planta bacterial growth of Clavibacter strains was measured by harvesting samples after inoculation with 108 colony forming units (cfu)/ml of C. michiganensis, and 4 ├Ś 108 cfu/ml of C. capsici at various time points. C. capsici- inoculated pepper plants and C. michiganensis-inoculated tomato plants were collected at 0, and 5 dai, and at 3, 7, and 10 dai, respectively. Three independent samples were ground, plated on KB medium plates, and then incubated at 26┬░C for 2 days. After incubation, the number of cfu per leaf discs (cfu/cm2) or leaf weight (cfu/g) by multiplying the average number of the colony were calculated.
With primer sets to amplify virulence genes or plasmid back-borne genes specific to Clavibacter species, PCR was performed to confirm the used strains. Type strains of each species, which are C. capsici PF008 and C. michiganensis LMG7333, were used as a positive control. Primers used in this study are listed in Supplementary Table 1. All PCR assay was performed using BioFACT 2├Ś Taq PCR Master Mix 2 (BioFACT, Daejeon, Korea).
Pepper defense-related genes, CaPR1 (accession no. AF053343), CaDEF (accession no. AF442388), CaPR4b (accession no. HM581975), CaPR10 (accession no. AF244121), CaLOX1 (accession no. FJ377808), and CaSAR8.2 (accession no. AF112868) and tomato defense-related genes, GluA (accession no. M80604), PR1a (accession no. M69247), Pin2 (accession no. AY129402), LoxA (accession no. U09026), PR2 (accession no. BT013355), Osmotin-like (accession no. m21346), Chitinase (accession no. BG629612), and Chitinase class 2 (accession no. U30465) were targeted to check their gene expression. Total RNA was isolated from infected plant samples at 0 (right after inoculation), 2, and 5 dai by EZ Total RNA Miniprep Kit (Enzynomics, Daejeon, Korea), and then reverse-transcribed to generate cDNA by TOPscript III Reverse Transcriptase (Enzynomics). Using these synthesized cDNA, qRT-PCR using TOPreal qPCR 2├Ś PreMIX (Enzynomics) was performed, and relative expression of target genes was normalized with CaActin (accession no. GQ339766) for pepper plants and SlActin (accession no. BT013524) for the tomato plants. The primer sets to amplify plant defense-related genes are listed in Supplementary Table 1.
Statistical analysis was performed using StatistiXL 2 (statistiXL, Broadway, Australia). Disease severity data were analyzed by the non-parametric ANOVA (Kruskal-Wallis test), and other data were analyzed by DuncanŌĆÖs multiple range test (P < 0.05). All experiments were repeated a minimum of three times with similar results.
In the previous study, we reported C. capsici natural isolates with differential virulence level in pepper plants (Hwang et al., 2020). To explore the role of defense mechanism in pepper plants, especially the expression of defense-related genes depending on virulence difference of C. capsici strains, we selected C. capsici type strain PF008 and natural isolate 822 as high-virulent strains and strain PF008ΔpCM1Cc and natural isolate 1101 as low-virulent strains due to the lack of the small plasmid pCM1Cc with important virulence genes such as chpE and chpG (Hwang et al., 2018). The virulence level of these selected strains were confirmed by inoculation into pepper leaves. Pepper leaves inoculated with strain 822 showed severe necrotic symptoms as observed in PF008-inoculated leaves. The disease symptoms by both strains in pepper leaves started to appear prior to 5 dai, and the entire inoculated leaves displayed necrotic symptoms at 7 to 10 dai (Fig. 1A). In contrast, PF008ΔpCM1Cc, and 1101 caused barely visible and weak necrotic symptoms at 7 dai. Their disease severity was quantified as shown in Supplementary Fig. 1.
We further checked the bacterial number of C. capsici strains in the infiltrated leaves. The result showed that bacterial numbers of low-virulent C. capsici strains were almost equivalent to high-virulent strains in pepper leaves, indicating that there is little correlation between the virulence level and bacterial population of these strains in plants (Fig. 1B). Collectively, these results imply that both high-virulent and low-virulent strains of C. capsici can survive and grow in plants regardless of the virulence level.
As we mentioned above, the high-virulent strains of C. capsici caused severe necrotic symptoms on pepper leaves within 7 dai. The expression of the virulence genes in high-virulent and/or low-virulent C. capsici strains was analyzed in the previous report (Hwang et al., 2020), showing that the transcript level of plasmid-borne virulence genes was differentially induced by the presence or the absence of the small plasmid, pCM1Cc. In particular, we found that the expression of most plasmid-borne virulence genes tended to decrease in low-virulent strains after inoculation in plants. So far, however, very little research has focused on changes in defenserelated genes in Clavibacter-inoculated plants. Thus, we analyzed how pepper defense-related genes respond to C. capsici infection. The expression profiles of defense-related genes such as CaPR1, CaDEF, CaPR4b, CaPR10, CaLOX1, and CaSAR8.2, which have been shown to respond to bacterial and/or fungal infection or various abiotic stresses (Choi and Hwang, 2015), were determined by qRT-PCR with high-virulent or low-virulent C. capsici strains at 0, 2 and 5 dai before the disease looks very severe.
Expression of three PR genes, CaPR1, CaPR4b, and CaPR10, was induced from 2 dai, unlike those genes were induced immediately after inoculation in pepper with Gram-negative bacteria, such as X. campestris pv. vesicatoria (Choi and Hwang, 2015). They displayed stronger and higher induction in plants inoculated with high-virulent strains, PF008 and 822, at 5 dai, which is the time when the disease symptom began to appear (Fig. 2). Although their expression significantly increased toward the later time points of infection with low-virulent strains, PF008ΔpCM1Cc and 110, it was not strong as much as that in plant leaves inoculated with high-virulent strains at 5 dai. Expression of CaDEF and CaLOX1 could significantly be detected right after inoculation, and their expression was highly induced at 5 dai (Fig. 2). Like PR genes, higher expression was observed in leaves inoculated with high-virulent strains than in leaves with low-virulent strains. While induction of these 5 genes after infection with C. capsici strains was notably increased during infection until 5 dai, expression of CaSAR8.2 was temporarily increased only at 2 dai regardless of virulence level of inoculated strains (Fig. 2). These results indicate that all examined defense-related genes exhibited increased expression during infection of C. capsici and the magnitude was bigger during infection of high-virulent strains than low-virulent strains.
C. michiganensis type strain LMG7333 known as a high-virulent strain against tomato plants was first selected as a high-virulent strain (Hwang et al., 2019). Then, two more C. michiganensis strains KACC16995 and LMG3685 were obtained, and their identities were checked by PCR with primers specific to C. michiganensis (Supplementary Fig. 2). To examine differences in virulence of these two C. michiganensis strains, tomato plants were inoculated by the leaf clipping method, and symptom development was observed for 4 weeks after inoculation. Tomato plants inoculated by strains either LMG7333 or KACC16995 developed wilting symptoms approximately 2 weeks after inoculation, whereas plants inoculated by LMG3685 remained mostly healthy over the two-week. After 3 weeks, strains LMG7333 and KACC16995 caused severe wilting and bacterial canker on their stems, resulting in eventual death of the plants (Fig. 3A and B). However, LMG3685-inoculated plants exhibited mild or no visual symptoms at the same time point (Fig. 3A and B). Therefore, KACC16995 and LMG3685 were categorized and selected as high-virulent and low-virulent strains, respectively.
To determine if the virulence of these strains is directly related to the bacterial population in plants, the bacterial growth assay was performed in planta. Regardless of virulence level of bacterial strains, all strains grew well, and there was no difference in bacteria titers even at 10 dai (Fig. 3C). These are consistent with those of C. capsici in pepper leaves (Fig. 1B). These results support that the disease severity caused by two Clavibacter species is not correlated with bacterial titers in plants, as shown in our previous report (Hwang et al., 2019).
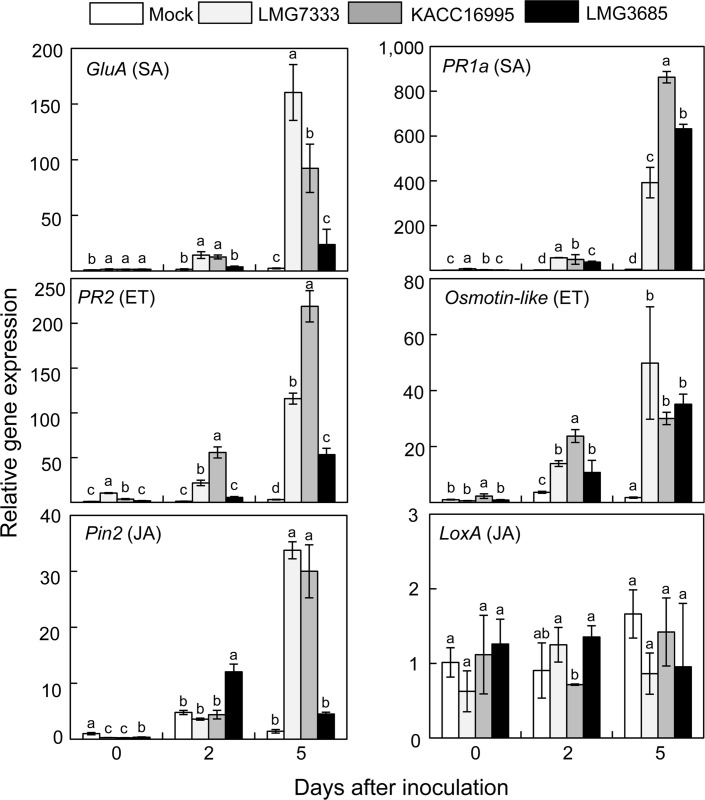
We hypothesized that the similar bacterial titers regardless of virulence level of C. michiganensis strains might affect to the expression of tomato defense-related genes during infection. To examine the change of the transcripts of defense-related genes in tomato plants inoculated with high-virulent or low-virulent C. michiganensis strains, we focused on the tomato genes, of which their expression was previously reported to be induced during C. michiganensis infection (Balaji et al., 2008). Among them, we selected GluA, encoding ╬▓-1,3 glucanase, and PR1a for the salicylic acid (SA) pathway, PR2 and osmotin-like gene for the ethylene (ET) pathway, and Pin2 (tomato protease inhibitor) and LoxA (lipoxygenase) genes for the jasmonic acid (JA) pathway.
Expression of GluA, PR2, and Pin2 was begun to be induced in response to infection of the high-virulent C. michiganensis strains, LMG7333 and KACC16995 at 2 dai and dramatically increased at 5 dai (Fig. 4). However, although expression of those genes was slightly increased by the low-virulent strain, LMG3685, their expression level was much lower than those by the high-virulent strains at 5 dai. Expression of PR1a and osmotin-like genes was also begun to be induced at 2 dai, but those expression was increased at the similar level by both the high-virulent and the low-virulent strains (Fig. 4). Similarly, expression of two chitinase genes related to the ET pathway was increased from 2 dai by both the high-virulent and the lowvirulent strains (Supplementary Fig. 3). Finally, expression of LoxA was not significantly different, compared to that of the control (Mock) plant, by both the high-virulent and the low-virulent strains (Fig. 4). These results indicate that both the high-virulent and low-virulent C. michiganensis strains could induce most of defense-related genes in tomato, but the expression level could be different due to the virulence level, but not because of bacterial titers inside plant tissues.
In this study, to gain molecular insights about molecular responses of pepper and tomato plants to Clavibacter infection, we analyzed gene expression profiles of pepper or tomato defense-related genes after infection of the highvirulent and low-virulent strains of Clavibacter species. Previously, only a few studies have been reported to show plant responses against C. michiganensis infection in tomato. During high-virulent C. michiganensis infection, typical basal defense responses in tomato plants were activated by induction of defense-related genes, production and scavenging of free oxygen radicals, enhanced protein turnover, and hormone synthesis (Balaji et al., 2008). Among the genes up-regulated by the high-virulent pathogen infection, PR1a, Pin2, Osmotin-like protein, PR2, and Chitinase class 2 were involved in the disease development (Balaji et al., 2008; Milling et al., 2011). Here, in common, infection with C. capsici and C. michiganensis in pepper and tomato plants, respectively, resulted in accumulation of transcripts of plant defense-related genes. Notably, we also found that host plants inoculated with the high-virulent Clavibacter had significantly higher expression of most defense-related genes and showed dramatically elevated transcript accumulation at 5 dai, compared to mock-inoculated plants (Figs. 2, and 4), although the expression of CaSAR8.2 in pepper and LoxA in tomato in response to Clavibacter infection only increased at 2 dai or no induction. The fact that most of pepper defense-related genes, CaPR1, CaDEF, CaPR4b, CaPR10, and CaLOX1, in this study were strongly induced at 5 dai indicates that pepper defense-related genes might contribute to defense responses to protect itself.
Moreover, we also showed that tomato plants responded to the high-virulent C. michiganensis infection regulated defense marker genes for the SA, JA, and ET defense pathways, which are responsible for plant defense responses (Fig. 4). Except for LoxA gene, other defense-related genes were strongly expressed, and its expression was peaked at 5 dai. In agreement with the reported data, defense-related genes in tomato were highly expressed during infection, indicating that tomato defense-related genes may participate in the activation of the defense responses against C. michiganensis infection through the expression of various defense-related genes related to plant hormone (Bari and Jones, 2009). Furthermore, when we analyzed the expression of the same defense-related genes during the lowvirulent Clavibacter infection, the patterns were different from ones during the high-virulent Clavibacter infection, and it compromised accumulation of defense-related genes. In the pepper plants, except CaSAR8.2, all tested defense genes showed reduced expression, compared to the pepper inoculated with the high-virulent strains. In the tomato plants, some of them, which are GluA, Pin2, and PR2, failed to express as much as those in plants infected with the high-virulent strains.
Differentially expressed defense-related genes depending on the level of virulence were extensively studied in the host plants infected with Gram-negative plant pathogenic bacteria (Choi and Hwang, 2015; Ciardi et al., 2000; Potnis et al., 2015). So far, pepper plants have been used as a model plant for the study of X. campestris pv. vesicatoriahost interactions and plant responses against bacterial infection. During infection with X. campestris pv. vesicatoria, the expression of CaPR1 gene, the representative defense gene in pepper, was highly induced from 6 hours after inoculation (hai) and reached a maximum at 18 to 25 hai (Kim et al., 2004). CaDEF gene was also expressed during X. campestris pv. vesicatoria infection, especially the compatible interaction, in pepper plants (Do et al., 2004). Other defense-related genes, such as CaPR4b, CaPR10, CaLOX1, and CaSAR8.2, tested in this study were strongly expressed and peaked before 24 hai after X. campestris pv. vesicatoria infection (Choi and Hwang, 2015; Hwang and Hwang, 2010). Other model systems, such as the interaction between Phytophthora capsici and a pepper plant, also showed that defense-related genes, such as CaPR1, Ca-DEF1, and CaSAR82, were significantly induced in pepper plants infected with P. capsici at relatively late time point, compared to those in the plants infected with X. campestris pv. vesicatoria (Jin et al., 2015). In most cases, these defense-related genes were more strongly expressed at the early time points in the incompatible interaction of the pepper with X. campestris pv. vesicatoria than in the compatible interaction, suggesting that the expression of these genes occurred differentially in the level of virulence-dependent manner (Choi and Hwang, 2015; Han and Hwang, 2017).
These Gram-negative bacterial pathogens have strong weapons, known as effectors secreted by type III secretion system (T3SS) to attack their host plants, however, the Gram-positive pathogen like Clavibacter lacks effectors delivered by T3SS (Eichenlaub and Gartemann, 2011; Nandi et al., 2018). Therefore, instead of effector proteins, Clavibacter species seem to use proteases, and these proteases act as virulence factors that play an important role in the virulence of this pathogen (Hwang et al., 2019; Thapa et al., 2019). To move within the plant, Clavibacter without flagella moves into the target area through the xylem vessels, implying that the expression of plant defenserelated genes seems to be related to the transport speed of Clavibacter and the ability to directly kill plant cells over time. Relatively late symptom development by Clavibacter pathogens compared to other Gram-negative bacterial pathogens with effectors led to the late transcript accumulation of defense-related genes in host plants. The unique lifestyle of this pathogen might prevent the plants from recognizing it, until plant cells were directly attacked, and interrupt the rapid turn-on of these defense-related genes. A notable finding that despite the reduced expression of plant defense gene during the interaction between the low-virulent Clavibacter and its host plants, the low-virulent Clavibacter can survive in the plants as an endophyte without restricting bacterial growth, suggesting that the presence of Clavibacter is difficult to detect by plants before this pathogen reveals its virulence, such as releasing virulence factors that can directly affect the plants. Otherwise, these lowvirulent Clavibacter strains might suppress plant defense responses activated by defense gene expression for survival in plants.
Overall, we showed the transcriptional changes of plant defense-related genes in response to the infection of Clavibacter with high or low virulence. The results of this work demonstrated that pepper or tomato plants have triggered the expression of defense-related genes against Clavibacter infection, even though gene expression was begun from the late time points with symptom development. We also revealed that these transcriptional changes between the highvirulent and the low-virulent inoculated host plants may result in its xylem-limited lifestyle of Clavibacter. We still do not yet know whether this late defense-related gene expression plays a significant role in the defense responses to suppress symptom development by Clavibacter infection, but this research may help us develop a potential option for understanding their interaction between Clavibacter and its host plants.
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
We thank Korean Agricultural Culture Collection and Dr. Jae-Soon Cha for providing C. michiganensis KACC16695 and LMG3685, respectively. This work was supported by the grant from Next-Generation BioGreen21 Program (project no. PJ013285), Rural Development Administration, Republic of Korea
REFERENCES
Abramovitch, R. B., Anderson, J. C. and Martin, G. B. 2006. Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7:601-611.
Balaji, V., Mayrose, M., Sherf, O., Jacob-Hirsch, J., Eichenlaub, R., Iraki, N., Manulis-Sasson, S., Rechavi, G., Barash, I. and Sessa, G. 2008. Tomato transcriptional changes in response to Clavibacter michiganensis subsp. michiganensis reveal a role for ethylene in disease development. Plant Physiol. 146:1797-1809.



Bari, R. and Jones, J. D. G. 2009. Role of plant hormones in plant defence responses. Plant Mol. Biol. 69:473-488.
Choi, H. W. and Hwang, B. K. 2015. Molecular and cellular control of cell death and defense signaling in pepper. Planta. 241:1-27.


Ciardi, J. A., Tieman, D. M., Lund, S. T., Jones, J. B., Stall, R. E. and Klee, H. J. 2000. Response to Xanthomonas campestris pv. vesicatoria in tomato involves regulation of ethylene receptor gene expression. Plant Physiol. 123:81-92.



Do, H. M. and Lee, S. C. 2004. Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in. Capsicum annuum. Plant Sci. 166:1297-1305.
Dodds, P. N. and Rathjen, J. P. 2010. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11:539-548.

Eichenlaub, R. and Gartemann, K.-H. 2011. The Clavibacter michiganensis subspecies: molecular investigation of grampositive bacterial plant pathogens. Annu. Rev. Phytopathol. 49:445-464.
Gartemann, K.-H., Abt, B., Bekel, T., Burger, A., Engemann, J., Fl├╝gel, M., Gaigalat, L., Goesmann, A., Gr├żfen, I., Kalinowski, J., Kaup, O., Kirchner, O., Krause, L., Linke, B., McHardy, A., Meyer, F., Pohle, S., R├╝ckert, C., Schneiker, S., Zellermann, E.-M., P├╝hler, A., Eichenlaub, R., Kaiser, O. and Bartels, D. 2008. The genome sequence of the tomatopathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J. Bacteriol. 190:2138-2149.

Han, S. W. and Hwang, B. K. 2017. Molecular functions of Xanthomonas type III effector AvrBsT and its plant interactors in cell death and defense signaling. Planta. 245:237-253.


Herman, M. A. B., Davidson, J. K. and Smart, C. D. 2008. Induction of plant defense gene expression by plant activators and Pseudomonas syringae pv. tomato in greenhouse-grown tomatoes. Phytopathology. 98:1226-1232.


Hong, J. K., Hwang, I. S. and Hwang, B. K. 2017. Functional roles of the pepper leucine-rich repeat protein and its interactions with pathogenesis-related and hypersensitive-induced proteins in plant cell death and immunity. Planta. 246:351-364.


Hwang, I. S. and Hwang, B. K. 2010. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 152:948-967.



Hwang, I. S., Lee, H. M., Oh, E.-J., Lee, S., Heu, S. and Oh, C.-S. 2020. Plasmid composition and the chpG gene determine the virulence level of Clavibacter capsici natural isolates in pepper. Mol. Plant Pathol. 21:808-819.

Hwang, I. S., Oh, E.-J., Kim, D. and Oh, C.-S. 2018. Multiple plasmid-borne virulence genes of Clavibacter michiganensis ssp. capsici critical for disease development in pepper. New Phytol. 217:1177-1189.


Hwang, I. S., Oh, E.-J., Lee, H. B. and Oh, C.-S. 2019. Functional characterization of two cellulase genes in the gram-positive pathogenic bacterium Clavibacter michiganensis for wilting in tomato. Mol. Plant-Microbe Interact. 32:491-501.

Jahr, H., Dreier, J., Meletzus, D., Bahro, R. and Eichenlaub, R. 2000. The endo-beta-1,4-glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol. Plant-Microbe Interact. 13:703-714.

Jin, J.-H., Zhang, H.-X., Tan, J.-Y., Yan, M.-J., Li, D.-W., Khan, A. and Gong, Z.-H. 2015. A new ethylene-responsive factor CaPTI1 gene of pepper (Capsicum annuum L.) involved in the regulation of defense response to Phytophthora capsici. Front. Plant Sci. 6:1217


Kim, N. H., Choi, H. W. and Hwang, B. K. 2010. Xanthomonas campestris pv. vesicatoria effector AvrBsT induces cell death in pepper, but suppresses defense responses in tomato. Mol. Plant-Microbe Interact. 23:1069-1082.

Kim, S. H., Hong, J. K., Lee, S. C., Sohn, K. H., Jung, H. W. and Hwang, B. K. 2004. CAZFP1, Cys2/His2-type zinc-finger transcription factor gene functions as a pathogen-induced early-defense gene in Capsicum annuum. Plant Mol. Biol. 55:883-904.


Lee, S. C. and Hwang, B. K. 2006. CASAR82A, a pathogeninduced pepper SAR8.2, exhibits an antifungal activity and its overexpression enhances disease resistance and stress tolerance. Plant Mol. Biol. 61:95-109.


Lee, S.-D., Yoon, C.-M., Lee, Y.-K., Choi, Y.-H. and Cho, Y.-S. 1999. Occurrence and distribution of bacterial canker of red pepper caused by Clavibacter michiganensis subsp. michignensis. Plant Dis. Agric. 5:105-110.
Li, X., Tambong, J., Yuan, K. X., Chen, W., Xu, H., L├®vesque, C. A. and De Boer, S. H. 2018. Re-classification of Clavibacter michiganensis subspecies on the basis of whole-genome and multi-locus sequence analyses. Int. J. Syst. Evol. Microbiol. 68:234-240.


Milling, A., Babujee, L. and Allen, C. 2011. Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS ONE. 6:e15853



Myung, I.-S., Kim, D. G., An, S. H., Lee, Y.-K. and Kim, W. G. 2008. First report of bacterial canker of tomato caused by Clavibacter michiganensis subsp. michiganensis in Korea. Plant Dis. 92:1472

Nandi, M., Macdonald, J., Liu, P., Weselowski, B. and Yuan, Z.-C. 2018. Clavibacter michiganensis ssp. michiganensis: bacterial canker of tomato, molecular interactions and disease management. Mol. Plant. Pathol. 19:2036-2050.



Oh, E.-J., Bae, C., Lee, H.-B., Hwang, I. S., Lee, H.-I., Yea, M. C., Yim, K.-O., Lee, S., Heu, S., Cha, J.-S. and Oh, C.-S. 2016. Clavibacter michiganensis subsp. capsici subsp. nov., causing bacterial canker disease in pepper. Int. J. Syst. Evol. Microbiol. 66:4065-4070.


Potnis, N., Timilsina, S., Strayer, A., Shantharaj, D., Barak, J. D., Paret, M. L., Vallad, G. E. and Jones, J. B. 2015. Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant. Pathol. 16:907-920.



Thapa, S. P., Davis, E. W., Lyu, Q., Weisberg, A. J., Stevens, D. M., Clarke, C. R., Coaker, G. and Chang, J. H. 2019. The evolution, ecology, and mechanisms of infection by gram-positive, plant-associated bacteria. Annu. Rev. Phytopathol. 57:341-365.


Thapa, S. P., Pattathil, S., Hahn, M. G., Jacques, M.-A., Gilbertson, R. L. and Coaker, G. 2017. Genomic analysis of Clavibacter michiganensis reveals insight into virulence strategies and genetic diversity of a gram-positive bacterial pathogen. Mol. Plant-Microbe Interact. 30:786-802.


Fig.┬Ā1
Virulence assay in pepper leaves with Clavibacter capsici natural isolates. (A) Virulence phenotypes on pepper leaves in response to C. capsici. The pepper leaves was bleached with ethanol to make symptom more obvious, and the picture was taken at 7 days after inoculation (dai). (B) Bacterial growth of C. capsici strains in plants at 0 and 5 dai. The data represent means ┬▒ standard deviation (n = 3). The different letters indicate statistically significant differences as determined by DuncanŌĆÖs multiple range test (P < 0.05).
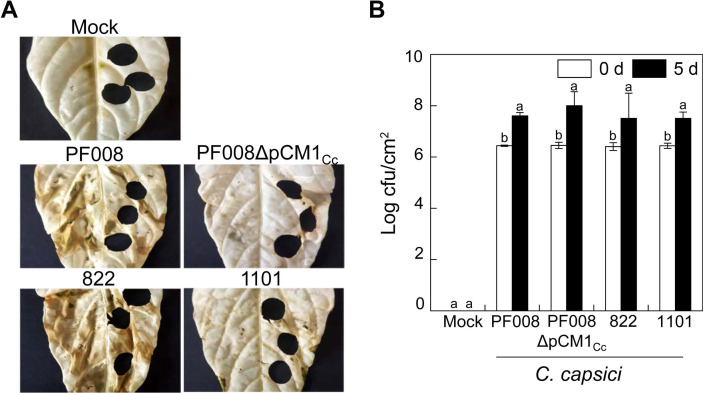
Fig.┬Ā2
Transcriptional changes of defense-related host genes in pepper plants after inoculation with less- or high-virulent isolates of Clavibacter capsici. Quantitative real-time PCR analysis of the indicated defense-related genes in pepper plants were measured at 0, 2, and 5 days after inoculation. The data represent means ┬▒ standard deviation (n = 3). The different letters indicate statistically significant differences as determined by DuncanŌĆÖs multiple range test (P < 0.05). Pepper Actin gene was used to normalize the expression of defense-related genes.
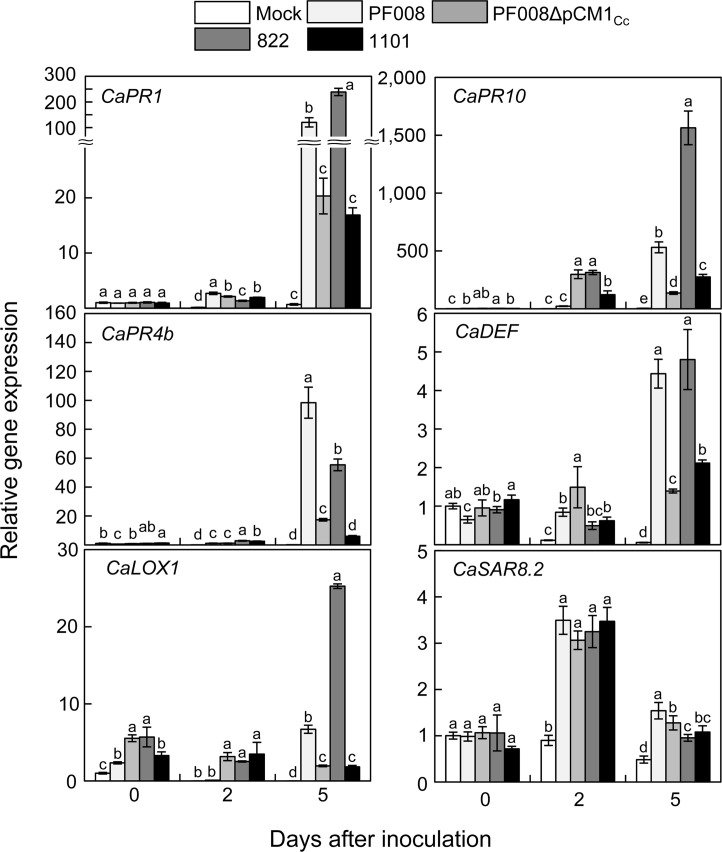
Fig.┬Ā3
Virulence assay in tomato plants with Clavibacter michiganensis isolates. (A) Phenotypes in tomato plants in response to C. michiganensis isolates. (B) Disease severity of C. michiganensis isolates in tomato plants at 14 days after inoculation. The data represent means ┬▒ standard deviation (n = 9). The different letters indicate statistically significant differences across treatments as per a non-parametric Kruskal-Wallis oneway ANOVA, followed by a posthoc test (DunnettŌĆÖs test, P < 0.05). (C) Bacterial growth of C. michiganensis isolates in tomato plants at the indicated time points. The data represent means ┬▒ standard deviation (n = 3). The different letters indicate statistically significant differences as determined by DuncanŌĆÖs multiple range test (P < 0.05).
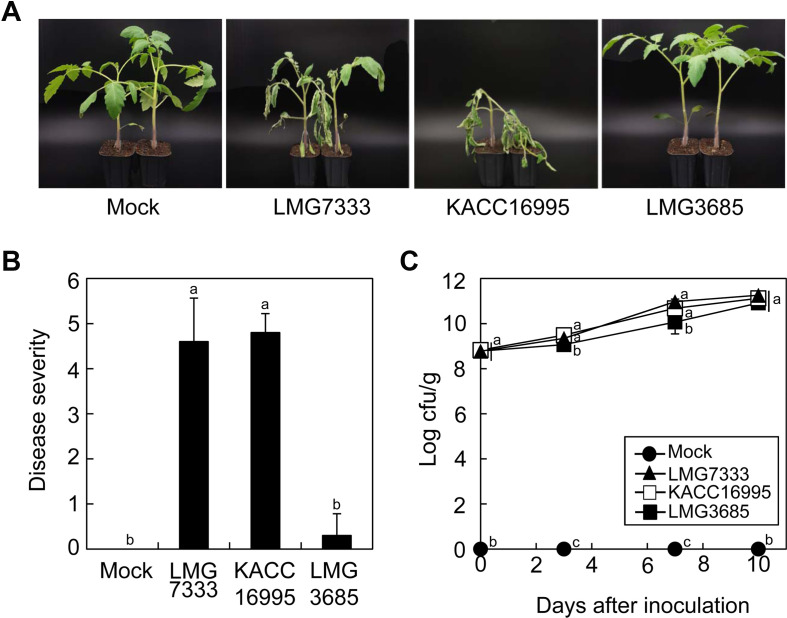
Fig.┬Ā4
Transcriptional changes of defense-related host genes in tomato plants after inoculation with Clavibacter michiganensis isolates. Quantitative real-time PCR analysis of the indicated defenserelated genes in tomato plants were measured at 0, 2, and 5 days after inoculation. The data represent means ┬▒ standard deviation (n = 3). The different letters indicate statistically significant differences as determined by DuncanŌĆÖs multiple range test (P < 0.05). Tomato Actin gene was used to normalize the expression of defense-related genes.




 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




