 |
 |
| Plant Pathol J > Volume 38(6); 2022 > Article |
|
Abstract
Root-knot nematode disease is a widespread and catastrophic disease of tobacco. However, little is known about the relationship between rhizosphere bacterial community and root-knot nematode disease. This study used 16S rRNA gene sequencing and PICRUSt to assess bacterial community structure and function changes in rhizosphere soil from Meloidogyne incognita-infected tobacco plants. We studied the rhizosphere bacterial community structure of M. incognita-infected and uninfected tobacco plants through a paired comparison design in two regions of tobacco planting area, Yuxi and Jiuxiang of Yunnan Province, southwest China. According to the findings, M. incognita infection can alter the bacterial population in the soil. Uninfested soil has more operational taxonomic unit numbers and richness than infested soil. Principal Coordinate Analysis revealed clear separations between bacterial communities from infested and uninfested soil, indicating that different infection conditions resulted in significantly different bacterial community structures in soils. Firmicutes was prevalent in infested soil, but Chloroflexi and Acidobacteria were prevalent in uninfested soil. Sphingomonas, Streptomyces, and Bradyrhizobium were the dominant bacteria genera, and their abundance were higher in infested soil. By PICRUSt analysis, some metabolism-related functions and signal transduction functions of the rhizosphere bacterial community in the M. incognita infection-tobacco plants had a higher relative abundance than those uninfected. As a result, rhizosphere soils from tobacco plants infected with M. incognita showed considerable bacterial community structure and function alterations.
The pathogen Meloidogyne spp. cause root-knot nematode (RKN) disease, a widespread tobacco-based soilborne disease, was infecting the roots of tobacco plants at various stages of growth. The tobacco RKN disease has been discovered in several tobacco-producing regions in China, including Heilongjiang and Liaoning in Northeast China, Henan and Hubei in Central China, and Yunnan and Sichuan in Southwest China on. The severity of the damage is increasing (Cui et al., 2021; Huang et al., 2020). Among them, the affected area in Yunnan Province alone is more than 26,000 hectares, with yield losses of 30% to 50% (Chen et al., 2015). More seriously, mechanical damage caused by Meloidogyne spp. It contributes to the infection of other pathogenic microorganisms. It causes various tobacco rhizome diseases, such as bacterial wilt, black shank, black root rot (Song et al., 2019), and Fusarium root rot (Zheng et al., 2021). In previous studies, tobacco RKN disease has been mainly prevented and controlled by planting resistant varieties (Li et al., 2017), non-host crop rotation (Wang and Kong, 2002), biological control (Ciancio et al., 2016; Norabadi et al., 2014) and chemical agents (Huang et al., 2020), that unquestionably exhibit certain limitations and side effects. Therefore, it is urgent to find other effective prevention and control measures.
Most plant-parasitic nematodes develop and reproduce in the rhizosphere of their host plants, unlike free-living nematodes. Their eggs infect plant roots, developing into juveniles that absorb nutrients from plant cells (Tian et al., 2015). The rhizosphere refers to the soil microzone affected by the root system and mainly involves the thin layer (1-2 cm) on the surface of roots (Philippot et al., 2013). This region hosts beneficial microbes, soilborne diseases, and competition, making it the most active place for microbial interactions (Kinkel et al., 2011; Raaijmakers et al., 2009). The initial mutual balance between rhizosphere soil microorganisms is destroyed in the “plant-soil-microorganism” ecosystem, a fundamental reason for disease outbreaks (Kim and Anderson, 2018). The complexity and diversity of microbial communities in the rhizosphere are essential for maintaining the dynamic balance of the ecosystem (She et al., 2017). Healthy soil with balanced soil microbial communities can better respond to stress, conducive to promoting plant growth and reducing soilborne diseases (Janvier et al., 2007; Nannipieri et al., 2003). Currently, the regulation of natural microbial communities is deemed one of the most promising strategies for improving soil health to achieve comprehensive and sustainable disease management (Chaparro et al., 2012).
The soil environment or plant species can quickly alter the composition of soil microbes, and different microorganisms perform diverse roles in the soil ecosystem (Huang et al., 2014). A few critical groups of rhizosphere microbial communities are essential for regulating disease occurrences (Berg and Smalla, 2009; Lawson et al., 2019). Some nematode-antagonistic microorganisms have been identified from the RKN-suppressive soil, such as Pseudomonas, Bacillus, Paecilomyces lilacinus, Trichoderma harzianum, etc. (Chen et al., 2022). Their suppressive mechanisms mainly include five aspects: (1) Predation and parasitic effects on Meloidogyne spp. (Liu, 2011; Stirling et al., 2011); (2) Induction of root exudates (Liu et al., 2019); (3) Production of active substances that kill Meloidogyne spp. (Yan, 2003); (4) Competition for nutrient and steric sites (Zuckerman and Kuhlman, 2000); (5) Induction of plant systemic resistance (Wang et al., 2010). This prior research demonstrates the antagonistic effect of soil microorganisms on plant-parasitic nematodes thoroughly and precisely. Despite this, little work has been done to identify specific groupings of soil bacteria linked with Meloidogyne spp. Both diseased and uninfested plants were found in the same plot in this study. We hypothesized that particular microorganisms in infected and uninfested soils regulated Meloidogyne spp. activity. Therefore, this study aims to analyze the bacterial community’s differences in tobacco rhizosphere soil among different infection conditions by 16S rRNA sequencing. PICRUSt further predicted the function of the bacterial community to provide a piece of scientific evidence for ecological control of RKN disease in tobacco.
The field experiment was conducted in Yunnan Province, China, in June 2018. There are two test areas: Yuxi City (24°30′N, 103°32′E) and Jiuxiang Township (25°1′43″N, 103°20′31″E). In the same plot, plants with typical symptoms (had nodules and necrosis with apparent symptoms on roots) of RKN disease (M. incognita) and healthy plants (average growth with no symptom of nodules and necrosis on roots) were sampled, with three plants per group. A total of 12 rhizosphere soils were collected in the two experimental sites. The samples were named Jiuxiang infested and uninfested soil (JX_D, JX_H), Yuxi infested and uninfested soil (YX_D, YX_H). Each soil sample was divided into two parts: quick-frozen with liquid nitrogen and stored at −80°C to analyze microbial communities; the other was dried naturally in the room and analyzed for soil chemical properties.
The soil pH was determined by a pH meter in a 1:2.5 soil/water (w/v) suspension (Wang et al., 2017). The content of soil available nitrogen (AN), available phosphorus (AP), available potassium (AK), and organic matter (OM) was respectively measured as previously described (Cai et al., 2021).
The total DNA extraction of 12 soil samples was done according to a FastDNA SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA). Polymerase chain reaction (PCR) amplification was done according to methods described by Xiaolong (2022) using Wang et al.’s (2017) mentioned primers for 16S rRNA, and 27 cycles were performed. The PCR products were purified using an AxyPrep DNA Gel Extraction Kit (Axygen, Union City, CA, USA) and sequenced by Shanghai Meiji Biological Medicine Technology Co., Ltd. (Shanghai, China) using MiSeq.
The original sequence was spliced by FLASH (Caporaso et al., 2011) software, and the chimeric sequences were removed to obtain the effective sequences by Usearch (Edgar, 2013) software. At a 97% sequence similarity level, the operational taxonomic units (OTUs) were clustered by UPARSE (Edgar, 2013; Stackebrandt and Goebel, 1994). The sequences were annotated by the RDP classifier (Wang et al., 2007) and the Silva database (Altschul et al., 1990). To minimize the effects of sequencing depth on alpha and beta diversity measure, the number of 16S rRNA gene sequences from each sample were rarefied to 22,225, which still yielded an average Good’s coverage of 99.36%, respectively. The coverage, Shannon, Simpson, ACE, and Chao1 indexes were calculated with Mothur software to evaluate the Alpha diversity. Principal coordinate analysis was performed with a Bray-Curtis dissimilarity matrix using Qiime. The Circos graph was built using Circos-0.67-7 software (Yan et al., 2020). Linear discriminant analysis (LDA) effect size (LEfSe) (Segata et al., 2011) analysis searched for statistically different microorganisms between different treatments using LEfSe software. Using pheatmap package in R (Kolde, 2019) to analyze the relationship between soil baterial community at the phylum and genus levels and soil chemical characteristics, and visualized through a heatmap. Data were statistically analyzed using analysis of variance (ANOVA) at P < 0.05, and means were compared using least significant difference (LSD) and Duncan’s multiple range test at P < 0.05.
Six hundred four thousand, seven hundred ninety-nine valid bacterial sequences were read from 12 field samples and divided into 3,216 bacterial OTUs, and 1,431 OTUs were found as common OTUs. According to the Venn diagram of the OTUs distribution of bacteria (Fig. 1A), the numbers of OTUs in two test plots showed a trend of M. incognita-infested soil (JX_D, 2,264; YX_D, 2,431) < uninfested soil (JX_H, 2,461; YX_H, 2,484). In particular, the number of OTUs of bacteria in M. incognita-infested soil from Jiuxiang (2,264) and Yuxi (2,431) was decreased by 8.00% and 2.13%, respectively, compared with that in uninfested soil from Jiuxiang (2,461) and Yuxi (2,484). Only 257 OTUs were shared by JX_D and JX_H, which accounted for only 5.44% of the total OTUs (4,725) in the areas. Only 293 common OTUs are found in YX_D and YX_H, and they make up 5.96% of the total OTUs (4,915) in these areas.
Fig. 1B-D shows the alpha-diversity index of bacteria in different samples. There was no significant difference in Shannon index between M. incognita-infested soil and uninfested soil in two test plot. For the richness estimator, the value of uninfested soil samples was higher than that of M. incognita-infested soil samples. Comparing JX_D with JX_H and YX_D with YX_H, the ACE index decreased by 6.59% and 0.23%, respectively; the Chao1 index decreased by 5.83% and 0.22%, respectively. Although there was no significant difference, it suggests that the RKN infection could lead to reduced bacterial richness in the rhizosphere.
Principal coordinate analysis (PCoA) was conducted using the Bray-Curtis distance method. The PC1 and PC2 accounted for 41.46% and 21.85% of the sample differences, respectively, with 63.31% (Fig. 2). Significant differences were found in the composition of bacterial communities in flue-cured tobacco rhizosphere soil between different experimental sites. JX_D and JX_H mainly occupied the negative x-axis, whereas YX_D and YX_H occupied the positive x-axis, and the distance between them was relatively great, indicating that there was a large difference in the bacterial communities at the two experimental sites. But it is worth noting that there were significant differences between M. incognita-infested soil and uninfested soil both in two sites. M. incognita-infested soil (JX_D, YX_D) mainly occupied the negative y-axis, whereas uninfested soil (JX_H, YX_H) mainly occupied the positive side. Overall, samples from the same field plants clustered together based on infestation states.
Fig. 3A shows the abundance of bacteria in the rhizosphere soil samples under each treatment at the phylum level. The bacteria in different rhizosphere soils were mainly composed of Actinobacteriota, Proteobacteria, Chloroflexi, Acidobacteriota, Firmicutes, and Gemmatimonadota. The contents were therein, relative abundance of Actinobacteriota in JX_D (34.98 ± 3.42%) was higher than in JX_H (30.04 ± 4.50%), there was no significant difference between YX_D (35.66 ± 1.15%) and YX_H (36.72 ± 1.51%). Relative abundance of Proteobacteria in JX_D (34.50 ± 3.31%) and JX_H (35.77 ± 1.06%) was no significant difference, YX_D (28.33 ± 2.13%) was higher than YX_H (23.40 ± 1.39%). The relative abundances of Chloroflexi and Acidobacteriota were higher in uninfected soil. Their relative abundances in JX_H were increased by 34.18% and 28.11%, respectively, compared to those in JX_D, whereas their relative abundances in YX_H increased by 19.58% and 64.23%, comparison with those in YX_D. In contrast, the relative abundance of Firmicutes was higher in M. incognita-infested soil. When comparing JX_D with JX_H and YX_D with YX_H, the relative abundances increased by 4.12% and 49.49%, respectively.
At the genus level (Fig. 3B), the dominant bacterial genera were unclassified_f_Micrococcaceae, Sphingomonas, Streptomyces, norank_o_Gaiellales, norank_c_Subgroup_6, and Bradyrhizobium. In both test pilots, the relative abundances of Sphingomonas, Streptomyces, and Bradyrhizobium was higher in the M. incognita-infested soil than in the uninfested soil. When comparing JX_D with JX_H, the relative abundances was increased by 12.16%, 51.95%, and 25.11%, respectively, and when comparing YX_D with YX_H, the relative abundances was increased by 8.58%, 25.72%, and 36.81%. These results indicate that RKN disease significantly affects the distribution and composition of bacterial communities in the rhizosphere soil of flue-cured tobaccos.
The samples from Jiuxiang and Yuxi were split into M. incognita-infested soil groups and uninfested soil groups to determine the flora driving the difference in bacterial communities between infected and healthy soil. The LEfSe program was then used to perform multilevel species difference analysis, identifying communities or species that caused substantial variations in sample division. The results revealed that among the species with an LDA value greater than 3.5 (Fig. 4), Ralstonia (genus), Acidobacteriia (class), Corynebacteriales (order), Streptosporangiales (order), Solibacteraceae_Subgroup_3 (family), Solibacterales (order), Frankiales (order) and Bryobacter (genus), etc. were enriched mainly in M. incognita-infested soil, whereas Subgroup_6 (the class to genus), KD4-96 (the class to genus), Chloroflexia (the class and its order), Roseiflexaceae (the family and its genus), Blastocatellia_Subgroup_4 (class), Pyrinomonadaceae (the order to family), and RB41 (genus) were located mainly in uninfested soil. These microorganisms, more abundant in M. incognita-infested soil than uninfested soil, could be essential players in community composition differences.
The soil chemical properties of different treatments are shown in Table 1, there was no significant difference in pH and AK between M. incognita-infested soil and uninfested soil in two test plots, and the content pH and AK of infected soil showed a decreasing trend compared to healthy soil (JX_D < JX_H, YX_D < YX_H). In contrast, AN and AP in the infected soil were higher than in healthy soil (JX_D > JX_H, YX_D > YX_H), but no significant difference was observed in them. When the two test sites are compared, the difference between the regions of JX and YX in AK and AP is clearly visible. Our results support that the influence of region on soil chemical properties is greater than that of disease infection.
Using the Spearman correlation heatmap, we also investigated the relationships between microbial preponderant phyla, genera, and main soil properties. Among the top 20 species in relative abundance at the phylum level (Fig. 5A), AN was positively related to the abundances of Halanaerobiaeota, Firmicutes, and WPS-2. In contrast, it was negatively related to Rokubacteria. The soil pH was positively related to the abundance of Latescibacteria. AK was positively related to the abundances of Nitrospirae, Chloroflexi, Acidobacteria, and Gemmatimonadetes, while it was negatively related to Patescibacteria, Fibrobacteres, Proteobacteria, and Bacteroidetes. AP was positively related to the abundances of Acidobacteria and Gemmatimonadetes, while it was negatively related to Cyanobacteria and Fibrobacteres. OM was positively related to the abundances of Verrucomicrobia, Planctomycetes, and Acidobacteria, while it was negatively related to Proteobacteria and Bacteroidetes.
Among the top 20 species in relative abundance at the genus level (Fig. 5B), the soil pH was negatively correlated to the abundances of Sphingomonas, Streptomyces, and Mycobacterium. In contrast, it was positively correlated to norank_c__Subgroup_6, and AP was positively correlated to Conexibacter and Rhodanobacter. AK and OM were negatively correlated to the abundances of Bradyrhizobium and Rhodanobacter. Overall, AK and AP had greater effects on soil bacterial communities than pH, AN and OM. The results suggest that soil chemical properties had significant influences on the structure of bacterial communities.
We predicted functional properties of the bacterial communities to understand better the micro-ecological activities of soil bacterial communities in M. incognita infected and uninfested soil (Fig. 6). The results showed that the bacterial functional features were related to infestation states. It could be observed the metabolic functions were enriched in soil samples and the bacterial metabolism tended to be vigorous. These functional features included: Global and overview maps; Carbohydrate metabolism; Amino acid metabolism; Energy metabolism; Nucleotide metabolism. At the pathway level 3, relative abundances of top-20 functions in M. incognita-infested soil were higher than uninfested soil in the two experimental sites (Supplementary Table 1).
Soil microbial populations are influenced by plant varieties, climate, soil conditions, and agricultural practices (Bowen et al., 2017; Wieland et al., 2001). Different microorganisms play different roles in the soil environment, but some microorganisms that play a crucial role are closely related to the occurrence of diseases. In this study, each pair of M. incognita-infected and uninfected tobacco plants had similar soil properties and local climate conditions, which supported that RKN disease was likely to be responsible for the changes in soil microbial community. Thus, we analyzed the bacterial diversity and community composition in M. incognita-infested and uninfested soil.
The diversity of soil microbial communities is critical to soil ecosystems’ integrity, stability, and sustainability (Zhang et al., 2020). Generally, high microbial diversity and activity can promote plant growth, enhance plant defense, and inhibit soilborne diseases (Mendes et al., 2015). This study found that uninfested soils always had more OTU numbers and higher richness than M. incognita-infested soils. Although the difference is slight, the RKN infection could lead to reduced bacterial richness in the rhizosphere. On the other hand, we looked at the bacterial community distribution in the tobacco rhizosphere soil. According to PCoA, the regions of Jiuxiang and Yuxi showed a distinction (distributed on both sides of x-axis), and under various infection stages, clear separations also revealed between infected soil and healthy soil (distributed on both sides of y-axis). It suggests that the bacterial communities in different areas may be more affected by soil chemical properties, but in the same area (with similar soil chemical properties) might be more driven by the state of plant infection. Our results on the relationships between bacterial community and chemical properties of soils also further supported this point, there was the obvious difference in soil chemical properties between regions of Jiuxiang and Yuxi, but there was no significant difference between infected and healthy soil in the same region.
Further analysis of the differences in bacterial community composition found that the relative abundances of Chloroflexi and Acidobacteriota were higher in uninfected soil. However, the relative abundance of Firmicutes was higher in M. incognita-infested soil. Chloroflexi can utilize organohalide compounds (Krzmarzick et al., 2012). Acidobacteria is mainly involved in the iron cycle and single-carbon compound metabolism and plays an essential role in degrading plant residues (Wang et al., 2016). A high abundance of Acidobacteria can boost the cycling of essential nutrients, thereby improving soil fertility and sustainability.
Firmicutes may digest cellulose, lignin, and lignocellulose by secreting hydrolase enzymes such-as glucosidase and xylanase, allowing them to use refractory carbon sources (Wei et al., 2018). Our results showed that the relative abundance of Firmicutes and the content of AN increased in the infected soil, and the two contents were positively correlated. The relative abundances of Sphingomonas, Streptomyces, and Bradyrhizobium was higher in the M. incognita-infested soil at the genus level. It has been reported that Sphingomonas is mainly involved in the degradation of aromatic compounds and the carbon cycle in soil. It can enhance the viability of plants under environmental stress by improving the soil environment and degrading toxic substances (Xie and Yokota, 2006). Bradyrhizobium can inhibit fungal pathogens and root-knot nematodes (Siddiqui and Shaukat, 2002). Streptomyces is known for their ability to produce antibiotics (Schlatter et al., 2009). The increase in the relative abundance of the above three genera in the infested soil may be related to the durability of plants to pathogen attack.
Compared with uninfected tobacco plants, some metabolism-related functions and signal transduction functions of the rhizosphere bacterial community in infected plants were enriched, such as amino acid metabolism; carbohydrate metabolism; energy metabolism; membrane transport; and signal transduction. Amino acids are constituents of proteins, which play a central role in many other physiological processes in plants. In addition, secondary metabolites formed from amino acids serve many vital functions in plants, such as signaling, defense, interactions with other organisms, and photoprotection (Florencio-Ortiz et al., 2018). Glycolysis and prokaryotes carbon fixation is important carbohydrate metabolism mechanisms that provide energy and metabolites to plants and bacteria for growth and development (Megguer et al., 2017). When M. incognita infection, the higher metabolic-related functions may be closely related to tobacco plant survival and stress response. The enhancement of signal transduction-related functions may be related to plant stress tolerance and scavenging of toxic compounds. However, there are still significant limitations in predicting the function of related bacteria only through PICRUSt. In the future, we will combine the traditional isolation and culture metagenomics methods to comprehensively analyze the bacterial community function in the process of M. incognita infection.
Our study highlighted the effect of M. incognita infection on bacterial community structure and diversity of tobacco rhizosphere soil. The results showed that the infection of M. incognita could cause significant changes in soil bacterial community and soil micro-ecological environment. The uninfested soil showed a richer and more diverse micro-ecological environment than the infested soil. Rhizosphere microbial community stabilization and diversification to improve rhizosphere micro-ecological resilience may help regulate RKN, while manipulating soil features to achieve the appropriate community structure remains difficult.
Acknowledgments
This study was supported by the Project of Yunnan Branch Company of China Tobacco Corporation (No. 2019530000241011, 2018, 1016, 2020, 2010).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
A Venn diagram shows the number of unique, shared, and common bacterial operational taxonomic units (A); Box plot showing the alpha diversity indices of bacterial communities under different treatments, Shannon index (B), ACE index (C), and Chao1 index (D). JX_D, Jiuxiang infested soil; JX_H, Jiuxiang uninfested soil; YX_D, Yuxi infested soil; YX_H, Yuxi uninfested soil. Least significant difference test was used and at a P < 0.05 marked as asterisk.
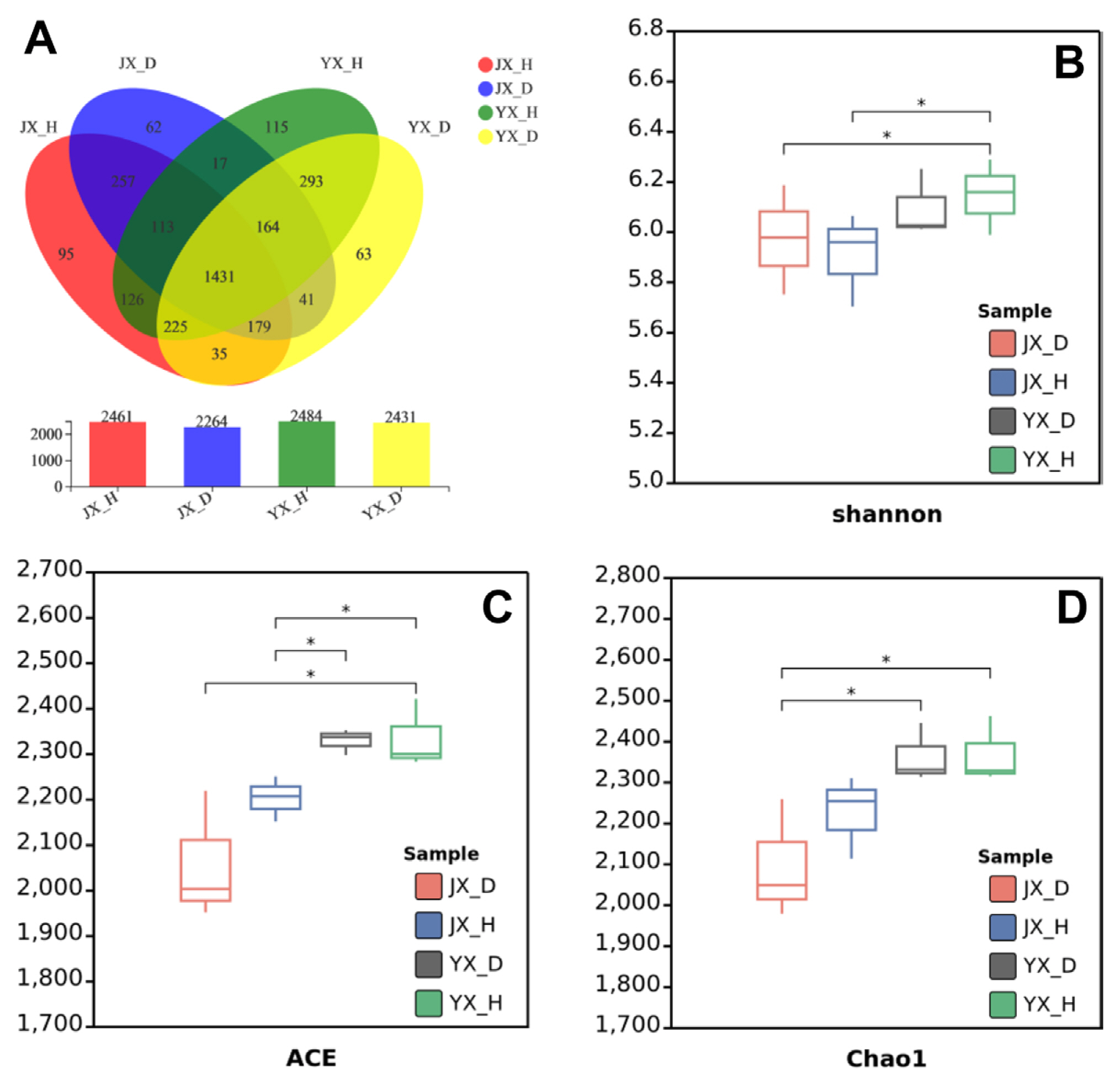
Fig. 2
Principal coordinate analysis (PCoA) plots based on the Bray-Curtis dissimilarity matrix showing the changes in the structure of bacterial communities. OTU, operational taxonomic unit; JX_D, Jiuxiang infested soil; JX_H, Jiuxiang uninfested soil; YX_D, Yuxi infested soil; YX_H, Yuxi uninfested soil.
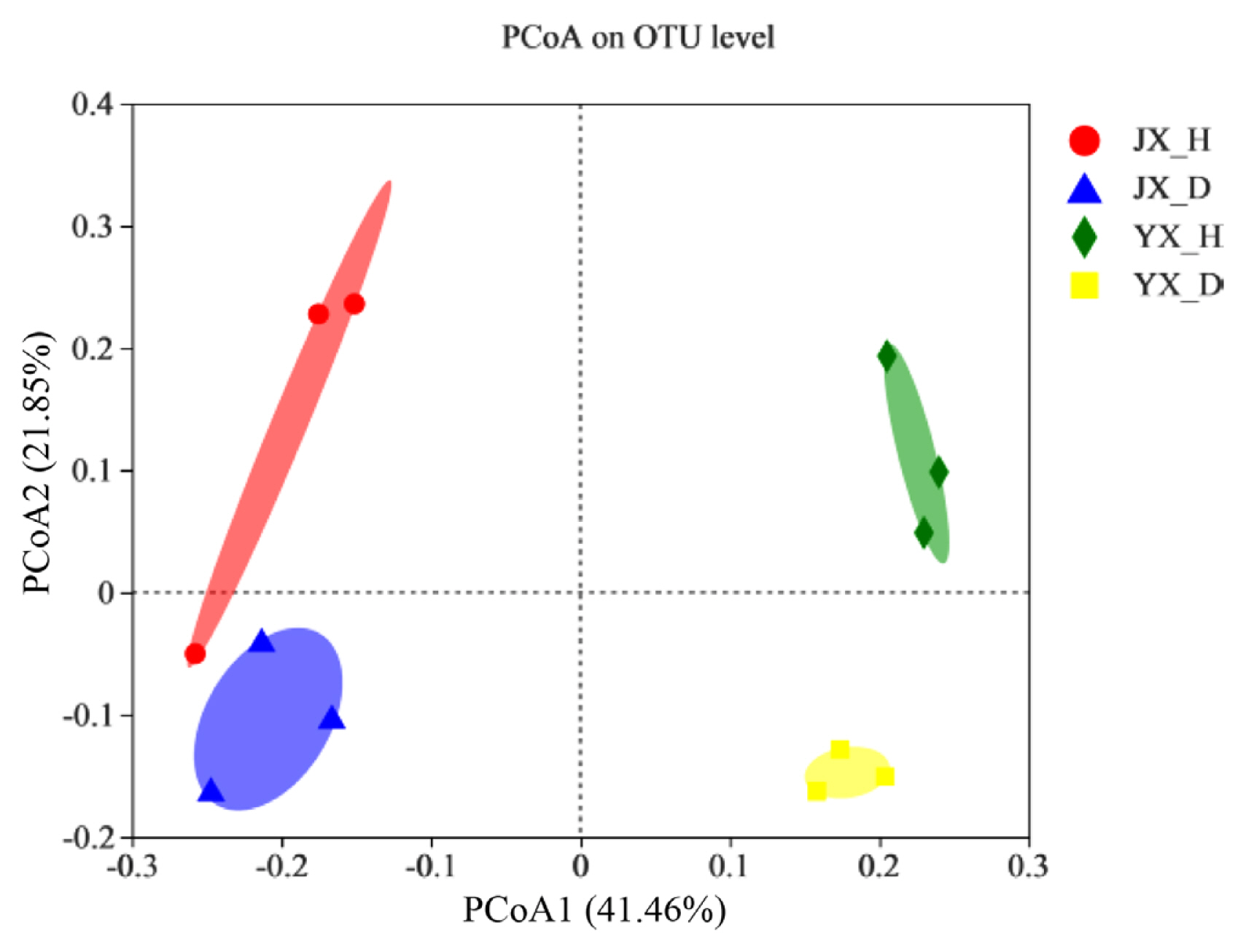
Fig. 3
The composition of most dominant endophytic bacterial communities at phylum (A) and genus level (B) under different treatments. JX_D, Jiuxiang infested soil; JX_H, Jiuxiang uninfested soil; YX_D, Yuxi infested soil; YX_H, Yuxi uninfested soil.
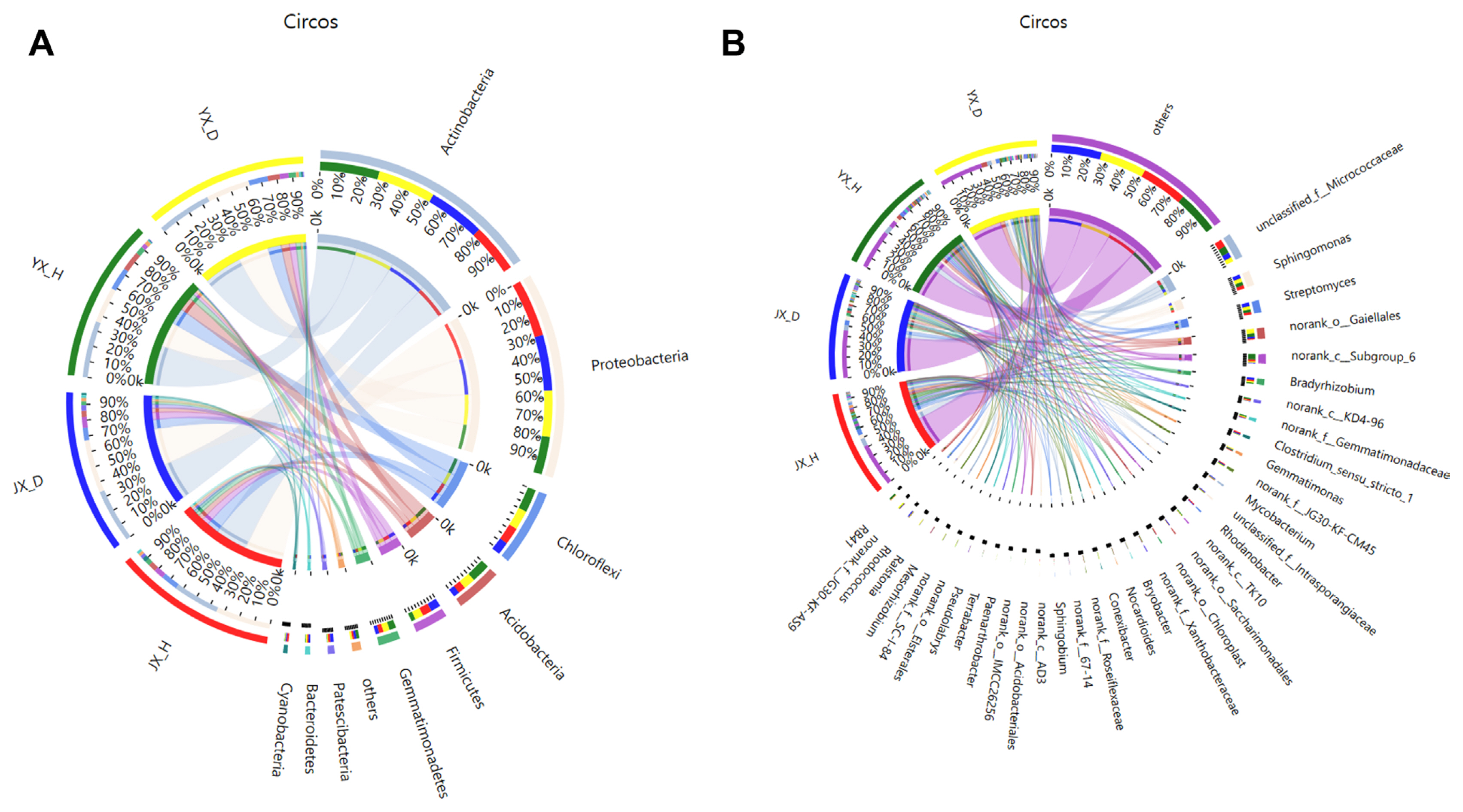
Fig. 4
Linear discriminant analysis (LDA) effect size (LEfSe) cladogram of the aggregated groups of bacterial community in root-knot nematode disease-infested soil (Disease) and healthy soil (Health) (taxa from phylum to genus level). Cladogram representing the abundance of the taxa (A), histogram of the microbiota with LDA = 2 (B).
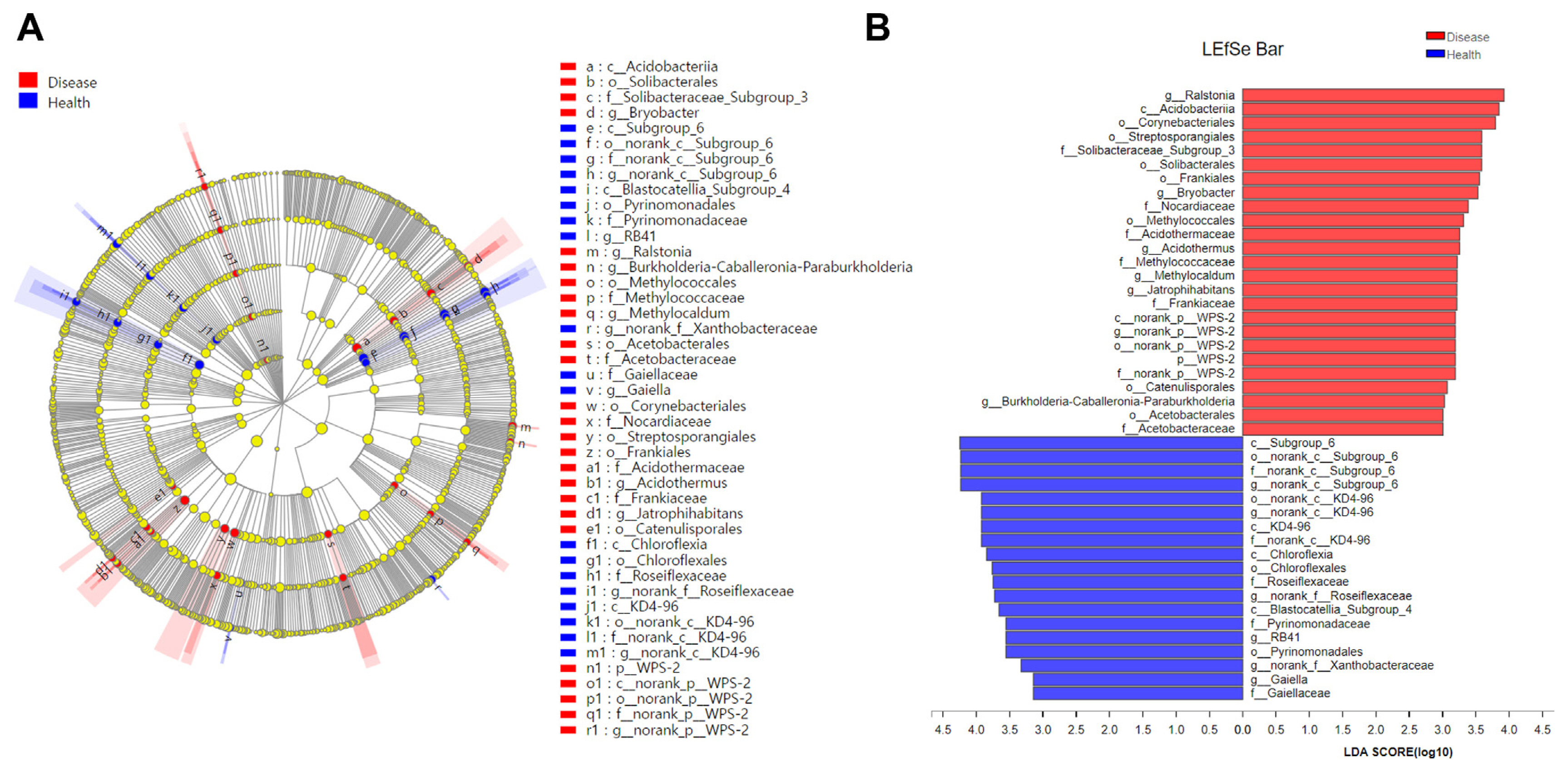
Fig. 5
The Spearman correlation heatmap between soil chemical characteristics and bacterial community composition at Phyla (A) and Genera (B) level. AN, available nitrogen; AP, available phosphorus; AK, available potassium; OM, organic matter. Asterisks represents the significant differences among treatments according to t-test at *0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001.
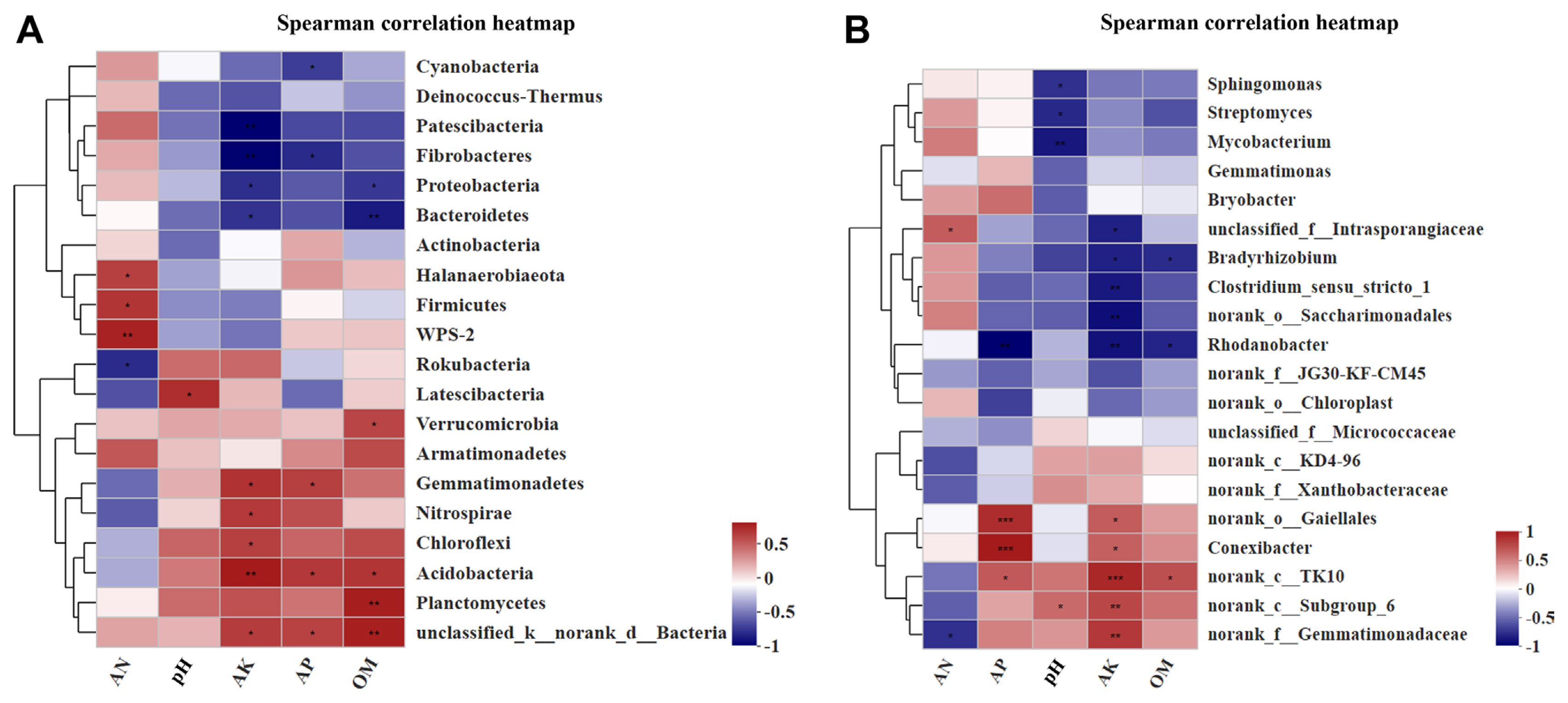
Fig. 6
Heatmap of the relative abundances of top-20 PICRUSt-predicted genes in the different treatments. The abscissa is the treatments, and the ordinate is the function name of pathway level 3. The color gradients of different color blocks represent the changes of different function abundances in the treatments. The legend is the value represented by the color gradient. JX_D, Jiuxiang infested soil; JX_H, Jiuxiang uninfested soil; YX_D, Yuxi infested soil; YX_H, Yuxi uninfested soil.
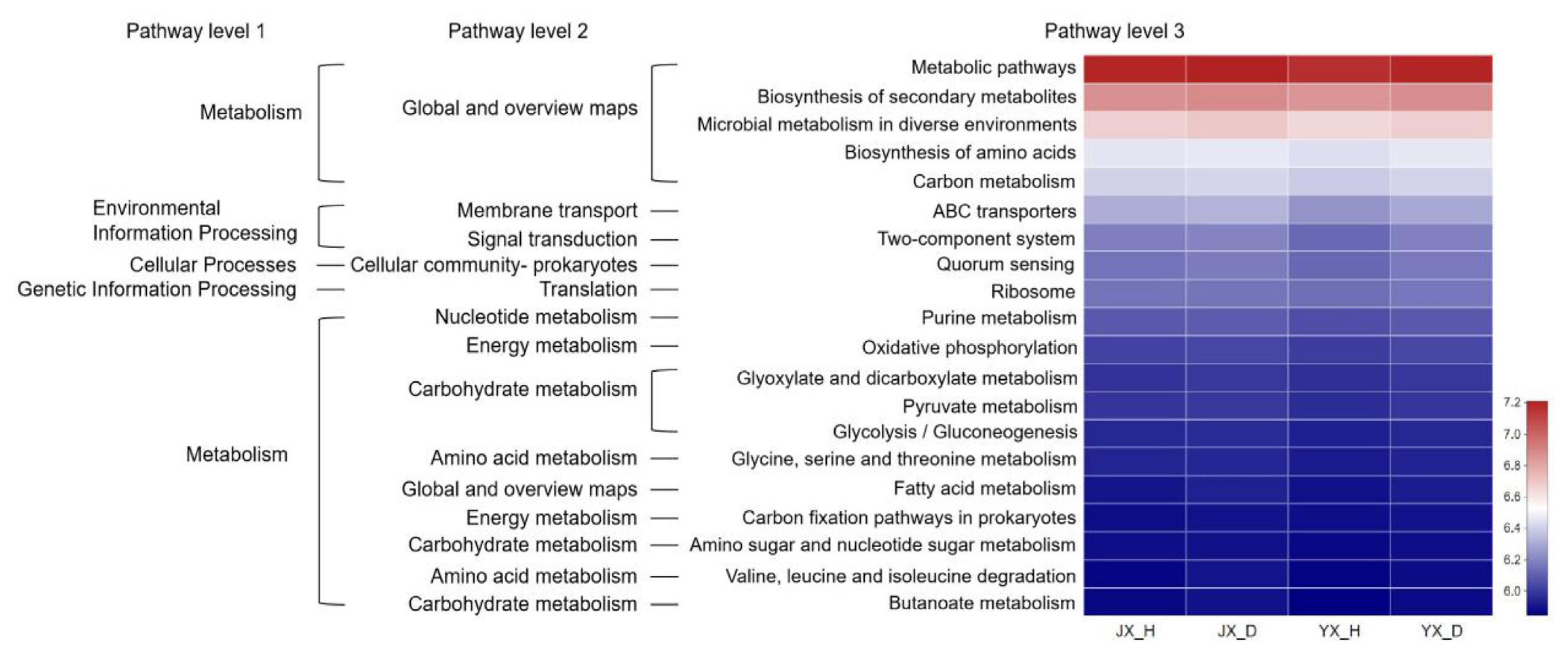
Table 1
Soil chemical properties in different treatments
References
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol 215:403-410.


Berg, G. and Smalla, K. 2009. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol 68:1-13.


Bowen, J.L., Kearns, P.J., Byrnes, J.E.K., Wigginton, S., Allen, W.J., Greenwood, M., Tran, K., Yu, J., Cronin, J.T. and Meyerson, L.A. 2017. Lineage overwhelms environmental conditions in determining rhizosphere bacterial community structure in a cosmopolitan invasive plant. Nat. Commun 8:433.




Cai, Q., Zhou, G., Ahmed, W., Cao, Y., Zhao, M., Li, Z. and Zhao, Z. 2021. Study on the relationship between bacterial wilt and rhizospheric microbial diversity of flue-cured tobacco cultivars. Eur. J. Plant Pathol 160:265-276.


Caporaso, J.G., Lauber, C.L., Walters, W.A., Berg-Lyons, D., Lozupone, C.A., Turnbaugh, P.J., Fierer, N. and Knight, R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci 108:Suppl 1. 4516-4522.



Chaparro, J.M., Sheflin, A.M., Manter, D.K. and Vivanco, J.M. 2012. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 48:489-499.


Chen, Z.B., Xia, Z.Y., Xu, S.G., Wang, H.Y., Zhao, X.N. and Li, J.W. 2015. Screening of tobacco endophytic bacteria resistant to Meloidogyne spp. and its control effect. Acta Tab. Sin 21:71-75 (in Chinese).
Chen, X., Gao, L., Deng, X., Yang, Y., Wang, J., Zhang, Z., Cai, Y., Huang, F., Yang, M., Tong, W. and Yu, L. 2022. Effects of Meloidogyne incognita on the fungal community in tobacco rhizosphere. Rev. Bras. Cienc. Solo 46:e0210127.
Ciancio, A., Colagiero, M., Pentimone, I. and Rosso, L. 2016. Soil microbial communities and their potential for root-knot nematodes management: a review. Environ. Eng. Manag. J 15:1833-1839.

Cui, J.-K., Ren, H.-H., Meng, H.-G., Chang, D. and Jiang, S.-J. 2021. Research progress on the occurrence and control of tobacco root knot nematode in China. Acta Phytopathol. Sin 51:663-682 (in Chinese).
Edgar, R.C. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10:996-998.



Florencio-Ortiz, V., Sellés-Marchart, S., Zubcoff-Vallejo, J., Jander, G. and Casas, J.L. 2018. Changes in the free amino acid composition of Capsicum annuum (pepper) leaves in response to Myzus persicae (green peach aphid) infestation. A comparison with water stress. PLoS ONE 13:e0198093.



Huang, K., Jiang, Q., Liu, L., Zhang, S., Liu, C., Chen, H., Ding, W. and Zhang, Y. 2020. Exploring the key microbial changes in the rhizosphere that affect the occurrence of tobacco root-knot nematodes. AMB Express 10:72.




Huang, X.-F., Chaparro, J.M., Reardon, K.F., Zhang, R., Shen, O. and Vivanco, J.M. 2014. Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92:267-275.

Janvier, C., Villeneuve, F., Alabouvette, C., Edel-Hermann, V., Mateille, T. and Steinberg, C. 2007. Soil health through soil disease suppression: which strategy from descriptors to indicators? Soil Biol. Biochem 39:1-23.

Kim, Y.C. and Anderson, A.J. 2018. Rhizosphere pseudomonads as probiotics improving plant health. Mol. Plant Pathol 10:2349-2459.


Kinkel, L.L., Bakker, M.G. and Schlatter, D.C. 2011. A coevolutionary framework for managing disease-suppressive soils. Annu. Rev. Phytopathol 49:47-67.


Kolde, R. 2019 pheatmap: pretty heatmaps URL https://CRAN.R-project.org/package=pheatmap
. [17 October 2022].
Krzmarzick, M.J., Crary, B.B., Harding, J.J., Oyerinde, O.O., Leri, A.C., Myneni, S.C. and Novak, P.J. 2012. Natural niche for organohalide-respiring Chloroflexi
. Appl. Environ. Microbiol 78:393-401.




Lawson, C.E., Harcombe, W.R., Hatzenpichler, R., Lindemann, S.R., Löffler, F.E., O’Malley, M.A., García Martín, H., Pfleger, B.F., Raskin, L., Venturelli, O.S., Weissbrodt, D.G., Noguera, D.R. and McMahon, K.D. 2019. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol 17:725-741.




Li, M., Jiao, F., Zeng, J., Wang, B., Song, Z., Wu, X., Li, Y., Gao, Y. and Li, W. 2017. Resistance identification of different types of new tobacco varieties (lines) to Meloidogyne incognita
. Tob. Sci. Technol 50:22-26 (in Chinese).
Liu, T.T., Lu, Q.F. and Wang, N.Q. 2019. The rhizosphere regulation mechanism and use of root exudates to inhibit continuous monocropping barrier by nematode disease. J. Plant Nutr. Fertil 25:1038-1046.
Liu, X.L. 2011. Infection mechanism of Paecilomyces lilacinus strains on Meloidogyne incognita
. Ph.D. thesis. Anhui Agricultural University, Hefei, China.
Megguer, C.A., Fugate, K.K., Lafta, A.M., Ferrareze, J.P., Deckard, E.L., Campbell, L.G., Lulai, E.C. and Finger, F.L. 2017. Glycolysis is dynamic and relates closely to respiration rate in stored sugarbeet roots. Front. Plant Sci 8:861.



Mendes, L.W., Tsai, S.M., Navarrete, A.A., de Hollander, M., van Veen, J.A. and Kuramae, E.E. 2015. Soil-borne microbiome: linking diversity to function. Microb. Ecol 70:255-265.



Nannipieri, P., Ascher, J., Ceccherini, M.T., Landi, L., Pietramellara, G. and Renella, G. 2003. Microbial diversity and soil functions. Eur. J. Soil Sci 54:655-670.


Norabadi, M.T., Sahebani, N. and Etebarian, H.R. 2014. Biological control of root-knot nematode (Meloidogyne javanica) disease by Pseudomonas fluorescens (Chao). Arch. Phytopathol. Plant Prot 47:615-621.

Philippot, L., Raaijmakers, J.M., Lemanceau, P. and van der Putten, W.H. 2013. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol 11:789-799.



Raaijmakers, J.M., Paulitz, T.C., Steinberg, C., Alabouvette, C. and Moënne-Loccoz, Y. 2009. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341-361.

Schlatter, D., Fubuh, A., Xiao, K., Hernandez, D., Hobbie, S. and Kinkel, L. 2009. Resource amendments influence density and competitive phenotypes of Streptomyces in soil. Microb. Ecol 57:413-420.



Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W.S. and Huttenhower, C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60.



She, S., Niu, J., Zhang, C., Xiao, Y., Chen, W., Dai, L., Liu, X. and Yin, H. 2017. Significant relationship between soil bacterial community structure and incidence of bacterial wilt disease under continuous cropping system. Arch. Microbiol 199:267-275.



Siddiqui, I.A. and Shaukat, S.S. 2002. Mixtures of plant disease suppressive bacteria enhance biological control of multiple tomato pathogens. Biol. Fertil. Soils 36:260-268.


Song, L., Zhou, P.-P., Li, Y., Zhang, L.-M., Zhao, J.-L., Zhang, G.-P., Luo, X.-L., Na, H.-Y., Liu, F., Gu, X.-H. and Ruan, W.-B. 2019. Distribution of Meloidogyne spp. in tobacco field of Yuxi, Yunnan Province and biological control against Meloidogyne spp. J. Agric. Resour. Environ 36:546-552 (in Chinese).
Stackebrandt, E. and Goebel, B.M. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol 44:846-849.

Stirling, G.R., Rames, E., Stirling, A.M. and Hamill, S. 2011. Factors associated with the suppressiveness of sugarcane soils to plant-parasitic nematodes. J. Nematol 43:135-148.


Tian, B.-Y., Cao, Y. and Zhang, K.-Q. 2015. Metagenomic insights into communities, functions of endophytes, and their associates with infection by root-knot nematode, Meloidogyne incognita, in tomato roots. Sci. Rep 5:17087.




Wang, G.H., Liu, J.J., Yu, Z.H., Wang, X.Z., Jin, J. and Liu, X.B. 2016. Research progress of Acidobacteria ecology in soils. Biotechnol. Bull 32:14-20 (in Chinese).
Wang, J. and Kong, F. 2002. Integrated control technique of tobacco root-knot nematodes diseases. Tob. Sci. Technol 9:43-44 (in Chinese).
Wang, Q., Garrity, G.M., Tiedje, J.M. and Cole, J.R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol 73:5261-5267.




Wang, R., Zhang, H., Sun, L., Qi, G., Chen, S. and Zhao, X. 2017. Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep 7:343.




Wang, W.N., Quan, X.J. and Xiao, C.G. 2010. Research advances on application and mechanism of induced resistance of plant. Hubei Agric. Sci 49:204-206.
Wei, H., Wang, L., Hassan, M. and Xie, B. 2018. Succession of the functional microbial communities and the metabolic functions in maize straw composting process. Bioresour. Technol 256:333-341.


Wieland, G., Neumann, R. and Backhaus, H. 2001. Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl. Environ. Microbiol 67:5849-5854.




Xie, C.-H. and Yokota, A. 2006.
Sphingomonas azotifigens sp. nov., a nitrogen-fixing bacterium isolated from the roots of Oryza sativa
. Int. J. Syst. Evol. Microbiol 56:889-893.


Yan, M., Chen, S., Huang, T., Li, B., Li, N., Liu, K., Zong, R., Miao, Y. and Huang, X. 2020. Community compositions of phytoplankton and eukaryotes during the mixing periods of a drinking water reservoir: dynamics and interactions. Int. J. Environ. Res. Public Health 17:1128.



Yan, X.N. 2003. Screening of Actinomycetes strains producing secondary metaboletes with nematicidal activity. PhD dissertation. Nanjing Agricultural University, Nanjing, China. (in Chinese).
Zhang, S., Wang, Y., Sun, L., Qiu, C., Ding, Y., Gu, H., Wang, L., Wang, Z. and Ding, Z. 2020. Organic mulching positively regulates the soil microbial communities and ecosystem functions in tea plantation. BMC Microbiol 20:103.




Zheng, Y.-X., Yang, M., Wu, J.-G., Wang, J.-M., Xu, Y.-L., Yan, D., Cai, X.-J., Tong, W.-J., Chen, X.-L., Cai, Y.-Z., Yu, L. and He, Y.-S. 2021. Study on interaction between bacterial community and microecological environment in rhizosphere soil of tobacco root rot caused by Fusarium solani
. Mater. Express 11:166-173.
- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 1,895 View
- 114 Download
- ORCID iDs
-
Xiaopeng Deng

https://orcid.org/0000-0003-4293-4066 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



