 |
 |
| Plant Pathol J > Volume 35(2); 2019 > Article |
Abstract
A duplex PCR method was developed for simultaneous detection and identification of tobacco root rot pathogens Phytophthora nicotianae and Thielaviopsis basicola. The specific primers for P. nicotianae were developed based on its internal transcribed spacer (ITS) regions of ribosomal gene, ras gene and hgd gene, while the specific primers for T. basicola were designed based on its ITS regions and β-tubulin gene. The specificity of the primers was determined using isolates of P. nicotianae, T. basicola and control samples. The results showed that the target pathogens could be detected from diseased tobacco plants by a combination of the specific primers. The sensitivity limitation was 100 fg/Οl of pure genomic DNA of the pathogens. This new assay can be applied to screen out target pathogens rapidly and reliably in one PCR and will be an important tool for the identification and precise early prediction of these two destructive diseases of tobacco.
Tobacco is an important economic crop worldwide. Black shank and black root rot of tobacco, caused by Phytophthora parasitica var. nicotianae Tucker and Thielaviopsis basicola (Berk. and Br.) Ferraris, respectively, are destructive soil-borne pathogens that continuously threaten the tobacco production in China and many other countries (Almario et al., 2013; Bian et al., 2017). The damages caused by these diseases have been rising in the past decades along with the expansion of planting scale and changes of cropping system in tobacco plantation (Anderson and Welacky, 1988; Shew, 1985). Yield losses have reached up to 80% in China (Chen et al., 1997; Zhu et al., 2002) due to changing weather conditions, soil quality, and tobacco cultivars (Shew, 1987; Zhang et al., 2008). Additionally, there is currently no efficient management strategy available (Xie et al., 2015). T. basicola has a wide range of hosts and survives for long periods in soil (Anderson and Welacky, 1988). Early detection and diagnosis of the pathogens in plants or soil are vital for successful disease management. The traditional detection methods are time consuming, labor intensive, and costly. Polymerase chain reaction (PCR) and other DNA-based techniques have offered reliable approaches to improve the accuracy and efficiency of plant pathogen detection and identification (Haudenshield et al., 2017; LĂŠvesque et al., 1998; Li et al., 2011).
In previous studies, two parA1 gene-specific primer pairs (Kong et al., 2003; Lacourt and Duncan, 1997) and three internal transcribed spacer (ITS) primer pairs (Grote et al., 2002; Ippolito et al., 2002; Tooley et al., 1997) for specific detection of P. nicotianae have been described. Some primer pairs were also designed for T. basicola based on a common PCR assay (Geldenhuis et al., 2004). However, to the best of our current knowledge, little study has reported the rapid detection for both pathogens in a single reaction using the duplex PCR method. The aim of this study was to develop a reliable and rapid method to detect P. nicotianae and T. basicola simultaneously by a single duplex PCR assay. The developed method could be a useful tool for early detection and identification of root rot pathogens in field control of tobacco fungal diseases.
In this work, diseased tobacco materials were collected from tobacco fields in Yiyang county of Henan province China (Supplementary Table 1). Isolates of P. nicotianae and T. basicola were obtained using the following methods: stems from diseased plants were collected, cut into small pieces, rinsed in tap water, and then air-dried in a sterile bench (biosafety cabinet). The interval between symptomatic and apparently healthy parts was cut into small pieces of approximately 4 mm. The selected pieces were sterilized with 70% ethanol for 15 to 20 s and then rinsed with 1% (wt/vol) of NaClO for 35 to 40 s, followed by sterile distilled water three times. After that, water residue was removed by blotting the sample with sterile filter paper, and the sample was transferred to a potato dextrose agar (PDA) plate containing streptomycin (50 Οg/ml). The plates with samples were incubated in the dark at 25°C until fungi sporulated. Fungal colonies were then subcultured on fresh PDA plates. Isolates were obtained by transferring hyphal tips from colony edges after 7-10 days growth. Isolates were maintained in PDA at 25°C. Soil samples from 1-20 cm depth of soil near tobacco rots were collected in three points from the same tobacco field in Yiyang county of Henan province, China, where tobacco is repeatedly cropped. The purified isolates of Alternaria alternata, Botrytis cinerea, and Cercospora nicotianae as controls were provided by the Institute of Plant Protection of Microbial Resources Preservation of Henan Agricultural University, China. The isolate of Fusarium oxysporum was provided by the Institute of Tobacco Research, Henan Academy of Agricultural Sciences, China. Peronophythora litchii and Rhizoctonia solani were provided by the Institute of Environment and Plant Protection, Chinese Academy of Tropical Agricultural Sciences and Henan Agricultural University, China (Supplementary Table 1).
Next, mycelia of each isolate were collected from 7-21 day cultures grown in PDA at 25°C in the dark. Genomic DNA from fungal mycelia was extracted according to Raeder and Broda (1985) method with some modifications. The genomic DNA of the plant and soil samples was extracted following the method described by Cullen et al. (2001). The quality of DNA samples was assessed by agarose gel electrophoresis. The quantity of isolated DNA was measured by Nano 2000 (Thermo Scientific, Chicago, US). Specific primers for the detection of P. nicotianae and T. basicola were designed using PrimerSelect software (DNASTAR, Madison, Wisconsin, USA) based on sequences previously generated in our lab, which were submitted to NCBI (National Center for Biotechnology Information, USA) (Supplementary Table 2, 3). The specificity of the primers was tested using the DNA template from P. nicotianae and T. basicola, along with other major tobacco pathogens, including Fusarium oxysporum, Alternaria alternata, Botrytis cinerea, Pseudomonas syringae pv. angula and Cercospora nicotianae. Peronophythora litchii and Rhizoctonia solani, which are widely distributed soil-borne pathogens in major tobacco-growing areas in China, were also used to test the specificity of the two target pathogens. Serial dilutions of DNA template from 100 ng/Οl to 10 fg/Οl were used to test the sensitivity of P. nicotianae and T. basicola in PCR detection.
PCR assays were performed in a total volume of 20 Οl containing 5 Οl 2 à Mix (Es Taq DNA Polymerase, 2 à Es Taq PCR Buffer, 4 mM MgCl2, and 0.4 mM dNTPs; Beijing ComWin Biotech Co., Ltd., Beijing, China), 0.5 Οl each primer, 1 Οl DNA template, and the total volume was made up to 20 Οl using nuclease-free water. Distilled water (CK) and healthy plant DNA (Nicotiana. tabacum L) were used as negative controls. PCRs consisted of an initial denaturing step at 95°C for 4 min, followed by 30 cycles of denaturing at 95°C for 30 s, annealing at 59°C for 30 s, and extension at 72°C for 20 s, with a final extension at 72°C for 10 min. The duplex PCR assays were performed in a total volume of 20 Οl containing 0.2 Οl Taq (5 u/Οl), 1.5 Οl dNTP (2.5 mM), 1 Οl MgCl2 (2.5 mM), 0.4/0.6/0.8 Οl primer 1, 0.4/0.6/0.8 Οl primer 2, and 2 Οl mixed DNA template. The total volume was made up to 20 Οl using nuclease-free water. Distilled water and healthy plant DNA (N. tabacum L) were used as a CK and negative control, respectively. Reaction conditions were the same as those described above. PCR products were resolved by electrophoresis in 1.5% agarose gel.
The specific detection primers were designed based on the gene sequence of isolates of F. oxysporum, A. alternata, B. cinerea, and C. nicotianae obtained from the NCBI database and the sequences of P. nicotianae and T. basicola sequenced in our lab (Supplementary Table 2, 3), the rDNA-ITS regions were selected to design primers for both P. nicotianae and T. basicola using PrimerSelect. The GenBank accession number of the rDNA-ITS sequences of the tested fungi were GQ121290.1 (F. oxysporum), JN880415.1 (A. alternata), FJ169664.2 (B. cinerea) and AY266159 (C. nicotianae). Similarly, the ras and hgd gene sequences were examined for the availability to define specific primers for P. nicotianae, and the β-tubulin gene sequence was used to define specific primers for T. basicola (Supplementary Table 2, 3). Primer specificity of P. nicotianae isolates was tested by using control isolates of T. basicola, A. alternariae, B. cinerea, C. nicotianae, P. syringae pv. angula, F. oxysporum, P. litchi and R. solani, which are the major tobacco diseases and common soilborne pathogens in the tobacco-planting area of China. Similarly, primer specificity of T. basicola isolates was tested against control isolates of P. nicotianae and the other seven isolates mentioned above.
In total, three primer pairs were designed for specific amplification of P. nicotianae, including one primer pair based on the ITS region, one primer pair based on the hgd gene sequence, and one primer pair based on the ras gene sequence (Table 1). The tests for P. nicotianae isolate showed, in each case, that a unique DNA fragment corresponding to the predicted size of 217 bp was resolved, while all of the other control isolates and the negative control did not yield any detectable PCR product using these three primer sets (Supplementary Fig. 1). Two primer pairs were designed for specific amplification of T. basicola based on the ITS region and β-tubulin gene sequence, respectively (Table 1). The results showed that tests for T. basicola isolates were able to obtain a unique DNA fragment corresponding to the predicted size of 132 bp for primer pair TBITS1419F/TBITS1419R and 76 bp for primer pair TBβ1419F/TBβ1419R, but no PCR product was detected from either the negative control or any other control isolates (Supplementary Fig. 2). Using serial dilution, the sensitivities of the primer combinations YYI-F/YYI-R, YYRAS1414-F2/YYRAS1414-R2 and YYH-F/YYH-R for P. nicotianae detection were determined to be 100 fg, 100 pg, and 10 pg, respectively (Supplementary Fig. 1). Similarly, the sensitivities of the primer combinations TBITS1419F/TBITS1419R and TBβ1419F/TBβ1419R for T. basicola were determined to be 100 fg and 10 pg, respectively (Supplementary Fig. 2). Primer set YYI-F/YYI-R and YYH-F/R for P. nicotianae showed a higher sensitivity than the other two primer sets. Thus, primer sets YYI-F/YYI-R and YYH-F/YYH-R of P. nicotianae, along with the primer sets TBITS1419-F/TBITS1419-R and TBβ1419-F/TBβ1419R for T. basicola, were selected for the development of the duplex PCR.
Furthermore, the amplification conditions of the PCR were optimized to reach better efficiency of the duplex PCR. For primer sets TBITS1419-F/TBITS1419-R and YYI-F/YYI-R or YYH-F/R, PCRs were performed in a total volume of 20 Οl. Each reaction consisted of 0.2 Οl Taq (5 u/Οl), 1.5 Οl dNTP (2.5 mM), 1 Οl MgCl2 (2.5 mM), 0.4 Οl primer 1, 0.6 Οl primer 2, and 1 Οl DNA template. For primer sets TBβ1419-F/TBβ1419R and YYI-F/YYIR or YYH-F/R, PCRs were performed in a total volume of 20 Οl. Each reaction consisted of 0.2 Οl Taq (5 u/Οl), 1.5 Οl dNTP (2.5 mM), 1 Οl MgCl2 (2.5 mM), 0.4 Οl primer 1, 0.4 Οl primer 2, and 1 Οl DNA template.
All pathogen isolates and the negative control were tested using the duplex PCR system to confirm the specificity of the primers. Using the selected primer sets (TBITS1419-F/TBITS1419-R and YYI-F/YYI-R, TBITS1419-F/TBITS1419-R and YYH-F/R, TBβ1419-F/TBβ1419R and YYI-F/YYI-R, TBβ1419-F/TBβ1419R and YYH-F/R), the duplex PCR system amplified a unique DNA fragment of P. nicotianae and T. basicola isolates (Fig. 1). The comparative assay indicated that no detectable PCR product was resolved from either the negative control or from other isolates (Supplementary Fig. 3). Different template ratios for the two targets of P. nicotianae and T. basicola were used in one reaction to check the duplex PCR efficiency. The results showed that the selected primer pair combination could resolve the target PCR products clearly at the given conditions, indicating its flexibility in potential application (Supplementary Fig. 4).
Based on the above experiment results, a duplex PCR assay was developed for the rapid and specific amplification of P. nicotianae and T. basicola from tobacco tissues. Using the stem base material, the duplex PCR with the selected primer sets amplified out two target DNA fragments in DNA templates of infected tobacco plant samples. The expected specific bands were detected in both latent and diseased plants. No fragment was detected in the healthy tobacco plants (Supplementary Fig. 5). Therefore, this duplex PCR method was successfully established in PCR detection for both latent and diseased tobacco tissues. No target PCR products for P. nicotianae or T. basicola were detected in the healthy tobacco tissues and soil samples. The duplex PCR method uses a mixture of locus-specific primer sets in a single reaction to amplify two target loci from one or more organisms (Markoulatos et al., 2002; Sudan et al., 2015). Both single and duplex PCRs could detect each pathogen in DNA mixtures of P. nicotianae and T. basicola. However, duplex PCR gives a significant advantage over single PCR detection systems when analyzing large numbers of samples and saves substantial time and cost. Although quantitative PCR (qPCR) has been reported to be an appropriate technique for the early detection of pathogens, it requires more expensive equipment and reagents. Thus, a developed detection assay should not be only rapid and sensitive but also economical and easy to conduct. In this study, a reliable method to identify and detect P. nicotianae and T. basicola simultaneously using specifically designed primer sets was developed.
Previously, a number of genes, including the coxI, coxII, β-tubulin, HS protein 90, nad1, nad9, rps10, secY, the elicitin gene ParA1, and Ypt1, were studied in the molecular identification of Phytophthora species (Blair et al., 2008; Kong et al., 2003; Lacourt and Duncan, 1997; Li et al., 2011; Li et al., 2015; Meng and Wang, 2010). A recent study indicated that the elicitin gene parA1 does not contain introns and, therefore, is unlikely to be sufficiently diverse to allow a broad range of species to be distinguished (Li et al., 2015). The ras, hgd, and β-tubulin genes seem to be more promising targets because their coding and noncoding regions have alternative structures. Furthermore, they are highly conserved, such that species-specific primers based on ras, hgd and β-tubulin genes in Phytophthora and the β-tubulin gene in Thielaviopsis would be more specific relative to other targets (Cheng et al., 2016; Grasko et al., 2009; Guiltinan et al., 1987). The BLAST results revealed that the nucleotide sequences in the rDNA-ITS regions of P. nicotianae and T. basicola are also highly conserved (Grote et al., 2002; Huang et al., 2010). In this study, the rDNA-ITS, together with ras and hgd gene sequences, were examined to define specific primers for P. nicotianae, and the rDNA-ITS, together with β-tubulin gene sequences, were used to define specific primers for T. basicola.
In conclusion, the duplex PCR method reported here could realize the accurate and rapid detection of these pathogens and be conducive to the diagnosis of diseases. Some traditional methods based on Southern blot or other molecular markers usually require 5-6 Îźg of target DNA to detect an isolate from pure cultures (Dodd et al., 2004). However, PCR-based methods have the advantage of allowing detection from mixed DNA samples (Leclerc-Potvin et al., 1999) and detection of a low amount of the target pathogen DNA in diseased plants and soil in a fast, reliable and affordable way (Dodd et al., 2004). The concentrations of the different primer sets used here in the duplex PCR were balanced in this study. Primer pairs TBITS1419-F/TBITS1419-R and YYH-F/R were the top candidate primer combinations in our study and could be successfully used in PCR detection for both latent and diseased plant materials. All in all, our study could be an effective tool not only for detecting both of the target pathogens simultaneously but also for surveying the diseased plant at a very early stage to prevent the spread of disease. The established duplex PCR method could be a helpful approach for the study of tobacco disease epidemics and the protection of tobacco production.
Acknowledgements
This work was supported by the Science and Technology Project of Henan Tobacco Company, China (Project No. 201741000027063) and the Self-innovation Project of Henan Academy of Agricultural Science, China (Project No. 2017ZC21). We are grateful to Professor Trevor C. Charles at the University of Waterloo Canada for reading through and editing our manuscript before submission to The Plant Pathology Journal.
Fig. 1
Duplex PCR results for Phytophthora nicotianae and Thielaviopsis basicola. (A, B) Duplex PCR for primer combination TBITS1419-F/TBITS1419-R of T. basicola with YYI-F/YYI-R and YYH-F/YYH-R of P. nicotianae. 1: DNA mixture of T. basicola and P. nicotianae; 2: DNA mixture of P. nicotianae and T. basicola; 3: DNA of T. basicola; 4: DNA of P. nicotianae; M: DM2000 DNA Marker; CK: control (distilled water). (C, D) Duplex PCR for primer TBβ1419-F/TBβ1419-R of T. basicola combined with YYI-F/YYI-R and YYH-F/YYH-R of P. nicotianae. 1: DNA mixture of T. basicola and P. nicotianae; 2: DNA mixture of P. nicotianae and T. basicola; 3: DNA of P. nicotianae; 4: DNA of T. basicola; M: DM2000 DNA Marker; CK: control (distilled water).
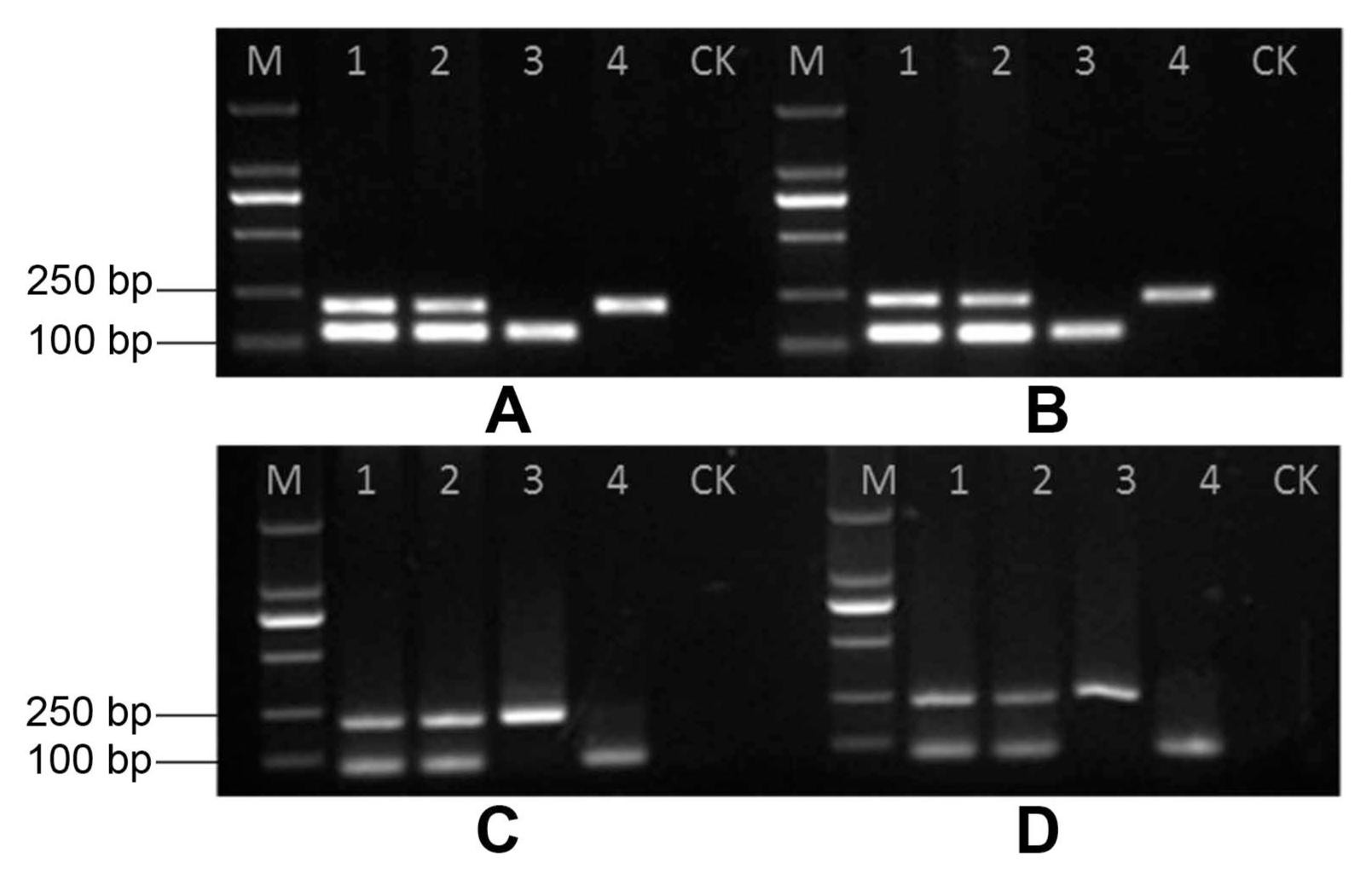
Table 1
Primers for the detection of Phytophthora nicotianae and Thielaviopsis basicola
References
Almario, J, KyselkovĂĄ, M, KopeckĂ˝, J, SĂĄgovĂĄ-MareÄkovĂĄ, M, Muller, D, Grundmann, GL and MoĂŤnne-Loccoz, Y 2013. Assessment of the relationship between geologic origin of soil, rhizobacterial community composition and soil receptivity to tobacco black root rot in Savoie region (France). Plant Soil. 371:397-408.

Anderson, TR and Welacky, TW 1988. Populations of Thielaviopsis basicola in burley tobacco field soils and the relationship between soil inoculum concentration and severity of disease on tobacco and soybean seedlings. Can J Plant Pathol. 10:246-251.

Bian, CH, Ding, YQ, Zhao, SM, Li, SJ and Kang, YB 2017 Molecular identification and pathogenicity analysis of Thielaviopsis basicola in Henan Province. Tobacco Sci Technol. 50:8-14 (in Chinese).
Blair, JE, Coffey, MD, Park, SY, Geiser, DM and Kang, S 2008. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol. 45:266-277.


Chen, RT, Zhu, XC, Wang, ZJ, Guo, ZY, Dong, HS, Wang, LZ, Liu, YR and Shi, JK 1997 A report of investigating and studying tobacco infectious diseases of 16 ma in tobacco producing provinces (regions) in China. Chin Tob Sci. 18:1-7 (in Chinese).
Cheng, Y, Wang, W, Yao, J, Huang, L, Voegele, RT, Wang, X and Kang, Z 2016. Two distinct Ras genes from Puccinia striiformis exhibit differential roles in rust pathogenicity and cell death. Environ Microbiol. 18:3910-3922.


Cullen, DW, Lees, AK, Toth, IK and Duncan, JM 2001. Conventional PCR and real-time quantitative PCR detection of Helminthosporium solani in soil and on potato tubers. Eur J Plant Pathol. 107:387-398.
Dodd, SL, Hill, RA and Stewart, A 2004. A duplex-PCR bioassay to detect a Trichoderma virens biocontrol isolate in non-sterile soil. Soil Biol Biochem. 36:1955-1965.

Geldenhuis, MM, Roux, J, Wingfield, MJ and Wingfield, BD 2004. Development of polymorphic markers for the root pathogen Thielaviopsis basicola using ISSR-PCR. Mol Ecol Notes. 4:547-550.

Grasko, JM, Hooper, AJ, Brown, JW, Mcknight, CJ and Burnett, JR 2009 A novel missense HGD gene mutation, K57N, in a patient with alkaptonuria. Clin Chim Acta. 403:254-256 (in Chinese).


Grote, D, Olmos, A, Kofoet, A, Tuset, JJ, Bertolini, E and Cambra, M 2002. Specific and sensitive detection of Phytophthora nicotianae by simple and nested-PCR. Eur J Plant Pathol. 108:197-207.
Guiltinan, MJ, Velten, J, Bustos, MM, Cyr, RJ, Schell, J and Fosket, DE 1987. The expression of a chimeric soybean beta-tubulin gene in tobacco. Mol Genet Genomics. 207:328-334.

Haudenshield, JS, Song, JY and Hartman, GL 2017. A novel, multiplexed, probe-based quantitative PCR assay for the soybean root- and stem-rot pathogen, Phytophthora sojae, utilizes its transposable element. PloS One. 12:e0176567



Huang, JL, Wu, JZ, Li, CJ, Xiao, CG and Wang, GX 2010. Detection of Phytophthora nicotianae in soil with realtime quantitative PCR. J Phytopathol. 158:15-21.

Ippolito, A, Schena, L and Nigro, F 2002. Detection of Phytophthora nicotianae and P. citrophthora in citrus roots and soils by nested PCR. Eur J Plant Pathol. 108:855-868.
Kong, P, Hong, C, Jeffers, SN and Richardson, PA 2003. A Species-specific polymerase chain reaction assay for rapid detection of Phytophthora nicotianae in irrigation water. Phytopathology. 93:822-831.


Lacourt, I and Duncan, JM 1997. Specific detection of Phytophthora nicotianae using the polymerase chain reaction and primers based on the DNA sequence of its elictin gene ParA1. Eur J Plant Pathol. 103:73-83.
Leclerc-Potvin, C, Balmas, V, Charest, PM and Jabaji-Hare, S 1999. Development of reliable molecular markers to detect non-pathogenic binucleate Rhizoctonia isolates (AG-G) using PCR. Mycol Res. 103:1165-1172.

LĂŠvesque, CA, Harlton, CE and de Cock, AW 1998. Identification of some oomycetes by reverse dot blot hybridization. Phytopathology. 88:213-222.


Li, B, Liu, P, Xie, S, Yin, R, Weng, Q and Chen, Q 2015. Specific and sensitive detection of Phytophthora nicotianae by nested PCR and loop-mediated isothermal amplification assays. J Phytopathol. 163:185-193.

Li, MZ, Asano, T, Suga, H and Kageyama, K 2011. A multiplex PCR for the detection of Phytophthora nicotianae and P. cactorum, and a survey of their occurrence in strawberry production areas of Japan. Plant Dis. 95:1270-1278.


Markoulatos, P, Siafakas, N and Moncany, M 2002. Multiplex polymerase chain reaction: a practical approach. J Clin Lab Anal. 16:47-51.



Meng, J and Wang, Y 2010. Rapid Detection of Phytophthora nicotianae in infected tobacco tissues and soil samples based on its Ypt 1 gene. J Phytopathol. 158:1-7.

Raeder, U and Broda, P 1985. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1:17-20.

Shew, HD 1985. Response of Phytophthora parasitica var. nicotianae to metalaxyl exposure. Plant Dis. 69:559-562.

Shew, HD 1987. Effect of host resistance on spread of Phytophthora parasitica var. nicotianae and subsequent development of tobacco black shank under field conditions. Phytopathology. 77:1090-1093.

Sudan, V, Jaiswal, AK, Parashar, R and Shanker, D 2015. A duplex PCR based assay for simultaneous detection of Trypanosoma evansi and Theileria annulata infections in water buffaloes. Trop Anim Health Prod. 47:915-919.


Tooley, PW, Bunyard, BA, Carras, MM and Hatziloukas, E 1997. Development of PCR primers from internal transcribed spacer region 2 for detection of Phytophthora species infecting potatoes. Appl Environ Microbiol. 63:1467-1475.



Xie, YH, Zhang, YG, Zhu, LQ, You, DG and Lu, Y 2015 Research advances in integrated management of tobacco black shank. Curr Biotechnol. 2015 41-46 (in Chinese).
Zhang, XG, Zheng, GS, Han, HY, Han, W, Shi, CK and Chang, CJ 2008. RAPD PCR for diagnosis of Phytophthora parasitica var. nicotianae isolates which cause black shank on tobacco. J Phytopathol. 149:569-574.

Zhu, CX, Wang, YT and Wang, ZF 2002. Tobacco diseases of China. China Agriculture Press, Beijing, China. 350.(in Chinese).



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




