Isolation and Characterization of the Colletotrichum acutatum ABC Transporter CaABC1
Article information
Abstract
Fungi tolerate exposure to various abiotic stresses, including cytotoxic compounds and fungicides, via their ATP-driven efflux pumps belonging to ATP-binding cassette (ABC) transporters. To clarify the molecular basis of interaction between the fungus and various abiotic stresses including fungicides, we constructed a cDNA library from germinated conidia of Colletotrichum acutatum, a major anthracnose pathogen of pepper (Capsicum annum L.). Over 1,000 cDNA clones were sequenced, of which single clone exhibited significant nucleotide sequence homology to ABC transporter genes. We isolated three fosmid clones containing the C. acutatum ABC1 (CaABC1) gene in full-length from genomic DNA library screening. The CaABC1 gene consists of 4,059 bp transcript, predicting a 1,353-aa protein. The gene contains the typical ABC signature and Walker A and B motifs. The 5′-flanking region contains a CAAT motif, a TATA box, and a Kozak region. Phylogenetic and structural analysis suggested that the CaABC1 is a typical ABC transporter gene highly conserved in various fungal species, as well as in Chromista, Metazoans, and Viridiplantae. We also found that CaABC1 was up-regulated during conidiation and a minimal medium condition. Moreover, CaABC1 was induced in iprobenfos, kresoxim-methyl, thiophanate-methyl, and hygromycin B. These results demonstrate that CaABC1 is necessary for conidiation, abiotic stress, and various fungicide resistances. These results will provide the basis for further study on the function of ABC transporter genes in C. acutatum.
Hot pepper (Capsicum annum L.) anthracnose has been a destructive disease in commercial pepper production fields (Harp et al., 2008; Lewis-Ivey et al., 2004; Park and Kim, 1992), and in Korea, the disease is estimated to cause a loss of more than US$100 million annually (Kim and Park, 1998). Furthermore, since the pepper anthracnose outbreak in 2002, this disease has received high priority (Kim et al., 2008). Several Colletotrichum spp., such as C. acutatum, C. coccodes, C. dematium, and C. gloeosporioides, thought to be causal agents of anthracnose (Park and Kim, 1992). However, more recently, C. acutatum was identified as the primary Colletotrichum species for anthracnose in pepper (Kim et al., 2008). Similar outbreaks of anthracnose on peppers have occurred in Ohio (Lewis-Ivey et al., 2004) and Florida, USA (Harp et al., 2008), and C. acutatum was identified as the primary Colletotricum species for anthracnose in pepper (Lewis-Ivey et al., 2004).
C. acutatum (teleomorph: Glomerella acutata) is an important anthracnose pathogen on a wide range of host plants, causing significant economic loss in various crops, including apple, almond, citrus, strawberry, tomato, and hot pepper (Sutton, 1992; Freeman et al., 1998; Peres et al., 2005). Various fungicides have been identified to control C. acutatum, including copper compounds (e.g., copper hydroxide), the quinone outside inhibitors (azoxystrobin, trifloxystrobin, or pyraclostrobin), triazoles, dithiocarbamates, and benzimidazole compounds (Harp et al., 2014; Wedge et al., 2007). Notably, C. acutatum is tolerant to benomyl and other benzimidazole fungicides (Adaskaveg and Hartin, 1997; Peres et al., 2002; Talhinhas et al., 2002; Talhinhas et al., 2005). The azole fungicides are the most effective in inhibiting in vitro growth of C. acutatum (Paredes and Munoz, 2002), but the rapid development of fungicide resistant strains has limited their use. An understanding of the fungicide-resistant mechanisms will help to enhance control of anthracnose in pepper.
Phytopathogenic fungi have developed various biological mechanisms that provide resistance to fungicides or abiotic stresses. Genes responsible for this resistance include ATP-binding cassette (ABC) transporters. For example, a gene deletion mutant of an ABC transporter, ABC1in Magnaporthe oryzae, showed hypersensitivity to several drugs (Urban et al., 1999). Moreover, ABC1 (Urban et al., 1999), ABC3 (Schneider and Hunke, 1998), ABC4 (Gupta and Chattoo, 2008), and ABC5 (Kim et al., 2013) from M. oryzae, GpABC1 from Gibberella pulicaris (Fleissner et al., 2002), Mgatr4 from Mycosphaerella graminicola (Stergiopoulos et al., 2003) and BcatrB from Botrytis cinerea (Schoonbeek et al., 2001) are required for pathogenicity.
ABC-transporter proteins utilize energy derived from the hydrolysis of ATP to “pump” the substrate across a membrane, thus effectively reducing intracellular concentration to less toxic levels. The proteins are defined by the presence of amino acid sequences such as the ABC-ATPase domain, ABC domain, or nucleotide-binding domain. This domain contains the two peptide motifs Walker A (p-loop) and a hydrophobic Walker B motif (Walker et al., 1982). Both motifs are involved in ATP-binding proteins and identified as ATP signatures (Hyde et al., 1990). In addition, transmembrane domains are embedded in cell membranes that consist of at least six transmembranes.
Until now, no ABC transporter genes have been isolated and characterized in C. acutatum. To begin defining the functional significance of the ABC transporter gene in C. acutatum, we are the first to identify a partial cDNA that encoded an ABC transporter, CaABC1, in C. acutatum. We also present the corresponding full-length gene structure of CaABC1 with the motifs. C. acutatum CaABC1 is most closely related to the ABC transporter XP_007590216 of C. fioriniae. CaABC1 also shares a high degree of homology with the other Colletotrichum spp., including C. higginsianum, C. sublineola, C. graminicola, C. orbiculare, and C. gloeosporioides. CaABC1 was up-regulated in conidiation, abiotic stresses, and multiple fungicides. To our knowledge, this is the first structural and functional analysis of an ABC transporter gene in C. acutatum. Our results will provide the basis for further study on the function of ABC transporter genes in fungicide resistance and pathogenicity in C. acutatum.
Materials and Methods
Fungal strains and culture conditions
C. acutatum strain JC24 was maintained on potato dextrose agar (PDA; Difco Laboratories, Sparks, MD, USA) at 25°C in constant dark. DNA and RNA were isolated from mycelia, germinated conidia, and conidia, which were grown in liquid potato dextrose medium or complete medium (CM) (Talbot et al., 1997) for 3–4 days.
Screening of an ABC transporter gene from cDNA library
To detect putative ABC transporter genes, a cDNA library generated from germinating conidia of C. acutatum JC24 (Kim et al., 2013) was used. A putative ABC transporter cDNA clone (Wb01014-C11) was selected by the process of end sequencing. To obtain more information on the Wb01014-C11 clone, primers were designed and applied for sequencing. The partial putative ABC transporter gene of the clone Wb01014-C11 was subsequently used as a probe.
Isolation and sequencing of the full-length C. acutatum ABC1 gene
To retain the full length of the ABC transporter gene in C. acutatum JC24, a genomic DNA library was constructed according to the manufacturer’s protocols using a commercial fosmid vector, pEpiFOS-5 (Epicentre Biotechnologies, Madison, WI, USA). The packaged library was transformed into Escherichia coli EPI-100, and E. coli transformants were selected on Luria–Bertani (LB) agar supplemented with chloramphenicol. The library clones were stored in a 96-well plate at −80°C.
To select genomic clones containing CaABC1 gene in the fosmid library, dot blot analysis and colony pooling polymerase chain reaction (PCR) were performed. Actively growing cells from the fosmid library were picked with a 96-well pin and transferred to a hybond-N+ membrane on an LB agar plate and then incubated at 37°C overnight. The colonies grown on an LB agar plate were treated with 0.4 N NaOH buffer 20 min for denaturation and then in a 5× SSC buffer 10 min for neutralization. Hybridization was performed with a a putative ABC transporter cDNA clone, Wb01014-C11, as a probe. Simultaneously, pooling PCR was also performed using a genomic fosmid library. We combined 96 colonies from a 96-well plate as a single unit, and then the pooled colonies were used as a template to amplify the putative ABC transporter gene.
Shotgun genome sequencing was performed with selected fosmid clones. The Promoter Scan software (Prestridge, 1995), ExPaSy program packages (http://www.expasy.org/spdbv), Lagergene software package program (DNASTAR, Madison, WI, USA), TMHMM server v.2.0 (http://www.cbs.duk.dk/services/TMHMM), and DDBJ/EMBL/GenBank database were used for sequence analysis and alignments.
Phylogenetic analysis
Homology searches of protein sequences were performed using the BLAST algorithms available at the National Center for Biotechnology Information (NCBI), Broad Institute (http://www.broadinstitute.org). Homologous genes of the CaACB1 amino acid sequences from other organisms were collected from Genoscope (http://www.genoscope.cns.fr), CFGP (http://cfgp.snu.ac.kr), JGI (http://jgi.doe.gov), Ensembl (http://www.ensembl.org), Wormbase (http://www.wormbase.org), and Flybase (http://www.flybase.org) (Table 1). The protein sequences were aligned with ClustalW using the MEGA6.03 software with default parameters (Tamura et al., 2013). A phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987) with 2,000 bootstrap replicates in the MEGA6.0 software. A protein structure of the ABC transporter genes was obtained from the InterPro database.
Developmental and stress conditions
Wild-type JC24 was used to collect fungal materials for developmental and stress samples. To collect developmental samples, conidia were harvested by flooding the plate with sterilized distilled water and germinated conidia were collected from 4-day-old cultures on liquid CM. Both conidia and germinated conidia was harvested using a 0.45-μm filter, after filtration of mycelia through Miracloth (Calbiochem, San Diego, USA).
For collection of abiotic stress samples, cultures of 100 ml liquid CM inoculated with 1 ml of a conidial suspension (5×104 conidia/ml) were incubated at 25°C for 3 days in an orbital shaker (120 rpm). The cultured mycelia were harvested, washed three times with 1 l of sterilized distilled water, then transferred to the following abiotic stress conditions: CM, minimal medium, carbon starvation, nitrogen starvation (Talbot et al., 1997), 1 M of sorbitol, KCl, and NaCl. The fungicides, including benomyl (50 μg/ml), iprobenfos (50 μg/ml), kresoxim-methyl (50 μg/ml), isoprothiolane (50 μg/ml), triflumizole (50 μg/ml), thiophanate-methyl (50 μg/ml), and hygromycin B (100 g/ml) were added to a treatment and then cultured for 4 h (Table 3). All the samples were harvested from three replicates of three biological repeats, immediately frozen using liquid nitrogen, and stored at −80°C until processed.
Analysis of transcript levels of CaABC1 in C. acutatum
Quantitative real-time PCR (qRT-PCR) was used to measure transcript levels. Total RNA samples and first-strand cDNA were prepared as described previously (Park et al., 2013). The qRT-PCR was conducted in a Hard-Shell 96-well semi-skirted PCR plate (Bio-Rad Laboratories, Hercules, CA, USA) and a Chromo4 Real-Time PCR Detector (Bio-Rad Laboratories). Each well contained 5 μl of 2× SYBR Green RT-PCR Reaction Mix (Bio-Rad Laboratories), 2 μl of cDNA (12.5 ng/μl), and 15 pmol of each primer (Table 2). All the reactions were performed in more than two biological replicates using three combined RNA samples extracted from independent fungal materials. A β-tubulin gene was included in the assays as an internal control for normalization (Table 2). All amplification curves were analyzed with a normalized reporter threshold of 0.25 to obtain the threshold cycle (Ct) values. The comparative ΔΔCt method was used to evaluate the relative quantities of each amplified product in the samples. Fold changes were calculated as 2−ΔΔCt (Livak and Schmittgen, 2001).
Results
Cloning of the putative ABC transporter gene from C. acutatum
A cDNA library prepared from germinating conidia of C. acutatum JC24 was screened to identify putative ABC transporter genes. The cDNA clone Wb01014-C11 was identified as containing a putative ABC transporter gene using cDNA end-sequencing. The corresponding gene will be referred to as CaABC1. The cDNA clone Wb01014-C11 contained a 1,322-bp long cDNA insert. To acquire full-length gene sequence, a genomic fosmid library was constructed from C. acutatum JC24. Colony dot blot hybridization with the cDNA clone as a probe and pooling PCR analysis were performed. Three positive fosmid clones, 56A12, 68H07, and 85D07, were identified. To obtain the full sequence, a fosmid clone 68H07 was selected, followed by shotgun sequencing.
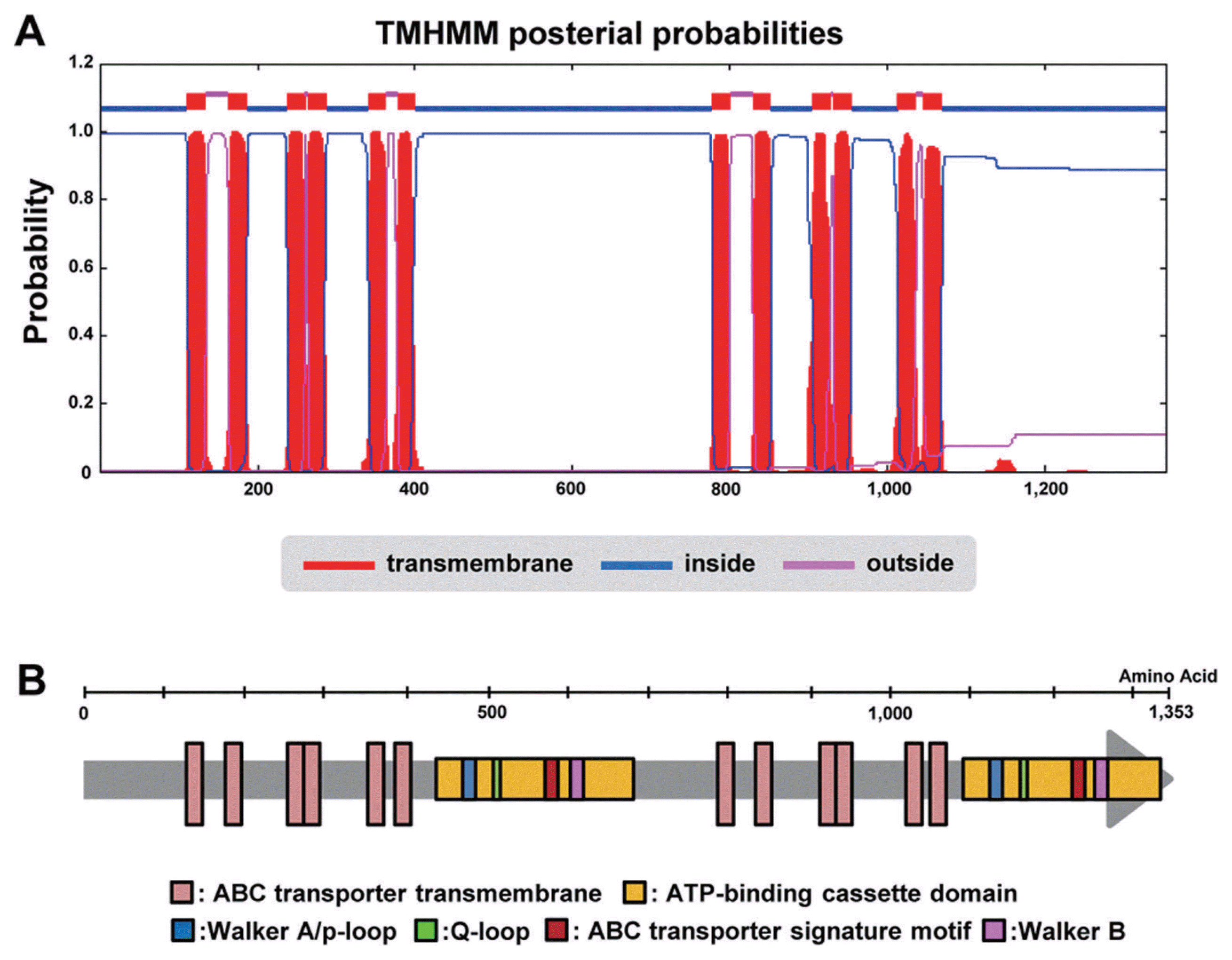
We obtained 6,937 bp sequences with a 5,061 bp predicted gene sequence (4,059 bp transcript). Sequence analysis revealed that the first methionine is located at nucleotide position 895, with a termination codon located at position 5,061 and a deduced polypeptide of 1,353 aa. The ORF is interrupted by just two introns of 63 bp (position 556–618) and 52 bp (position 1,882–1,934) in length. Comparison of the deduced protein with sequences in the GENSCAN (http://genes.mit.edu/GENSCAN.html) protein sequence database revealed significant similarity with members of the ABC transporter superfamily. The TMHMM Web-based programs identified 12 potential membrane-spanning regions (Fig. 1A and B). We also found that both Walker A (p-loop) and Walker B motifs, as well as the ABC signature motif, were present in the sequence (Fig. 1B) (Walker et al., 1982). We deposited this nucleotide and the protein sequences in GenBank under the accession no. KM264299.
Phylogenetic analysis
To find homologous genes in other organisms, the predicted ABC transporter gene CaABC1 sequence encoded by the 1,353 bp aa sequences was used for BLAST P homology searches of GenBank in the NCBI database (Fig. 1B). Numerous hits were returned with high similarity scores for ascomycetous fungi. The CaABC1 protein shares highest amino acid conservation with C. fiorina (99% identity), but also shares between 43% and 97% identity with other ascomycetous and basidiomycetous fungi (Table 1 and Fig. 2). Phylogenetic analysis was performed to determine the evolutionary relationship of the homologous genes. The resulting phylogenetic tree showed that homologous genes of CaABC1 from Colletotrichum spp. were more closely related to each other than to the other homologous genes (Fig. 2).

Molecular phylogeny of the C. acutatum CaABC1 transporter gene and 35 homologous genes from various organisms. The depicted phylogram was obtained by neighbor-joining using MEGA6.0 software and reflect the relationship between 36 aa sequences of homologous CaABC1 proteins. Results from bootstrapping with 2,000 replicates are indicated when higher than 50%.
All identified homologous genes of CaABC1 had two AAA ATPase domains (IPR003593), ABC transporter-like domains (IPR003593), ABC transporter’s transmembrane domains (IPR011527), ABC transporter’s conserved sites (IPR 017871), and ABC transporter’s integral membrane type 1 (IPR017940) (Fig. 2), but not in Melampsora larici-populina. This resulted in a separate position of other genes in the topology of the phylogenetic tree (Fig. 2).
The ATP-binding domain of an ABC transporter is the transmembrane ABC transporters. Walker et al. (Walker et al., 1982) reported that the Walker A and Walker B motifs are protein sequence motifs of ATP-binding proteins. To find these conserved motifs in CaABC1 and five representatives derived from phylogenetic analysis, the six protein sequences were aligned by ClustalW. Relatively lower amino acid conservation exists between the ABC half transporters in the N-terminal region in contrast to the high degree of conservation marked in the region containing the Walker A (P-loop), Walker B, and the ABC signature (Fig. S2).
CaABC1 expression in response to cell development, abiotic stresses, and fungicides
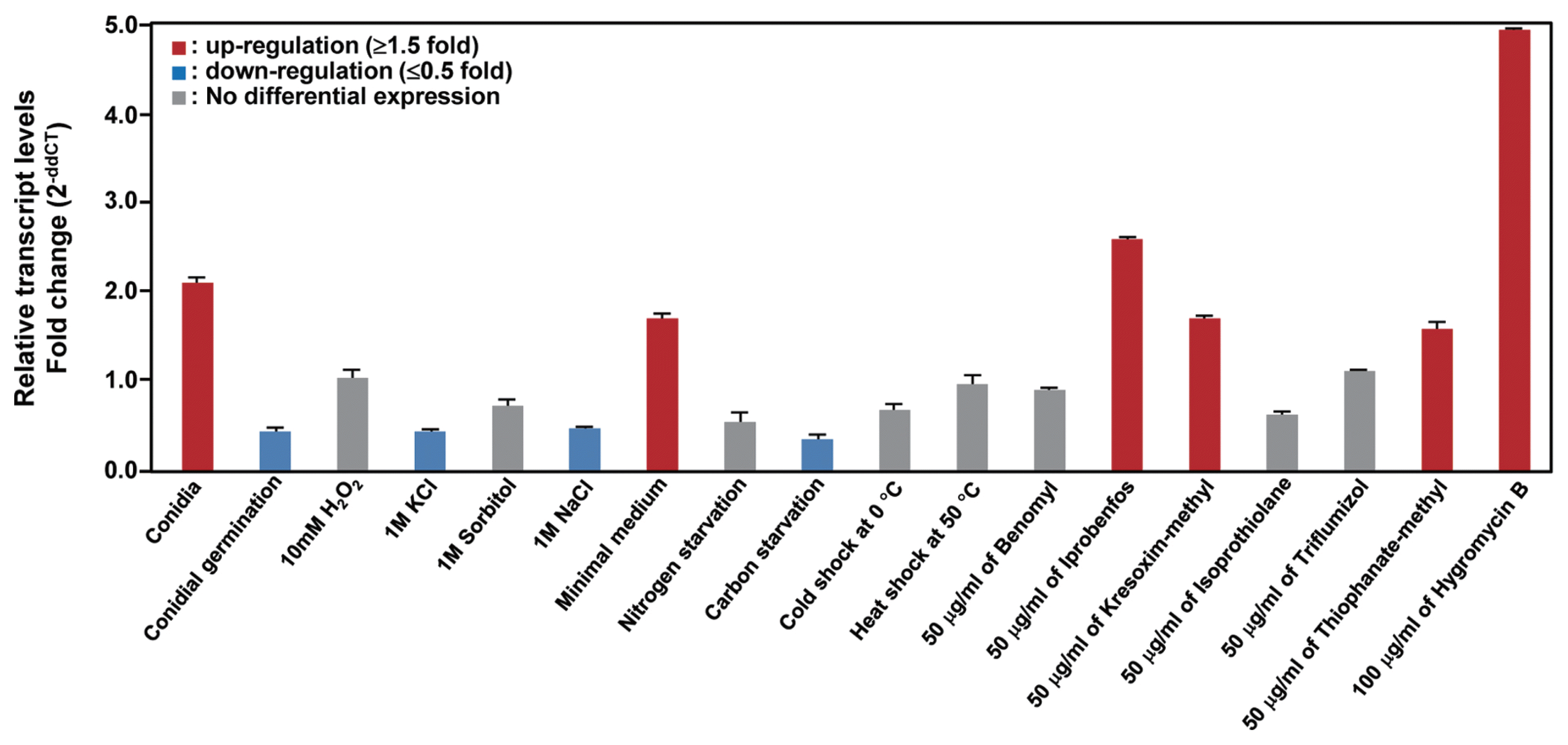
To obtain insight into the physiological roles of the CaABC1 transporter, we conducted expression analysis in various conditions, including cell developments, various abiotic conditions, and fungicide treatments (Table 3). We found that CaABC1 was specifically up-regulated in conidia and mycelia under a minimal medium condition (Fig. 3). We also observed that CaABC1 was induced in fungicides, including iprobenfos, kresoxim-methyl, and thiophanate-methyl. Furthermore, CaABC1 was highly up-regulated in a hygromycin treatment (Fig. 3).
Discussion
In this study, we identified a gene (CaABC1) that encodes ATP-binding elements of C. acutatum. Based on DNA and encoded amino acid sequences, this gene is potentially an ABC transporter. Since ABC transporter genes have been involved in tolerance and resistance to toxic substances, we hypothesized that the CaABC1 gene may be responsible. Supporting this hypothesis was the observation that expression of CaABC1 resulted in increased transcripts under cell development and abiotic stresses, including fungicides (Fig. 3). This expression data suggested a role of the CaABC1 transporters in tolerating these abiotic stress conditions and fungicide resistance.
We also found that CaABC1 encoded a “full-length” ABC transporter protein of 12 transmembrane regions and two nucleotide binding sites (Fig. 1). Based on sequence homology and the conservation of two intron positions, CaABC1 is an apparent homolog of XP_007590216 in C. fioriniae. We observed that these two genes share 99% protein sequence identity (Table 1). Shivas and Tan (Shivas and Tan, 2009) examined molecular differences by internal transcribed spacer (ITS) and β-tubulin sequence data using taxonomically identified 48 Australian C. acutatum. They found that 48 C. acutaum isolates were divided into three different species, including C. acutatum, C. fioriniae, and C. simmondisii. This indicates that the three species might be difficult to identify with morphological characteristics. This result also implies that these species may have recently diverged compared to other Colletotrichum spp. High amino acid sequence similarity between CaABC1 in C. acutatum and XP_007590216 in C. fioriniae might explain this close relatedness.
In the human pathogenic fungus Candida albicans, overexpression of the drug efflux pump encoding genes CDR1 and CDR2 belonging to the ABC transporter is one of the principal mechanisms of azole resistance (Pao et al., 1998; White et al., 1998). In the phytopathogenic fungus Fusarium graminearum, FGSC_06771, which is a homologous gene in CaABC1, was induced under azole fungicide treatment (Becher et al., 2011). To control C. acutatum, the benzimidazole fungicides, including benomyl, have been frequently used (Adaskaveg and Hartin, 1997; Peres et al., 2002; Talhinhas et al., 2002; Talhinhas et al., 2005). Since many studies reported that C. acutatum is less sensitive to banzimidazole fungicides, we anticipated that the CaABC1 gene may be responsible for this fungicide resistance. However, we could not detect induction of CaABC1 transcripts with treatment of benomyl (Fig. 3). These results indicate that CaABC1 may not be responsible for the resistance to benomyl in C. acutatum. Many ABC transporter genes in C. acutatum remain to be explored. A draft genome sequence of C. acutatum was recently reported (Baroncelli et al., 2014) and it could represent a challenge to examine the rest of the ABC transporter genes in C. acutatum.
However, we found that transcripts were induced in iprobenfos, kresoxim-methyl, and thiophanate-methyl, suggesting that the CaABC1 gene may contribute to resistance to these fungicides. Moreover, transcripts of CaABC1 were highly up-regulated with the treatment of hygromycin B, which is involved in cell wall stress (Fig. 3), indicating that CaABC1 may be also involved in other abiotic stress responses. Numerous studies have reported that genes homologous to CaABC1 in other organisms have different roles. For example, a homologous gene NCU07546 in Neurospora crassa was up-regulated in the death-inducer staurosporine (Fernandes et al., 2011), and C47A10.1 in Caenorhabditis elegans was induced in exposure to cadmium. In addition, future studies involving gene disruption would be useful in determining the role of CaABC1 in fungicide resistance and abiotic stresses, as well as pathogenicity, in pepper.
Acknowledgments
This research was supported by Dongeui University Research Grant 2014AA218. Blue-Bio industry RIC (RIC 08-06-07) under Ministry of Knowledge Economy and Busan City contributed for experimental equipments.