 |
 |
| Plant Pathol J > Volume 40(1); 2024 > Article |
|
Abstract
The Colletotrichum gloeosporioides species complex includes many phytopathogenic species, causing anthracnose disease on a wide range of host plants and appearing to be globally distributed. Seventy-one Colletotrichum isolates in the complex from different plants and geographic regions in Korea were preserved in the Korean Agricultural Culture Collection (KACC). Most of them had been identified based on hosts and morphological features, this could lead to inaccurate species names. Therefore, the KACC isolates were re-identified using DNA sequence analyses of six loci, comprising internal transcribed spacer, gapdh, chs-1, his3, act, and tub2 in this study. Based on the combined phylogenetic analysis, KACC strains were assigned to 12 known species and three new species candidates. The detected species are C. siamense (n = 20), C. fructicola (n = 19), C. gloeosporioides (n = 9), C. aenigma (n = 5), C. camelliae (n = 3), C. temperatum (n = 3), C. musae (n = 2), C. theobromicola (n = 2), C. viniferum (n = 2), C. alatae (n = 1), C. jiangxiense (n = 1), and C. yulongense (n = 1). Of these, C. jiangxiense, C. temperatum, C. theobromicola and C. yulongense are unrecorded species in Korea. Host plant comparisons showed that 27 fungus-host associations are newly reported in the country. However, plant-fungus interactions need to be investigated by pathogenicity tests.
The Colletotrichum gloeosporioides species complex is the most diverse group in the genus Colletotrichum, causing anthracnose disease in many economically important crops worldwide and also occurring as asymptomatic endophytes in a broad range of host plants (Hyde et al., 2009; Liu et al., 2022; Lu et al., 2004). In Korea, the species in this complex have been reported as destructive diseases on widely cultivated crops such as C. siamense and C. fructicola on apple (Kim et al., 2018a; Park et al., 2018), peach (Lee et al., 2020a, 2020b) and strawberry (Nam et al., 2013, 2022), and C. gloeosporioides on citrus, apple, grape, persimmon and strawberry (Korean Society of Plant Pathology, 2022).
C. gloeosporioides was first introduced by Penzig (1882), and then became a common phytopathogenic fungus with a global distribution. Prior to the availability of DNA sequence data, C. gloeosporioides and other taxa in the genus Colletotrichum were identified based on host species and morphological characteristics. However, micro-morphological features such as the size and shape of conidia, appressoria and setae or teleomorph characters overlapped in many species and these features could change under different growing conditions (Weir et al., 2012). For instance, cylindrical conidia with round ends were typical characteristics of species of the C. gloeosporioides species complex, but these characteristics were also observed in many members in the C. dracaenophilum, C. magnum, and C. orchidearum species complexes, or in some singleton species (Damm et al., 2019; Liu et al., 2022). Cultural characteristics of different strains of a species in the C. gloeosporioides species complex were often significant variations (Weir et al., 2012). On the other hand, most members of the C. gloeosporioides species complex have a broad host range and a plant could be associated with more than one Colletotrichum species (Jayawardena et al., 2021; Johnston, 2000; Johnston et al., 2005). Therefore, the host reference and morphology are insufficient to distinguish species members in this species complex.
A large number of Colletotrichum species have been re-identified based on DNA sequence analyses of multiple loci such as the nuclear ribosomal internal transcribed spacer (ITS) region, glyceraldehyde-3-phosphate dehydrogenase (gapdh), chitin synthase 1 (chs-1), histone-3 (his3), actin (act), and beta-tubulin 2 (tub2) (Cannon et al., 2012; Jayawardena et al., 2016; Liu et al., 2022; Weir et al., 2012). To date, at least 61 accepted species have been introduced in the C. gloeosporioides species complex (Liu et al., 2022; Zhang et al., 2023; Zheng et al., 2022). They shared high similarity of ITS sequences but could separate using multi-locus analyses of informative loci, especially gapdh and tub2 genes (Weir et al., 2012).
Isolates of the C. gloeosporioides species complex have been collected in Korea since the 1990s and stored in the Korean Agricultural Culture Collection (KACC), National Institute of Agricultural Sciences. However, these isolates had been identified based mainly on the host species and morphological characteristics by depositors. Hence, this study aims to (1) reassess the identification of the C. gloeosporioides species complex isolates in KACC using multi-locus analyses of six loci (ITS, gapdh, chs-1, his3, act, and tub2); (2) re-arrange the combination between host plants and fungal species.
Seventy-one isolates in the C. gloeosporioides species complex that originated in Korea and stored in KACC were used in this study. Details of the isolates are listed in Table 1. The living cultures preserved in liquid nitrogen were retrieved on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) and were used for DNA extraction and phylogenetic analyses.
The mycelium of each isolate was taken from a 5-day-old culture on a PDA medium. Genomic DNA extraction was performed with the DNeasy plant mini kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. Six loci of Colletotrichum isolates, including ITS, gapdh, chs-1, his3, act, and tub2 were amplified with the primer pairs ITS1/ITS4 (White et al., 1990), GDF1/GDR1 (Guerber et al., 2003), CHS-79F/CHS-345R (Carbone and Kohn, 1999), CYLH3F/CYLH3R (Crous et al., 2004), ACT-512F/ACT-783R (Carbone and Kohn, 1999), and T1/BT2b (Glass and Donaldson, 1995; O’Donnell and Cigelnik, 1997), respectively. Each polymerase chain reaction (PCR) was performed in a volume of 25 μl containing 12.5 μl PCR Master Mix (2×), 8.5 μl nuclease-free water, 1 μl (4.5 pMol) of each primer, and 2 μl DNA template (100 ng/μl). PCR conditions of six loci were set up as described by Thao et al. (2023). PCR amplifications were carried out using a MJ Research PTC-200 Thermal Cycler (MJ Research, Ramsey, MN, USA). PCR products were purified with the QIAquick PCR Purification Kit (Qiagen) and checked by gel electrophoresis before being sent to the Macrogen Company (Seoul, Korea) for sequencing with the amplification primers.
Raw sequences generated from forward and reverse primers in this study were trimmed and paired by MEGA 11 (Tamura et al., 2021). Assemble sequences were deposited to RDA-GeneBank (http://genebank.rda.go.kr) with accession numbers in Table 1. The closest reference sequences in GenBank were obtained by BLASTN search and were used for phylogenetic analyses with sequences from 71 KACC isolates and a species in the different species complex as an outgroup. The dataset of each locus was separately aligned using the multiple alignment program MAFFT version 7 (https://mafft.cbrc.jp/alignment/server/) with the G-INS-1 option. The poor alignments at both ends were cut and the alignments were improved by visual inspection in MEGA 11 and concatenated afterward in this software. A maximum likelihood (ML) phylogenetic tree was inferred based on the combined six datasets (ITS, gapdh, chs-1, his3, act, and tub2), using IQ-TREE with the best-fit model “TN+F+G4” and 1,000 ultrafast bootstrap replicates. The substitution model options were auto-evaluated according to the provided alignment files. The phylogenetic trees were viewed in MEGA11 and depicted in Adobe Illustrator.
The phylogenetic analysis included 71 KACC isolates that originated in Korea (Table 1), 58 reference strains from 56 previously accepted species and C. arecacearum (MH0003) as the outgroup (Supplementary Table 1). The concatenated dataset alignment consisted of 2,504 characters (1,419 constant, 1,002 variable, and 502 parsimony-informative characters), including gaps. Of these, 565 characters were from ITS, 282 characters from gapdh, 251 characters from chs-1, 376 characters from his3, 264 characters from act, and 766 characters from tub2.
The multi-locus phylogenetic analysis resolved all KACC isolates into 15 clades. Each of the 12 clades was well clustered with an ex-type or holotype strain of the previously described species, supported by 86% to 100% ML bootstrap values (Fig. 1). Of which, 20 isolates clustered with C. siamense, 19 isolates with C. fructicola, nine isolates with C. gloeosporioides sensu stricto, five isolates with C. aenigma, three isolates with C. camelliae, three isolates with C. temperatum, two isolates with C. musae, two isolates with C. theobromicola, two isolates with C. viniferum, one isolate with C. alatae, one isolate with C. jiangxiense and one isolate with C. yulongense. The isolate KACC 47776 formed a sister clade to C. makassarense and C. tropicale. However, this isolate was distinguished from C. makassarense at gapdh (93.97% identity), act (98.76% identity), chs-1 (98.8% identity), and ITS (98.97% identity), and differentiated from C. tropicale by gapdh (94.03% identity), act (98.77% identity), and chs-1 (97.61% identity). The isolate KACC 47035 was phylogenetically related to C. endophyticum and C. artocarpicola, but shared low sequence similarity at gapdh (92.13% and 90.75%, respectively), act (97.93% and 98.35%), his3 (97.2% with C. endophyticum), chs-1 (98.01% with both species), and tub2 (98.34% with both species). Exception of ITS, all other loci of KAC 48700 shared low sequence similarity with the most closely related species, consisting of gapdh (91.3% identity shared with C. siamense), his3 (92.2% identity with C. siamense), act (95.45% identity with C. conoides, C. aenigma and C. siamense), tub2 (95,71% identity with C. endophyticum), and chs-1 (97.61% identity with C. siamense). According to phylogenetic analysis and BLASTN search results, each of the isolates, KACC 47776, KACC 47035, and KACC 48700, were genetically distinct from the other taxa.
Twenty combinations between hosts and accepted fungal species found in this study have not been reported in any countries. However, the fungi were just isolated from the hosts and their pathogenicity was not confirmed. They are C. aenigma on Ampelopsis glandulosa f. citrulloides, Boehmeria japonica, Ilex rotunda, Laurus nobilis, and Zanthoxylum schinifolium; C. alatae on Dioscorea polystachya; C. camelliae on Cayratia japonica; C. fructicola on Pinus koraiensis, Pinus strobus, and Nepeta tenuifolia; C. gloeosporioides on Amaranthus hybridus, Boehmeria japonica, and Rhododendron micranthum; C. jiangxiense on Atractylodes ovata; C. siamense on Limonium sinuatum and Malus coronaria; C. temperatum on Malus domestica and Tricyrtis macropoda; C. theobromicola on Actinidia arguta; and C. yulongense on Camellia japonica. Seven other combinations, including C. fructicola on Capsicum annuum, Castanea sp., and Limonium sinuatum; C. gloeosporioides on Euonymus japonicus, Morus alba, and Portulaca oleracea; and C. siamense on Vitis vinifera are first reported in Korea (Table 2).
Based on DNA sequence analyses of six loci, 43 KACC isolates were renamed, three isolates were newly identified at the species level, 21 strains remained as original names and three isolates were considered as three new species candidates. Nineteen isolates in this study had previously been identified as C. gloeosporioides sensu lato by Kim et al. (2006) and one isolate had been identified as C. acutatum by Lee et al. (2007) using morphological features, ITS and tub2 sequence analyses. However, these isolates were resolved by Kim et al. (2020) and by the present study into four different species, C. siamense (n = 10), C. fructicola (n = 7), C. theobromicola (n = 2), and C. temperatum (n = 1), based on multi-locus analyses of ITS, gapdh, chs-1, tub2 and (his3, act) or (cal and ApMat). The results of this study also revealed four fungal species, C. jiangxiense, C. temperatum, C. theobromicola, and C. yulongense, were first recorded in Korea.
C. siamense (n = 20), C. fructicola (n = 19), and C. gloeosporioides sensu stricto (n = 9) are the most dominant species among KACC isolates in the C. gloeosporioides species complex. According to previous studies and this work, C. gloeosporioides, C. siamense, C. fructicola, C. aenigma, C. jiangxiense, C. theobromicola, C. viniferum, and C. temperatum have a wide range of hosts, whereas C. musae has a narrow host range, on Musa spp. and Mangifera indica, and C. alatae is a host-specific fungus, on the genus Dioscorea (Farr and Rossman, 2021; Jayawardena et al., 2021; Li et al., 2019; Liu et al., 2022; Weir et al., 2012; Zhang et al., 2023). In addition, C. yulongense was originally described by Wang et al. (2019a) on Vaccinium dunalianum (Ericaceae family) and this fungus was found on Camellia japonica (Theaceae family) in this study as the second host plant. Another species, C. camelliae was also reported only on the genus Camellia of the family Theaceae (Liu et al., 2022), but this fungal species was herein from the different family Vitaceae (Cayratia japonica).
C. aenigma was introduced by Weir et al. (2012) and this fungus later became a very popular species, causing anthracnose on the grapevine, Asian pear, and walnut in China (Fu et al., 2019; Wang et al., 2021; Yan et al., 2015), on dragon fruits in Thailand (Meetum et al., 2015), on avocado in Israel (Sharma et al., 2017), on miracle in Japan (Truong et al., 2018), on grape and orange stonecrop in Korea (Choi et al., 2017; Kim et al., 2021). In our study, this fungus was isolated from five new hosts (Ampelopsis glandulosa f. citrulloides, Boehmeria japonica, Ilex rotunda, Laurus nobilis, and Zanthoxylum schinifolium) belonging to the five different families.
Apple (Malus domestica) is one of the most economically important crops in Korea. Kim et al. (2006) and Lee et al. (2007) implicated that C. gloeosporioides and C. acutatum were serious pathogens causing bitter rots of apple fruits in Korea. However, the identified species were unreliable as mentioned above. Later, causal agents of apple bitter rot disease in the country were authentically identified as C. siamense, C. fructicola, C. nymphaeae, C. fioriniae, C. gloeosporioides, and C. aenigma based on multi-locus sequence analyses (Cheon et al., 2016; Kim et al., 2020; Lee et al., 2021; Oo et al., 2018). The Colletotrichum isolates from apple in this work and in our earlier study (Thao et al., 2023) have been collected since 1997 from many different geographic locations in Korea, and the most prevalent species on apple were C. siamense (10 isolates), followed by C. fructicola (seven), C. nymphaeae (five) and C. fioriniae (four), while only one isolate of C. gloeosporioides sensu stricto was from apple and no C. acutatum sensu stricto isolates were found in KACC. Additionally, a fungal species C. temperatum, originally described by Doyle et al. (2013) on Vaccinium macrocarpon, was first detected on apple in this study.
Grape (Vitis vinifera) is an extensively cultivated crop worldwide and in Korea. The crop was seriously damaged by the infection of C. acutatum and C. gloeosporioides (Hong et al., 2008), C. viniferum (Oo and Oh, 2017), and C. aenigma (Kim et al., 2021). In this study, two isolates on grape (Vitis sp.) were identified as C. viniferum, whereas the other two isolates on Vitis vinifera were identified as C. siamense.
Kiwiberry (Actinidia spp.) has been widely cultivated in the country and worldwide (Kim et al., 2017; Latocha, 2017). The anthracnose symptoms caused by C. nymphaeae on kiwiberry fruits were observed in Jeonnam province (Kim et al., 2018b). Many other Colletotrichum species also infected kiwi plants worldwide such as C. aenigma on Actinidia arguta in China (Wang et al., 2019b), C. fioriniae, C. karsti, C. gloeosporioides, C. fructicola, and C. siamense on Actinidia spp. in Japan (Poti et al., 2023). A new combination of kiwiberry and C. theobromicola was found in this study.
The main causal fungi of chili pepper (Capsicum annuum) anthracnose in Korea were known as C. gloeosporioides sensu lato, C. acutatum s. lat., C. dematium s. lat., and C. trucatum s. lat. (Oo and Oh, 2016, 2020; Oo et al., 2017; Park and Kim, 1992). However, only one strain of C. fructicola in the C. gloeosporioides species complex from chili pepper is stored in KACC. While 16 strains of the C. acutatum species complex from chili pepper are preserved in KACC (Thao et al., 2023). It suggests that C. gloeosporioides s.c. is not a main causal fungus on chili pepper anthracnose in Korea.
Machilus thunbergii is a medicinal plant and it was widely distributed in the west-southern islands of Korea and some other Asian countries (Park et al., 1990). Only Glomerella cingulate and C. fructicola were formerly known as pathogens on this plant in Japan (Kobayashi, 2007) and China (Liu et al., 2022), respectively. A new species candidate (KACC47776) from Machilus thunbergii in Jeju, Korea was identified in this study. The second new species candidate (KACC48700) was from Paederia foetida, this host was also infected by many different Colletotrichum species (Liu et al., 2022). The third new species candidate (KACC47035) was the first report of a Colletotrichum fungus on coastal hogfennel (Peucedanum japonicum), an edible wild vegetable and medicinal resource.
A reassessment of members of the C. gloeosporioides species complex in Korea revealed new records of four Colletotrichum species and 27 combinations between plants and fungi in Korea. Of which, new findings of C. fructicola on pepper, C. siamense on grape, C. temperatum on apple, and C. theobromicola on kiwiberry could play an important role in the agricultural sciences and practices of Korea. However, the new combinations found in this study need to be clarified by pathogenicity tests in further studies.
Acknowledgments
This study was supported by a grant (PJ017286) from the National Institute of Agricultural Sciences and was a part of the “2023 KoRAA Long-term Training Program”, Rural Development Administration, Republic of Korea.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Maximum likelihood tree based on multi-locus sequences of ITS, tub2, his3, gapdh, chs-1, and act. Species names are followed by strain accession numbers and their hosts (blue). Isolates in this study are in bold and listed by the original names. Bootstrap values below 70% are not present. Ex-type strains are emphasised by asterisks (*). The tree is rooted to Colletotrichum arecacearum (MH003).
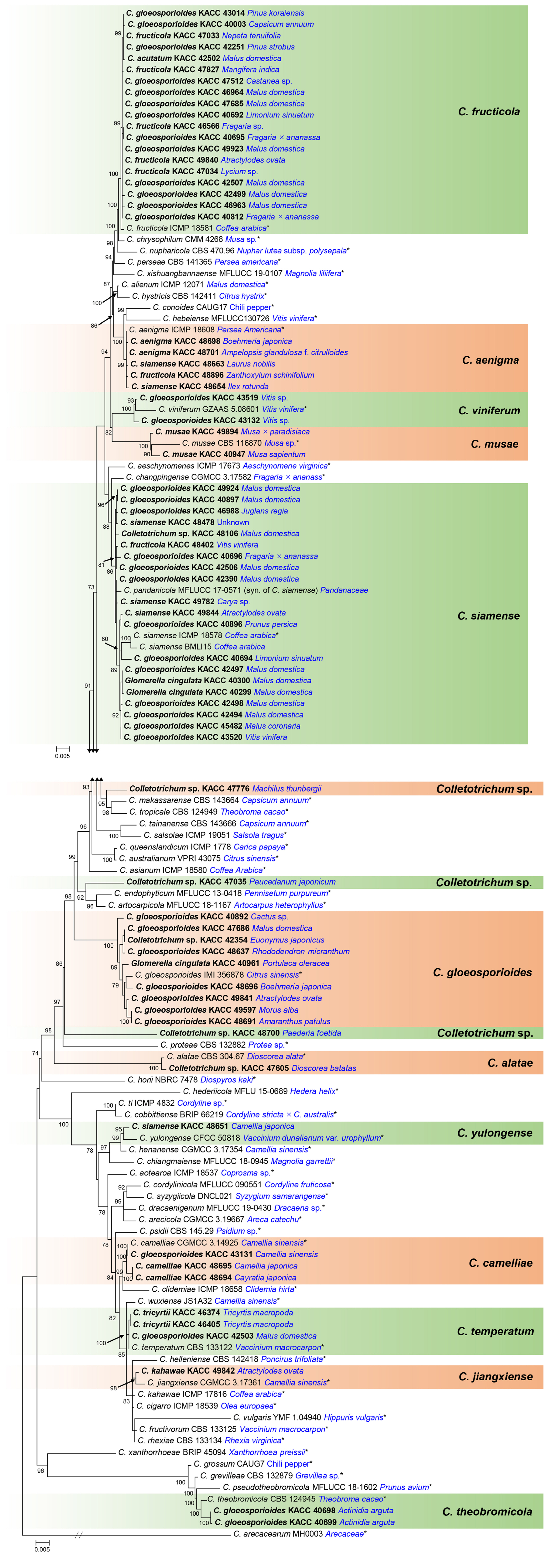
Table 1
Isolates of the Colletotrichum gloeosporioides species complex used in this study
| Species (this study) | Isolate no. | Location | Collection date | Hosta | Previous identifications by depositors | Re-identifications in previous studies | RDA-GeneBank accession no. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| ITS | gapdh | chs-1 | his3 | act | tub2 | |||||||
| C. aenigma | KACC 48663 | Wando | 2018 | Laurus nobilis | C. siamense | - | RDA0062101 | RDA0062486 | RDA0067036 | RDA0067150 | RDA0066965 | RDA0062292 |
| KACC 48701 | Seogwipo | 2018 | Ampelopsis glandulosa f. citrulloides | C. aenigma | - | RDA0062110 | RDA0062500 | RDA0067035 | RDA0067151 | RDA0066966 | RDA0062306 | |
| KACC 48698 | Jeju | 2018 | Boehmeria japonica | C. aenigma | - | RDA0062108 | RDA0062498 | RDA0067038 | RDA0067148 | RDA0066964 | RDA0062304 | |
| KACC 48896 | Yeosu | 2019 | Zanthoxylum schinifolium | C. fructicola | - | RDA0062113 | RDA0062504 | RDA0067039 | RDA0067147 | RDA0066944 | RDA0062309 | |
| KACC 48654 | Wando | 2018 | Ilex rotunda | C. siamense | - | RDA0062099 | RDA0062484 | RDA0067037 | RDA0067149 | RDA0066987 | RDA0062290 | |
| C. alatae | KACC 47605 | Gongju | 2013 | Dioscorea polystachya | Colletotrichum sp. | - | RDA0062083 | RDA0062456 | RDA0067092 | RDA0067176 | RDA0067034 | RDA0062252 |
| C. camelliae | KACC 43131 | Boseong | 2006 | Camellia sinensis | C. gloeosporioides | - | RDA0062064 | RDA0062421 | RDA0067095 | RDA0067180 | RDA0067025 | RDA0062216 |
| KACC 48695 | Jeju | 2018 | Camellia japonica | C. camelliae | - | RDA0062105 | - | RDA0067100 | RDA0067182 | RDA0067024 | RDA0062301 | |
| KACC 48694 | Jeju | 2018 | Cayratia japonica | C. camelliae | - | RDA0062104 | RDA0062495 | RDA0067099 | RDA0067183 | RDA0067027 | RDA0062300 | |
| C. fructicola | KACC 43014 | Daejeon | 2006 | Pinus koraiensis | C. gloeosporioides | - | RDA0062063 | RDA0062408 | - | - | RDA0067015 | RDA0062202 |
| KACC 40003 | Daejeon | 1993 | Capsicum annuum | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006) | RDA0062019 | RDA0062330 | - | - | RDA0067017 | RDA0062123 | |
| KACC 42251 | Daejeon | Unknown | Pinus strobus | C. gloeosporioides | - | RDA0062049 | RDA0062377 | RDA0067062 | RDA0067117 | RDA0067012 | RDA0062173 | |
| KACC 47033 | Eumseong | 2012 | Nepeta tenuifolia | C. fructicola | - | RDA0062078 | RDA0062449 | RDA0067073 | RDA0067111 | RDA0067009 | RDA0062244 | |
| KACC 42502 | Seoul | 2006 | Malus domestica | C. acutatum | C. acutatum (Lee et al., 2007) | RDA0062057 | RDA0062396 | RDA0067066 | RDA0067115 | RDA0067011 | RDA0062191 | |
| KACC 46566 | Gyeryong | 2009 | Fragaria sp. | C. fructicola | - | RDA0062072 | RDA0062439 | RDA0067057 | RDA0067119 | RDA0067020 | RDA0062232 | |
| KACC 40695 | Geochang | 1999 | Fragaria × ananassa | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006 ) | RDA0060939 | RDA0062351 | RDA0067045 | RDA0067143 | RDA0067021 | RDA0062142 | |
| KACC 40812 | Damyang | 1998 | Fragaria × ananassa | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006) | RDA0062041 | RDA0062364 | - | - | RDA0067016 | RDA0062162 | |
| KACC 40692 | Namwon | 1998 | Limonium sinuatum | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006) | RDA0060934 | RDA0062349 | RDA0067058 | RDA0067118 | RDA0067013 | RDA0062141 | |
| KACC 42507 | Jeongeup | 2006 | Malus domestica | C. gloeosporioides | C. gloeosporioides (Lee et al., 2007) | RDA0062061 | RDA0062401 | RDA0067053 | RDA0067121 | RDA0067014 | RDA0062196 | |
| KACC 49840 | Sangju | 2020 | Atractylodes ovata | C. fructicola | - | RDA0062116 | RDA0062511 | RDA0067048 | RDA0067122 | RDA0067018 | RDA0062315 | |
| KACC 47034 | Cheongyang | 2012 | Lycium sp. | C. fructicola | - | RDA0062079 | RDA0062450 | RDA0067074 | RDA0067110 | RDA0067008 | RDA0062245 | |
| KACC 49923 | Yeongdong | 2016 | Malus domestica | C. gloeosporioides | - | RDA0062121 | RDA0062521 | RDA0067046 | RDA0067142 | RDA0067019 | RDA0062325 | |
| KACC 47512 | Gongju | 2013 | Castanea sp. | C. gloeosporioides | - | RDA0062082 | RDA0062455 | RDA0067054 | RDA0067120 | RDA0066989 | RDA0062251 | |
| KACC 46964 | Gunwi | 2012 | Malus domestica | C. gloeosporioides | - | RDA0062076 | RDA0062444 | RDA0067072 | RDA0067112 | RDA0066988 | RDA0062239 | |
| KACC 47685 | Gunwi | 2013 | Malus domestica | C. gloeosporioides | - | RDA0062084 | RDA0062458 | RDA0067043 | RDA0067144 | RDA0066990 | RDA0062263 | |
| KACC 47827 | Iksan | 2014 | Mangifera indica | C. fructicola | - | RDA0062088 | RDA0062465 | RDA0067063 | RDA0067116 | RDA0066991 | RDA0062271 | |
| KACC 46963 | Gimcheon | 2012 | Malus domestica | C. gloeosporioides | - | RDA0062075 | RDA0062443 | RDA0067070 | RDA0067113 | RDA0067010 | RDA0062238 | |
| KACC 42499 | Cheongsong | 2006 | Malus domestica | C. gloeosporioides | C. gloeosporioides (Lee et al., 2007) | RDA0062055 | RDA0062393 | RDA0067068 | RDA0067114 | RDA0067022 | RDA0062188 | |
| C. gloeosporioides | KACC 40892 | Jeju | Unknown | Cactus sp. | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006) | RDA0062042 | RDA0062367 | RDA0067081 | RDA0067101 | RDA0067003 | RDA0062163 |
| KACC 47686 | Mungyeong | 2013 | Malus domestica | C. gloeosporioides | - | RDA0062085 | RDA0067187 | RDA0067085 | RDA0067103 | RDA0066997 | - | |
| KACC 42354 | Namyangju | 2006 | Euonymus japonica | Colletotrichum sp. | - | RDA0062050 | RDA0062380 | RDA0067083 | RDA0067104 | RDA0066999 | RDA0062176 | |
| KACC 48637 | Goesan | 2017 | Rhododendron micranthum | C. gloeosporioides | - | RDA0062097 | RDA0062482 | - | - | RDA0067001 | RDA0062288 | |
| KACC 40961 | Sancheong | 2001 | Portulaca oleracea | Glomerella cingulata | G. cingulata (Kim et al., 2006) | RDA0062047 | RDA0062373 | RDA0067086 | RDA0067102 | RDA0066996 | RDA0062169 | |
| KACC 48696 | Jeju | 2018 | Boehmeria japonica | C. gloeosporioides | - | RDA0062106 | RDA0062496 | RDA0067079 | RDA0067108 | RDA0067004 | RDA0062302 | |
| KACC 49841 | Mungyeong | 2020 | Atractylodes ovata | C. gloeosporioides | - | RDA0062117 | RDA0062512 | RDA0067080 | RDA0067107 | RDA0067002 | RDA0062316 | |
| KACC 49597 | Jangseong | 2019 | Morus alba | C. gloeosporioides | - | RDA0062114 | RDA0062506 | RDA0067084 | RDA0067109 | RDA0066998 | RDA0062311 | |
| KACC 48691 | Okcheon | 2018 | Amaranthus hybridus | C. gloeosporioides | - | RDA0062103 | RDA0062493 | RDA0067082 | RDA0067105 | RDA0067000 | RDA0062298 | |
| C. jiangxiense | KACC 49842 | Sangju | Atractylodes ovata | C. kahawae | - | RDA0062118 | RDA0062513 | RDA0067093 | RDA0067179 | RDA0067028 | RDA0062317 | |
| C. musae | KACC 49894 | Wanju | 2021 | Musa × paradisiaca | C. musae | - | RDA0062120 | RDA0062515 | RDA0067078 | RDA0067171 | RDA0067005 | RDA0062319 |
| KACC 40947 | Daejeon | 2001 | Musa × paradisiaca | C. musae | - | RDA0062046 | RDA0062372 | RDA0067077 | RDA0067172 | RDA0067006 | RDA0062168 | |
| C. siamense | KACC 49924 | Yeongju | 2016 | Malus domestica | C. gloeosporioides | - | RDA0062122 | RDA0062522 | RDA0067047 | RDA0067166 | RDA0066969 | RDA0062326 |
| KACC 40897 | Suwon | Unknown | Malus domestica | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006) | RDA0062045 | RDA0062370 | RDA0067044 | RDA0067167 | RDA0066970 | RDA0062166 | |
| KACC 46988 | Chungju | 2011 | Juglans regia | C. gloeosporioides | - | RDA0062077 | RDA0062445 | RDA0067050 | RDA0067164 | RDA0066981 | RDA0062240 | |
| KACC 48478 | Sangju | 2015 | Unknown | C. siamense | - | RDA0062096 | RDA0062479 | RDA0067060 | RDA0067159 | RDA0066986 | RDA0062285 | |
| KACC 48106 | Chuncheon | 2015 | Malus domestica | Colletotrichum sp. | - | RDA0062092 | RDA0062474 | RDA0067067 | RDA0067155 | RDA0066963 | RDA0062279 | |
| KACC 48402 | Hwaseong | 2017 | Vitis vinifera | C. fructicola | - | RDA0062095 | RDA0062477 | RDA0067056 | RDA0067161 | RDA0066976 | RDA0062283 | |
| KACC 40696 | Miryang | 1999 | Fragaria × ananassa | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006) | RDA0060941 | RDA0062352 | RDA0067052 | RDA0067169 | RDA0066979 | RDA0062151 | |
| KACC 42506 | Paju | 2005 | Malus domestica | C. gloeosporioides | C. gloeosporioides (Lee et al., 2007) | RDA0062060 | RDA0062400 | RDA0067075 | - | RDA0067007 | RDA0062195 | |
| KACC 42390 | Sangju | 2004 | Malus domestica | C. gloeosporioides | C. gloeosporioides (Lee et al., 2007) | RDA0062051 | RDA0062381 | RDA0067076 | - | RDA0066980 | RDA0062177 | |
| KACC 49782 | Miryang | 2020 | Carya sp. | C. siamense | - | RDA0062115 | RDA0062508 | RDA0067049 | RDA0067165 | RDA0066982 | RDA0062312 | |
| KACC 49844 | Mungyeong | 2020 | Atractylodes ovata | C. siamense | - | RDA0062119 | RDA0062514 | RDA0067071 | RDA0067153 | RDA0066971 | RDA0062318 | |
| KACC 40896 | Yeongi | 2000 | Prunus persica | C. gloeosporioides | - | RDA0062044 | RDA0062369 | RDA0067064 | RDA0067157 | RDA0066974 | RDA0062165 | |
| KACC 40694 | Suwon | 1998 | Limonium sinuatum | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006) | RDA0060936 | RDA0062350 | RDA0067042 | RDA0067168 | RDA0066978 | RDA0066985 | |
| KACC 40299 | Uiseong | 1997 | Malus domestica | Glomerella cingulata | G. cingulata (Kim et al., 2006) | RDA0062028 | RDA0062344 | RDA0067091 | RDA0067152 | RDA0066943 | RDA0062136 | |
| KACC 40300 | Gunwi | 1997 | Malus domestica | Glomerella cingulata | G. cingulata (Kim et al., 2006) | RDA0062029 | RDA0062345 | RDA0067051 | RDA0067163 | RDA0066968 | RDA0062137 | |
| KACC 42497 | Gunwi | 2006 | Malus domestica | C. gloeosporioides | C. gloeosporioides (Lee et al., 2007); C. siamense (Kim et al., 2020) | RDA0062053 | RDA0062391 | RDA0067059 | RDA0067160 | RDA0066967 | RDA0062186 | |
| KACC 42498 | Yeongju | 2006 | Malus domestica | C. gloeosporioides | C. gloeosporioides (Lee et al., 2007); C. siamense (Kim et al., 2020) | RDA0062054 | RDA0062392 | RDA0067069 | RDA0067154 | RDA0066972 | RDA0062187 | |
| KACC 42494 | Cheongsong | 2006 | Malus domestica | C. gloeosporioides | C. gloeosporioides (Lee et al., 2007); C. siamense (Kim et al., 2020) | RDA0062052 | RDA0066942 | RDA0067055 | RDA0067162 | RDA0066977 | RDA0066983 | |
| KACC 45482 | Jinju | 2010 | Malus coronaria | C. gloeosporioides | - | RDA0062069 | RDA0062434 | RDA0067065 | RDA0067156 | RDA0066973 | RDA0062227 | |
| KACC 43520 | Cheongju | 2006 | Vitis vinifera | C. gloeosporioides | - | RDA0062067 | RDA0062424 | RDA0067061 | RDA0067158 | RDA0066975 | RDA0062219 | |
| C. temperatum | KACC 42503 | Yeongju | 2004 | Malus domestica | C. gloeosporioides | C. gloeosporioides (Lee et al., 2007) | RDA0062058 | RDA0062397 | RDA0067096 | RDA0067178 | RDA0067023 | RDA0062192 |
| KACC 46405 | Osan | 2011 | Tricyrtis macropoda | C. tricyrtii | - | RDA0062071 | RDA0062438 | RDA0067097 | RDA0067177 | RDA0067030 | RDA0062231 | |
| KACC 46374 | Gapyeong | 2011 | Tricyrtis macropoda | C. tricyrtii | - | RDA0062070 | RDA0062437 | RDA0067094 | RDA0067181 | RDA0067026 | RDA0062230 | |
| C. theobromicola | KACC 40698 | Muan | 1998 | Actinidia arguta | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006) | RDA0060942 | RDA0062353 | RDA0067089 | RDA0067185 | RDA0067032 | RDA0062152 |
| KACC 40699 | Muan | 1998 | Actinidia arguta | C. gloeosporioides | C. gloeosporioides (Kim et al., 2006) | RDA0060943 | RDA0062354 | RDA0067088 | RDA0067186 | RDA0067033 | RDA0062153 | |
| C. viniferum | KACC 43519 | Cheonan | 2006 | Vitis sp. | C. gloeosporioides | - | RDA0062066 | RDA0062423 | - | - | RDA0066993 | RDA0062218 |
| KACC 43132 | Cheonan | 2006 | Vitis sp. | C. gloeosporioides | - | RDA0062065 | RDA0062422 | RDA0067040 | RDA0067146 | RDA0066994 | RDA0062217 | |
| C. yulongense | KACC 48651 | Wando | 2018 | Camellia japonica | C. siamense | - | RDA0062098 | RDA0062483 | RDA0067098 | RDA0067184 | RDA0067029 | RDA0062289 |
| Colletotrichum sp. | KACC 47776 | Jeju | 2014 | Machilus thunbergii | Colletotrichum sp. | - | RDA0062087 | RDA0066941 | RDA0067041 | RDA0067170 | RDA0066992 | RDA0066984 |
| Colletotrichum sp. | KACC 47035 | Eumseong | 2012 | Peucedanum japonicum | Colletotrichum sp. | - | RDA0062080 | RDA0062451 | RDA0067087 | RDA0067173 | RDA0066995 | RDA0062246 |
| Colletotrichum sp. | KACC 48700 | Jeju | 2018 | Paederia foetida | Colletotrichum sp. | - | RDA0062109 | RDA0062499 | RDA0067090 | RDA0067175 | RDA0067031 | RDA0062305 |
Table 2
Comparison of host plants of KACC isolates and previous reports
| Species | Hosta | ||||
|---|---|---|---|---|---|
|
|
|||||
| KACC | The List of Plant Diseases in Korea | Previous reports in Korea | Global reports | References | |
| C. aenigma | Ampelopsis glandulosa f. citrulloides* | - | - | - | - |
| Boehmeria japonica* | - | - | - | - | |
| Ilex rotunda* | - | - | - | - | |
| Laurus nobilis* | - | - | - | - | |
| Zanthoxylum schinifolium* | - | - | - | - | |
| C. alatae | Dioscorea polystachya* | - | - | - | - |
| C. camelliae | Camellia sinensis | Camellia sinensis | Camellia sinensis | Camellia sinensis | Hassan et al. (2023) |
| Camellia japonica | Camellia japonica | Camellia japonica | Camellia japonica | Korean Society of Plant Pathology (2022) | |
| Cayratia japonica* | - | - | - | - | |
| C. fructicola | Atractylodes ovata | - | Atractylodes ovata | Atractylodes ovata | Hassan et al. (2022) |
| Capsicum annuum | - | - | Capsicum annuum | Liu et al. (2016) | |
| Castanea sp. | - | - | Castanea sp. | Jiang et al. (2021) | |
| Fragaria × ananassa | - | Fragaria × ananassa | Fragaria × ananassa | Nam et al. (2013) | |
| Limonium sinuatum | - | - | Limonium sinuatum | Zhang et al. (2023) | |
| Lycium sp. | - | Lycium chinense | Lycium chinense | Paul et al. (2014) | |
| Malus domestica | Malus domestica | Malus domestica | Malus domestica | Kim et al. (2018a) | |
| Mangifera indica | Mangifera indica | Mangifera indica | Mangifera indica | Joa et al. (2016) | |
| Pinus koraiensis* | - | - | - | - | |
| Pinus strobus* | - | - | - | - | |
| Nepeta tenuifolia* | - | - | - | - | |
| C. gloeosporioides | Amaranthus hybridus* | - | - | - | - |
| Atractylodes ovata | Atractylodes ovata | Atractylodes ovata | Atractylodes ovata | Hassan et al. (2022) | |
| Boehmeria japonica* | - | - | - | - | |
| Cactus sp. | - | Cactus sp. | Cactus sp. | Kim et al. (2000) | |
| Euonymus japonicus | - | - | Euonymus japonicus | Huang et al. (2016) | |
| Malus domestica (syn. M. pumila) | Malus pumila | Malus pumila | Malus pumila | Cheon et al. (2016) | |
| Morus alba | - | - | Morus alba | Sharma et al. (2013) | |
| Portulaca oleracea | - | - | Portulaca oleracea | Simmonds (1966) | |
| Rhododendron micranthum* | - | - | - | - | |
| C. jiangxiense | Atractylodes ovata* | - | - | - | - |
| C. musae | Musa × paradisiaca | Musa × paradisiaca | Musa × paradisiaca | Musa × paradisiaca | Lim et al. (2002) |
| C. siamense | Atractylodes ovata | - | Atractylodes ovata | Atractylodes ovata | Hassan et al. (2022) |
| Carya sp. | - | Carya sp. | Carya sp. | Oh et al. (2021) | |
| Fragaria × ananassa | - | Fragaria × ananassa | Fragaria × ananassa | Nam et al. (2022) | |
| Juglans regia | Juglans regia | Juglans regia | Cho et al. (2023) | ||
| Limonium sinuatum* | - | - | - | - | |
| Malus coronaria* | - | - | - | - | |
| Malus domestica (syn. M. pumila) | Malus pumila | Malus pumila | Malus pumila | Park et al. (2018) | |
| Prunus persica | Prunus persica | Prunus persica | Prunus persica | Lee et al. (2020a) | |
| Vitis vinifera | - | - | Vitis vinifera | Zhang et al. (2023) | |
| C. temperatum | Malus domestica* | - | - | - | - |
| Tricyrtis macropoda* | - | - | - | - | |
| C. theobromicola | Actinidia arguta* | - | - | - | - |
| C. viniferum | Vitis sp. | Vitis vinifera | Vitis vinifera | Vitis vinifera | Oo and Oh (2017) |
| C. yulongense | Camellia japonica* | - | - | - | - |
| Colletotrichum sp. (KACC 47776, new species candidate) | Machilus thunbergii* | - | - | - | - |
| Colletotrichum sp. (KACC 47035, new species candidate) | Peucedanum japonicum* | - | - | - | - |
| Colletotrichum sp. (KACC 48700, new species candidate) | Paederia foetida* | - | - | - | - |
References
Cannon, P. F., Damm, U., Johnston, P. R. and Weir, B. S. 2012.
Colletotrichum: current status and future directions. Stud. Mycol. 73:181-213.



Carbone, I. and Kohn, L. M. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553-556.

Cheon, W., Lee, S. G. and Jeon, Y. 2016. First report on fruit spot caused by Colletotrichum gloeosporioides in apple (Malus pumila Mill.) in Korea. Plant Dis. 100:210.

Cho, S.-E., Oh, J. Y., Lee, D.-H. and Kim, C.-W. 2023. First report of anthracnose on Juglans regia caused by Colletotrichum siamense in Korea. Plant Dis. 107:218.

Choi, H.-W., Lee, Y. K. and Hong, S. K. 2017. First report of Colletotrichum aenigma causing anthracnose on Sedum kamtschaticum in Korea. Plant Dis. 101:2150.

Crous, P. W., Groenewald, J. Z., Risède, J.-M., Simoneau, P. and Hywel-Jones, N. L. 2004.
Calonectria species and their Cylindrocladium anamorphs: species with sphaeropedunculate vesicles. Stud. Mycol. 50:415-430.
Damm, U., Sato, T., Alizadeh, A., Groenewald, J. Z. and Crous, P. W. 2019. The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Stud. Mycol. 92:1-46.



Doyle, V. P., Oudemans, P. V., Rehner, S. A. and Litt, A. 2013. Habitat and host indicate lineage identity in Colletotrichum gloeosporioides s.l. from wild and agricultural landscapes in North America. PLoS ONE 8:e62394.



Farr, D. F. and Rossman, A. Y. 2021 Fungal databases, systematic mycology and microbiology laboratory ARS, USDA. URL http://nt.ars-grin.gov/fungaldatabases/ [27 September 2023].
Fu, M., Crous, P. W., Bai, Q., Zhang, P. F., Xiang, J., Guo, Y. S., Zhao, F. F., Yang, M. M., Hong, N., Xu, W. X. and Wang, G. P. 2019.
Colletotrichum species associated with anthracnose of Pyrus spp. in China. Persoonia 42:1-35.



Glass, N. L. and Donaldson, G. C. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61:1323-1330.




Guerber, J. C., Liu, B., Correll, J. C. and Johnston, P. R. 2003. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95:872-895.


Hassan, O., Kim, J. S., Romain, B. B. N. D. and Chang, T. 2022. An account of Colletotrichum species associated with anthracnose of Atractylodes ovata in South Korea based on morphology and molecular data. PLoS ONE 17:e0263084.



Hassan, O., Kim, S.-H., Kim, K.-M. and Chang, T. 2023. First report of leaf anthracnose caused by Colletotrichum camelliae on tea plants (Camellia sinensis) in South Korea. Plant Dis. Online publication. https://doi.org/10.1094/PDIS-11-22-2622-PDN.


Hong, S. K., Kim, W. G., Yun, H. K. and Choi, K. J. 2008. Morphological variations, genetic diversity and pathogenicity of Colletotrichum species causing grape ripe rot in Korea. Plant Pathol. J. 24:269-278.

Huang, L., Li, Q.-C., Zhang, Y., Li, D.-W. and Ye, J.-R. 2016.
Colletotrichum gloeosporioides sensu stricto is a pathogen of leaf anthracnose on evergreen spindle tree (Euonymus japonicus). Plant Dis. 100:672-678.


Hyde, K. D., Cai, L., Cannon, P. F., Crouch, J. A., Crous, P. W., Damm, U., Goodwin, P. H., Chen, H., Johnston, P. R., Jones, E. B. G., Liu, Z. Y., McKenzie, E. H. C., Moriwaki, J., Noireung, P., Pennycook, S. R., Pfenning, L. H., Prihastuti, H., Sato, T., Shivas, R. G., Tan, Y. P., Taylor, P. W. J., Weir, B. S., Yang, Y. L. and Zhang, J. Z. 2009.
Colletotrichum: names in current use. Fungal Divers. 39:147-182.
Jayawardena, R. S., Bhunjun, C. S., Hyde, K. D., Gentekaki, E. and Itthayakorn, P. 2021.
Colletotrichum: lifestyles, biology, morpho-species, species complexes and accepted species. Mycosphere 12:519-669.

Jayawardena, R. S., Hyde, K. D., Damm, U., Cai, L., Liu, M., Li, X. H., Zhang, W., Zhao, W. S. and Yan, J. Y. 2016. Notes on currently accepted species of Colletotrichum. Mycosphere 7:1192-1260.

Jiang, N., Fan, X. and Tian, C. 2021. Identification and characterization of leaf-inhabiting fungi from Castanea plantations in China. J. Fungi 7:64.



Joa, J. H., Lim, C. K., Choi, I. Y., Park, M. J. and Shin, H. D. 2016. First report of Colletotrichum fructicola causing anthracnose on mango in Korea. Plant Dis. 100:1793.

Johnston, P. R. 2000. The importance of phylogeny in understanding host relationships within Colletotrichum. In: Colletotrichum: host specificity, pathogenicity, and host-pathogen interactions, eds. by D. Prusky, S. Freeman and M. B. Dickman, pp. 21-28. APS Press, St. Paul, MN, USA.
Johnston, P. R., Pennycook, S. R. and Manning, M. A. 2005. Taxonomy of fruit-rotting fungal pathogens: what’s really out there? N. Z. Plant Prot. 58:42-46.


Kim, C., Hassan, O., Lee, D. and Chang, T. 2018a. First report of anthracnose of apple caused by Colletotrichum fructicola in Korea. Plant Dis. 102:2653.

Kim, C. H., Hassan, O. and Chang, T. 2020. Diversity, pathogenicity, and fungicide sensitivity of Colletotrichum species associated with apple anthracnose in South Korea. Plant Dis. 104:2866-2874.


Kim, D.-H., Jeon, Y.-A., Go, S.-J., Lee, J.-K. and Hong, S.-B. 2006. Reidentification of Colletotrichum gloeosporioides and C. acutatum isolates stored in Korean Agricultural Culture Collection (KACC). Res. Plant Dis. 12:168-177 (in Korean).

Kim, G. H., Choi, D. H., Park, S. Y. and Koh, Y. J. 2018b. First report of anthracnose caused by Colletotrichum nymphaeae on kiwiberry in Korea. Plant Dis. 102:1455.

Kim, G. H., Kim, D. R., Park, S.-Y., Lee, Y. S., Jung, J. S. and Koh, Y. J. 2017. Incidence rates of major diseases of kiwiberry in 2015 and 2016. Plant Pathol. J. 33:434-439.



Kim, J. S., Hassan, O., Go, M. J. and Chang, T. 2021. First report of Colletotrichum aenigma causing anthracnose of grape (Vitis vinifera) in Korea. Plant Dis. 105:2729.

Kim, Y. H., Jun, O. K., Sung, M. J., Shin, J.-S., Kim, J. H. and Jeoung, M.-I. 2000. Occurrence of Colletotrichum stem rot caused by Glomerella cingulata on graft-cactus in Korea. Plant Pathol. J. 16:242-245.
Kobayashi, T. 2007. Index of fungi inhabiting woody plants in Japan. Host, distribution and literature. Kyokai Publishing Co., Ltd, Zenkoku-Noson-Kyoiku, Japan. pp. 1227.(in Japanese).
Korean Society of Plant Pathology 2022. List of plant diseases in Korea. 6th ed. Korean Society of Plant Pathology, Seoul, Korea. pp. 630.
Latocha, P. 2017. The nutritional and health benefits of kiwiberry (Actinidia arguta): a review. Plant Foods Hum. Nutr. 72:325-334.




Lee, D. H., Kim, D.-H., Jeon, Y.-A., Uhm, J. Y. and Hong, S.-B. 2007. Molecular and cultural characterization of Colletotrichum spp. causing bitter rot of apples in Korea. Plant Pathol. J. 23:37-44.

Lee, D. M., Hassan, O. and Chang, T. 2020a. Identification, characterization, and pathogenicity of Colletotrichum species causing anthracnose of peach in Korea. Mycobiology 48:210-218.


Lee, D. M., Hassan, O., Kim, C. H. and Chang, T. 2020b. First report of anthracnose of peach (Prunus persica) caused by Colletotrichum fructicola in Korea. Plant Dis. 104:1556.

Lee, S.-Y., Ten, L. N., Ryu, J.-J., Kang, I.-K. and Jung, H.-Y. 2021.
Colletotrichum aenigma associated with apple bitter rot on newly bred cv. RubyS Apple. Res. Plant Dis. 27:70-75.


Li, Q., Bu, J., Shu, J., Yu, Z., Tang, L., Huang, S., Guo, T., Mo, J., Luo, S., Solangi, G. S. and Hsiang, T. 2019.
Colletotrichum species associated with mango in southern China. Sci. Rep. 9:18891.




Lim, J., Lim, T. H. and Cha, B. 2002. Isolation and identification of Colletotrichum musae from imported bananas. Plant Pathol. J. 18:161-164.
Liu, F., Ma, Z. Y., Hou, L. W., Diao, Y. Z., Wu, W. P., Damm, U., Song, S. and Cai, L. 2022. Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud. Mycol. 101:1-56.



Liu, F., Tang, G., Zheng, X., Li, Y., Sun, X., Qi, X., Zhou, Y., Xu, J., Chen, H., Chang, X., Zhang, S. and Gong, G. 2016. Molecular and phenotypic characterization of Colletotrichum species associated with anthracnose disease in peppers from Sichuan Province, China. Sci. Rep. 6:32761.




Lu, G., Cannon, P. F., Reid, A. and Simmons, C. M. 2004. Diversity and molecular relationships of endophytic Colletotrichum isolates from the Iwokrama Forest Reserve, Guyana. Mycol. Res. 108:53-63.


Meetum, P., Leksomboon, C. and Kanjanamaneesathian, M. 2015. First report of Colletotrichum aenigma and C. siamense, the causal agent of anthracnose disease of dragon fruit in Thailand. J. Plant Pathol. 97:402.
Nam, M. H., Park, M. S., Lee, H. D. and Yu, S. H. 2013. Taxonomic re-evaluation of Colletotrichum gloeosporioides isolated from strawberry in Korea. Plant Pathol. J. 29:317-322.



Nam, M. H., Park, M. S., Yoo, J. H., Lee, B. J. and Lee, J. N. 2022. First report of anthracnose crown rot caused by Colletotrichum siamense on strawberry in Korea. Korean J. Mycol. 50:235-241.
O’Donnell, K. and Cigelnik, E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7:103-116.


Oh, J. Y., Heo, J.-I. and Lee, D.-H. 2021. First report of anthracnose on pecan (Carya illinoiensis) caused by Colletotrichum siamense in Korea. Plant Dis. 105:3296.
Oo, M. M., Lim, G., Jang, H. A. and Oh, S.-K. 2017. Characterization and pathogenicity of new record of anthracnose on various chili varieties caused by Colletotrichum scovillei in Korea. Mycobiology 45:184-191.




Oo, M. M. and Oh, S.-K. 2016. Chilli anthracnose (Colletotrichum spp.) disease and its management approach. Korean J. Agric. Sci. 43:153-162.

Oo, M. M. and Oh, S.-K. 2017. Identification and characterization of new record of grape ripe rot disease caused by Colletotrichum viniferum in Korea. Mycobiology 45:421-425.




Oo, M. M. and Oh, S.-K. 2020. First report of anthracnose of chili pepper fruit caused by Colletotrichum truncatum in Korea. Plant Dis. 104:564.

Oo, M. M., Yoon, H.-Y., Jang, H. A. and Oh, S.-K. 2018. Identification and characterization of Colletotrichum species associated with bitter rot disease of apple in South Korea. Plant Pathol. J. 34:480-489.




Park, J. C., Kim, B. W. and Young, H. S. 1990. Further study on the flavonoids from the leaves of Machilus thunbergii in Korea. Korean J. Pharmacogn 21:197-200.
Park, K. S. and Kim, C. H. 1992. Identification, distribution and etiological characteristics of anthracnose fungi of red pepper in Korea. Korean J. Plant Pathol. 8:61-69.
Park, M. S., Kim, B.-R., Park, I.-H. and Hahm, S.-S. 2018. First report of two Colletotrichum species associated with bitter rot on apple fruit in Korea: C. fructicola and C. siamense. Mycobiology 46:154-158.



Paul, N. C., Lee, H. B., Lee, J. H., Shin, K. S., Ryu, T. H., Kwon, H. R., Kim, Y. K., Youn, Y. N. and Yu, S. H. 2014. Endophytic fungi from Lycium chinense Mill and characterization of two new Korean records of Colletotrichum. Int. J. Mol. Sci. 15:15272-15286.



Penzig, A. G. O. 1882. Fungi agrumicoli. Contribuzione allo studio dei funghi parassiti degli agrumi. Michelia 2:385-508 (in Italian).
Poti, T., Kisaki, G., Arita, K. and Akimitsu, K. 2023. Identification and characterization of Colletotrichum species causing kiwifruit anthracnose in Kagawa Prefecture, Japan. J. Gen. Plant Pathol. 89:84-90.


Sharma, G., Maymon, M. and Freeman, S. 2017. Epidemiology, pathology and identification of Colletotrichum including a novel species associated with avocado (Persea americana) anthracnose in Israel. Sci. Rep. 7:15839.




Sharma, G., Pinnaka, A. K. and Shenoy, B. D. 2013. ITS-based diversity of Colletotrichum from India. Curr. Res. Environ. Appl. Mycol. 3:194-220.

Simmonds, J. H. 1966. Host index of plant diseases in Queensland. Queensland Department of Primary Industries, Brisbane, Australia. pp. 111.
Tamura, K., Stecher, G. and Kumar, S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38:3022-3027.




Thao, L. D., Choi, H., Choi, Y., Mageswari, A., Lee, D. and Hong, S.-B. 2023. Re-identification of Colletotrichum acutatum species complex in Korea and their host plants. Plant Pathol. J. 39:384-396.




Truong, H. H., Sato, T., Ishikawa, S., Minoshima, A., Nishimura, T. and Hirooka, Y. 2018. Three Colletotrichum species responsible for anthracnose on Synsepalum dulcificum (miracle fruit). Int. J. Phytopathol 7:89-101.


Wang, Q., Liu, X., Ma, H., Shen, X. and Hou, C. 2019a.
Colletotrichum yulongense sp. nov. and C. rhombiforme isolated as endophytes from Vaccinium dunalianum var. urophyllum in China. Phytotaxa 394:285-298.


Wang, X., Liu, X., Wang, R., Fa, L., Chen, L., Xin, X., Zhang, Y., Tian, H., Xia, M. and Hou, X. 2021. First report of Colletotrichum aenigma causing walnut anthracnose in China. Plant Dis. 105:225.

Wang, Y., Qin, H. Y., Liu, Y. X., Fan, S. T., Sun, D., Yang, Y. M., Li, C. Y. and Ai, J. 2019b. First report of anthracnose caused by Colletotrichum aenigma on Actinidia arguta in China. Plant Dis. 103:372.

Weir, B. S., Johnston, P. R. and Damm, U. 2012. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 73:115-180.



White, T. J., Bruns, T., Lee, S. and Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications, eds. by M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White, pp. 315-322. Academic Press, New York, USA.

Yan, J-Y, Jayawardena, M. M. R. S., Goonasekara, I. D., Wang, Y., Zhang, W., Liu, M., Huang, J.-B., Wang, Z.-Y., Shang, J.-J., Peng, Y.-L., Bahkali, A., Hyde, K. D. and Li, X.-H. 2015. Diverse species of Colletotrichum associated with grapevine anthracnose in China. Fungal Divers. 71:233-246.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




