 |
 |
| Plant Pathol J > Volume 40(2); 2024 > Article |
|
Abstract
Citrus yellow vein clearing virus (CYVCV) is a member of the Alphaflexiviridae family that causes yellow vein clearing symptoms on citrus leaves. A total of 118 leaf samples from nine regions of six provinces in Korea were collected from various citrus species in 2020 and 2021. Viral diagnosis using next-generation sequencing and reverse transcription polymerase chain reaction (RT-PCR) identified four viruses: citrus tristeza virus, citrus leaf blotch virus, citrus vein enation virus, and CYVCV. A CYVCV incidence of 9.3% was observed in six host plants, including calamansi, kumquat, Persian lime, and Eureka lemon. Among the citrus infected by CYVCV, only three samples showed a single infection; the other showed a mixed infection with other viruses. Eureka lemon and Persian lime exhibited yellow vein clearing, leaf distortion, and water-soak symptom underside of the leaves, while the other hosts showed only yellowing symptoms on the leaves. The complete genome sequences were obtained from five CYVCV isolates. Comparison of the isolates reported from the different geographical regions and hosts revealed the high sequence identity (95.2% to 98.8%). Phylogenetic analysis indicated that all the five isolates from Korea were clustered into same clade but were not distinctly apart from isolates from China, Pakistan, India, and Türkiye. To develop an efficient diagnosis system for the four viruses, a simultaneous detection method was constructed using multiplex RT-PCR. Sensitivity evaluation, simplex RT-PCR, and stability testing were conducted to verify the multiplex RT-PCR system developed in this study. This information will be useful for developing effective disease management strategies for citrus growers in Korea.
Citrus refers to citrus fruits from trees belonging to the genera Citrus, Fortunella, and Poncirus of the Rutaceae family. Among these, the most widely cultivated species in Korea is the satsuma mandarin (Citrus unshiu Marc.). Recently, citrus fruits such as lemon (C. limon), lime (C. latifolia), and calamansi (C. microcarpa), which were not traditionally grown in Korea, have gained popularity as new income crops and are being cultivated nationwide (Abobatta, 2019; Song, 2023). According to the survey conducted by agriX (https://edu.agrix.go.kr/), the cultivation areas for lemon and late-maturing citrus have been gradually increasing from 2015 to 2020. Additionally, due to climate change, citrus cultivation is expanding beyond Jeju Island to inland southern regions like Jeonnam and Gyeongnam, and this trend is expected to continue. However, except for lemon, these crops are not yet cultivated in large quantities but are partially grown in houses. Therefore, it is challenging to calculate the exact amount of cultivation for these crops.
Currently, about 23 species of viruses that infect citrus trees have been identified worldwide (Umer et al., 2019). Viral diseases are a worldwide problem, and emerging viruses such as citrus yellow vein clearing virus (CYVCV) have become serious problems for citrus production (Li et al., 2017; Liu et al., 2020). Among these viruses, citrus tristeza virus (CTV), citrus tatter leaf virus (CTLV), satsuma dwarf virus (SDV), citrus sadwamosaic virus (CiMV), citrus vein enation virus (CVEV), and citrus leaf blotch virus (CLBV) have been previously reported in Korea (Hyun et al., 2017; Kim et al., 1999; Park et al., 2019; Yang et al., 2019).
CYVCV belongs to the subgenus Mandarivirus of the Alphaflexiviridae family. CYVCV was first discovered in Pakistan. It has since been reported in Türkiye, India, Iran, and China (Alshami et al., 2003; Catara et al., 1993; Chen et al., 2014; Hashmian and Aghajanzadeh, 2017; Önelge et al., 2010). Most citrus plants infected with CYVCV are asymptomatic. However, a few citrus hosts, such as lemon and lime, are susceptible hosts that exhibit typical symptoms of yellow vein clearing disease. Virus-infected hosts showed venial yellowing, distortion on leaves, and venial water soaking on the underside of the leaves (Liu et al., 2020). CYVCV is transmitted through Aphis spiraecola, A. craccivora, Dialeurodes citri, grafting, mechanical inoculation, and contaminated tools (Önelge et al., 2011; Zhang et al., 2018, 2019a, 2019b). It has been reported to infect most citrus species, grapevine (Vitis vinifera) (Afloukou and Önelge, 2020), herbaceous plants such as Phaseolus vulgaris var. Dermason, Vigna unguiculata (L.) and wild weed species (Önelge et al., 2016). CYVCV has not yet been reported in Korea. The genetic characterization of the virus was discovered in previous studies with complete genome. CYVCV is known to have a sequence identity of up to 95% between isolates, and the genetic variation between hosts is low (Liu et al., 2020). The isolates from China were phylogenetically distinct from the isolates reported in Türkiye, India, and Pakistan, indicating that genetic differences according to geographic origin are not large and do not form distinct clusters (Zhou et al., 2017).
Citrus trees infected with viruses show various symptoms, including malformation and curling of the leaves, stem pitting, and fruit damage such as poor coloration, malformation, and early drop. It is difficult to diagnose viral diseases based on symptoms alone, as the symptom expression can vary depending on the species and/or variety of citrus plant and co-infection. To control a virus effectively, it is necessary to examine the presence of virus in plants, identify the specific pathogen, and understand how it spreads. Next-generation sequencing (NGS) is an innovative method for confirming the presence of plant viruses, offering advantages over Sanger sequencing. Unlike Sanger sequencing, NGS is not limited by the length of a nucleotide sequence that can be read at a time and it doesn’t require any prior information about nucleotide sequences. This provides NGS with a significant edge in the primary detection of plant viruses (Kesanakurti et al., 2016; Pecman et al., 2017; Roossinck et al., 2015).
Research on citrus viruses in Korea began in 1999 with the first studies focusing on the occurrence of CTV, SDV, and CTLV (Kim et al., 1999). Subsequently, CiMV, was identified in satsuma mandarin on Jeju Island (Hyun et al., 2017), and the presence of CLBV was confirmed thorough NGS (Park et al., 2019). CVEV, which was previously designated as a causal agent of disease managed by quarantine, was found to have spread widely across Korea (Kim et al., 2019). A multiplex reverse transcription polymerase chain reaction (mRT-PCR) assay for CTV, CiMV, and CTLV was designed at the Citrus Research Institute (National Institute of Horticultural and Herbal Science) for simultaneous detection of viruses (Hyun et al., 2017). Although the occurrence, partial genome information, and diagnostic methods of viruses in satsuma mandarin have been studied, the distribution and genetic diversity of viruses in other citrus species are still unclear.
In this study, the distribution of citrus viruses infecting various citrus plants including newly cultivated citrus species was determined analyzing 118 samples collected from Korea during 2020 and 2021. CYVCV was identified for the first time in Korea, and it showed a low infection rate (9.3%). In addition, a mRT-PCR system was developed for diagnosis of four viruses present in citrus species newly cultivated in Korea.
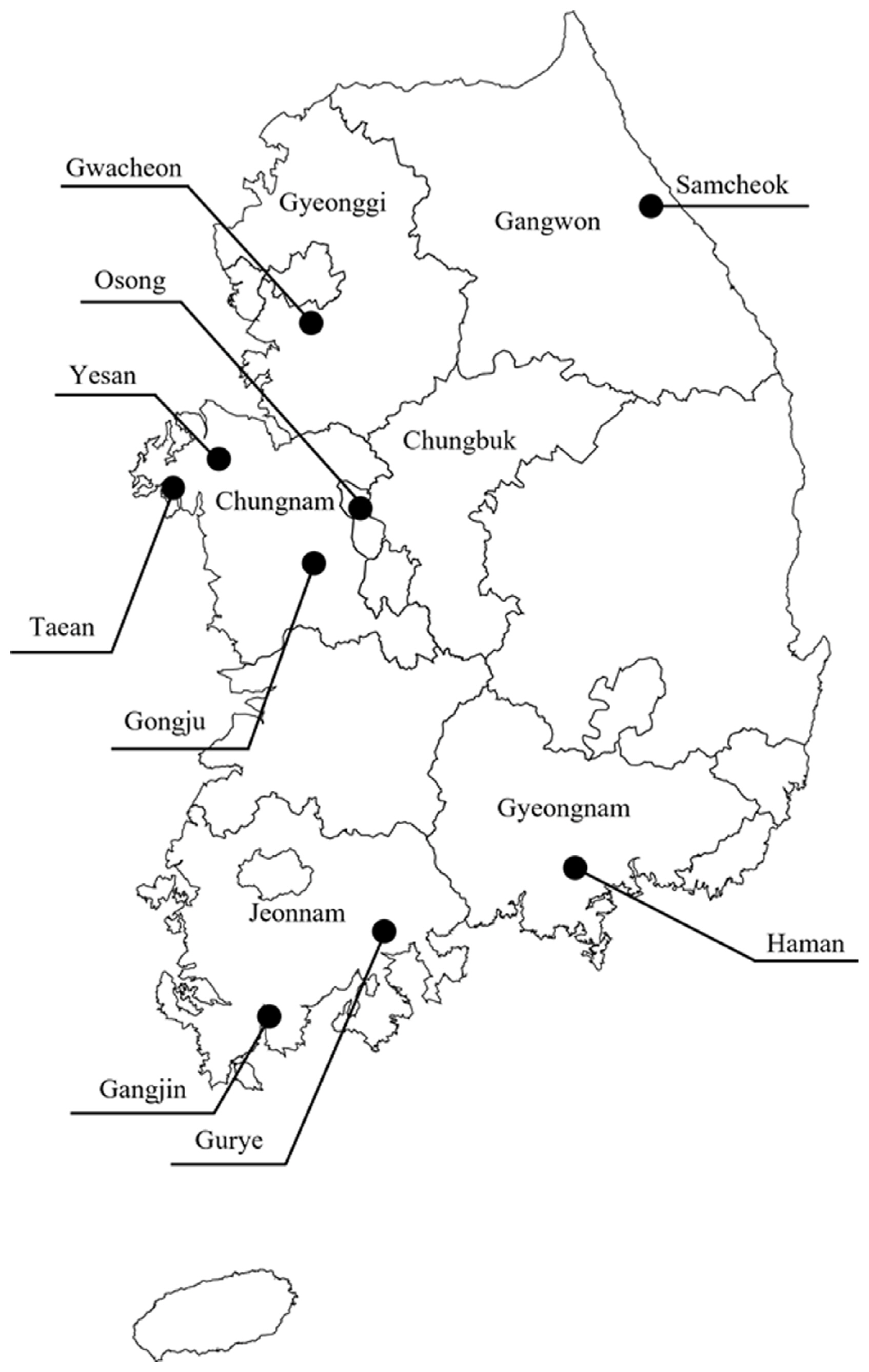
From February 2020 to September 2021, a total of 118 citrus leaf samples were collected from the Chungbuk, Chungnam, Gangwon, Gyeonggi, Gyeongnam, and Jeonnam provinces (Fig. 1). Fifteen varieties of citrus cultivars, including Eureka lemon (Citrus limon (L.) Burm. f.), Persian lime (C. latifolia Tanaka), calamansi (C. microcarpa), pummelo (C. grandis (L.) Osbeck), were surveyed (Table 1).
Total RNA was extracted from leaf samples of citrus using easy-spin Total RNA Extraction kit (iNtRON Biotechnology, Seongnam, Korea). RNA quality was assessed using the Tapestation RNA Screen Tape (Agilent, Santa Clara, CA, USA). Subsequently, cDNA libraries were prepared with TruSeq Stranded Total RNA and a Ribo-Zero Plant kit (Illumina, San Diego, CA, USA), removing plant-derived ribosomal RNA to boost the viral nucleic acid in NGS reads. Sequencing was performed on an Illumina NovaSeq 6000 system following the manufacturer’s instructions at Macrogen Inc. (Seoul, Korea). For NGS analysis, total RNA was extracted from kumquat, Persian lime, Eureka lemon, and calamansi in Gongju, and mixed to prepare the cDNA library. In Taean, total RNA was extracted from natsudaidai, satsuma mandarin, ‘Kanpei’, ‘Shiranui’, and lemon. The reverse transcription polymerase chain reaction (RT-PCR) experiment was performed using total RNA isolated from each citrus host. Gene-specific primer sets (Table 2) were used for the four viruses (CTV, CYVCV, CVEV, and CLBV) identified from the NGS results. One-step RT-PCR using SuPrimeScript RT-PCR Premix 2× (Genetbio, Daejeon, Korea) was conducted to diagnose viral disease. After purifying the product using an Expin Combo GP kit (GeneAll, Seoul, Korea), nucleotide sequences were analyzed by Sanger sequencing (BIONICS, Daejeon, Korea).
Complete genome sequencing analysis was performed on five CYVCV isolates identified in diagnosis by RT-PCR. Primer sets for whole-genome sequencing of CYVCV were designed and used in the experiment, and long RT-PCR was performed to amplify the nucleotides of CYVCV by dividing them into a total of three fragments (Table 3). Fragment 1 was produced with a combination of the CYVCV 1F and 2660R primers, fragment 2 was a combination of 2412F and 5124R primers, and fragment 3 was a combination of 4893F and 7531R primers in long RT-PCR. All the PCR products were subsequently cloned into the pGEM-T easy vector (Promega, Madison, WI, USA), and three clones of each fragment were randomly selected and sequenced by Sanger sequencing. Sequenced complete nucleotide and amino acid were aligned using the MUSCLE method to discover the genetic trait of CYVCV isolates from Korea. Pairwise comparison was conducted using Geneious Prime software (version 2020.1.2) on the nucleotide sequence of whole genome. Phylogenetic trees were constructed by maximum likelihood (ML) method using MEGA X software (version 10. 1. 8) using the complete genome region of CYVCV isolates.
To design the mRT-PCR primer sets of CTV, CYVCV, CVEV, and CLBV, more than 10 isolates reported from different geographical regions were retrieved from the NCBI database and aligned using MUSCLE method. A primer binding site was selected as conserved regions. The primer sets for mRT-PCR of CTV were designed on p61 region; CYVCV were designed on RNA-dependent RNA polymerase (RdRp) region; CVEV were designed on coat protein (CP) region and CLBV were designed on CP region. The amplified product sizes of each primer set were designed as different lengths, and melting temperature of all primer sets were adjusted to 57℃ for efficiency of this method. The sequence of primer sets for mRT-PCR developed in this study was detailed in Table 4. A mRT-PCR system was established by designing primer sets to identify the four viruses. One-step RT-PCR was performed using SuPrimeScript RT-PCR Premix 2× (Genetbio), and four viruses were simultaneously detected using the CTV 13169F/13907R, CYVCV 102F/698R, CVEV 4831F/5303R, and CLBV 7120F/7505R primer sets designed in this study.
A total of 118 samples from nine regions of six provinces were analyzed to determine the presence of citrus leaf viruses. Among the collected samples from Gongju, and Chungnam, Persian lime and Eureka lemon showed yellow vein clearing, distortion on the leaves, and water-soak symptom on the underside of the leaves. Of the citrus samples collected in Taean, and Chungnam, natsudaidai showed virus-like symptoms of yellowing and vein banding, while satsuma mandarin did not show any symptoms. ‘Kanpei’, ‘Shiranui’, and lemon exhibited yellowing across the midvein, mild mottle on leaves (Fig. 2). Only yellowing or latent infection was observed in pummelo and lemon in Yesan, as well as in C. reticulata × sinensis collected in Gangjin, Gurye, and Haman. Citrus leaves collected in Cheongju, Samcheok, and Gwacheon exhibited chlorosis and yellowing across the vein of leaf samples of C. reticulata × sinensis, ‘Ehime Kashi 28 gou’, ‘Shiranui’, satsuma mandarin, calamansi, lemon, lime, citron, ‘Setoka’, ‘Kanpei’, excepting ‘Winter Prince’ and ‘YN-26’ showed asymptomatic. Two groups of citrus leaves exhibiting different symptoms were detected: (1) yellow vein clearing, severe distortion on the leaves, and water-soaking symptoms on the underside of the leaves were observed in Gongju and Chungnam; and (2) chlorosis, vein banding, and yellowing across the midvein symptoms on the leaves of various citrus hosts were observed in Taean, Yesan, Gangjin, Haman, Cheongju, and Samcheok (Fig. 2).
Library 1 was constructed using five citrus samples collected from Gongju (group 1 above) to screen for the viruses present in the samples. After de novo assembly using BLAST-n/x of a total 87,344,454 reads sequenced through RNA sequencing, 115,269 transcript contigs were identified (Table 5). Among the assembled contigs, 28 were CTV contigs, 1 was a CYVCV contig, 1 was a CVEV contig, and 1 was a CLBV contig. The total counts of reads sequenced by NGS for the four viruses were 80,095 for CTV, 7,078 for CYVCV, 7,945 for CVEV, and 292 for CLBV. The assembled viral contigs were highly homologous (94-98%) to previously reported isolates registered in the NCBI database (https://www.ncbi.nlm.nih.gov/). RT-PCR with viral gene-specific primer sets confirmed the NGS results, identifying CTV, CVEV, CLBV, and CYVCV (Table 2). Library 2 was constructed using 12 citrus samples collected from Taean showing chlorosis, vein banding, yellowing across the midvein symptoms on the leaves. On de novo assembly of the 78,481,350 reads analyzed in RNA sequencing through BLAST-n/x, 191,636 transcript contigs were identified (Table 5). Among the analyzed contigs, viral contigs included 40 of CTV, 1 of CVEV, and 2 of CLBV. The number of reads that combined with each contigs of the CTV, CVEV, and CLBV were 57,535, 12,088, and 2,255, respectively. Sequenced viral contigs showed a 93.38-99.38% identity with isolates from NCBI (NC001661, NC021564, NC003877, and NC021564). RT-PCR was performed to confirm the findings of NGS, and three viruses were identified: CTV, CVEV, and CLBV. Seventeen leaf samples of citrus collected from Gongju (library 1), and Taean (library 2) were tested by RT-PCR for CTV, CYVCV, CVEV, and CLBV. CTV was present in 11 samples. CYVCV, which is reported for the first time in Korea, was detected in five samples collected in Gongju. CVEV was diagnosed in 2 samples, and CLBV was infected with 2 samples. Except for CYVCV, the three viruses were present in both libraries collected from Gongju and Taean.
To investigate the distribution of viruses identified by NGS in Korea, a total of 118 leaves were collected from citrus trees in Korea. The final infection rate was 70/118 (59.32%); overall, the virus infection rate was highest for CTV, then CLBV, then CVEV (Table 6). CYVCV infection was confirmed in six host plants, including calamansi, kumquat, lime, pummelo, and lemon. Of the infected hosts, calamansi and kumquat were first identified as host of CYVCV in this study. The CYVCV infection rate was 11/118 (9.3%) on RT-PCR. Of the citrus infected by CYVCV, only three samples were single infection: the other showed mixed infection with other viruses. Among the CYVCV-infected hosts, only Eureka lemon and Persian lime showed yellow vien clearing disease (YVCD) symptoms, while the other hosts showed only yellowing symptoms on the leaves. Among the citrus hosts surveyed, lemon, lime, calamansi, pummelo, C. reticulata × sinesis, and ‘Shiranui’ were the most frequently collected and tested for virus identification, with 15, 12, 12, 12, 22, and 12 leaves tested, respectively. Interestingly, all samples collected from lime and ‘Shiranui’ were confirmed to be infected with at least one virus by RT-PCR, while calamansi and C. reticulata x sinensis had infection rates of 55% and 50%, respectively, indicating that approximately half of the samples were infected. Lemon and pummelo showed the lowest virus infection rate among the above six major collected crops, with 20% and 8.3%, respectively. Among infected samples, 55.71% had a single infection, 37.14% had a double infection (two viruses), and 7.14% had a triple infection (three viruses) (Table 7). Of the 12 samples of calamansi infected with the virus, 92.1% were infected with two or more viruses. This is significantly higher than the multiple infection rate of 8.3-36.3% in all virus-infected samples in lime, ‘Shiranui’, and C. reticulata × sinensis (Table 7). Total 70 samples were infected with the viruses, 45.7% were infected with two or more viruses, of which 37.14% were double infected with viruses and 7.14% were complex infections caused by the three viruses. In case of double infection of viruses, the co-infection with CTV and CLBV were confirmed as 21.43%, followed by infection with CTV and CYVCV as 8.57%, and infection with CTV and CVEV as 5.71%. In the case of triple infection of viruses, samples infected with CTV, CVEV, and CLBV (5.71%) were relatively high. One interesting point is that no single infections were observed with CVEV or CLBV.
Five isolates of CYVCV (GJ1-5, KACC nos CV201126-1-4, and CV211125-6) were collected in Gongju, Chungnam, and sequenced to obtain the 7,530 nt of complete genome of CYVCV (GenBank accession nos. OL581610, OL441344, OQ174723, and OL441344). Out of the five isolates, GJ1 was isolated from calamansi, GJ2-3 from Persian lime, GJ4 from Eureka lemon, and GJ5 from kumquat. As a result of the whole-genome sequencing, the genome structure of the CYVCV was identified as six open reading frames (ORFs), with ORF 1 coding for the RdRP protein in 79-5,028 nt. ORF 2 to 4 were analyzed as 5,035-5,712, 5,690-6,016, and 5,943-6,125 nt, respectively, coding for the 25 kDa triple gene block. ORF 5 was confirmed to code for the CP region in 6,149-7,126 nt, and ORF 6 coded for the 23 kDa nucleic acid binding protein and contained a sequence of 6,826-7,494 nt. The five GJ isolates of CYVCV showed 99.3-99.8% sequence homogeneity with each other (Table 8). When compared to 17 isolates reported in China, India, Pakistan, and Türkiye, nucleotide sequence identity ranged from 95.2-98.8%. There was a slight difference in sequence identity between isolates from China (97.7 to 98.8%) and isolates from India, Pakistan, Türkiye (95.2 to 97.4%), but the difference was not significant. Among the isolates, the lowest identity was observed with isolates reported in etrog citron in India (KT696510) at 95.2-95.4%. At the amino acid level, the 26 isolates of CYVCV also shared high identity in CP region (96.1 to 98.8%). Similar to the above result, isolates from China shared slightly higher identity (97.3 to 98.8%) than isolates from India, Iran, Pakistan, and Türkiye (96.1-97.9%). The isolates GJ 1-5 showed high identity (>95%) in nucleotide and amino acid level, showing no significant genetic difference.
A phylogenetic tree was constructed with CYVCV isolates reported from China, Pakistan, Iran, India, Türkiye, including the five isolates from Korea. Using the amino acid sequences of the CP regions, it was confirmed that all five isolates from Korea were phylogenetically distinct from isolates reported from China, Pakistan, India, and Türkiye. However, no clear clustering was observed using the Tamura-Nei model, Hasegawa-Kishino-Yano model, Jukes-Cantor model, or Kimura 2-parameter model of ML method. Additionally, isolates reported in etrog citron from India (KT696510) were not phylogenetically close to the GJ 1-5 isolates (Fig. 3).
mRT-PCR was performed with the primers selected in simplex RT-PCR of CTV 13169F/13907R, CYVCV 102F/698R, CVEV 4831F/5303R, and CLBV 7120F/7505R. Products of the expected size of the primer sets of CTV, CYVCV, CVEV, and CLBV (738, 596, 472, and 385 bp, respectively), was observed, while no product amplified from the negative control. Furthermore, the amplified bands of each virus were identified by sequencing. Only the specific band of designed primer sets was amplified, despite using a mixed template of total RNA including four viral RNAs. These results demonstrated the specificity the specificity of the designed primer sets and their successful incorporation into mRT-PCR. To test the stability of mRT-PCR, assays were conducted with different combination of total RNA, including viral genomic RNA of the four viruses, under the optimized mRT-PCR conditions. All four viruses (CTV, CYVCV, CVEV, and CLBV) could be distinguished by their band size on electrophoresis in the 11 cases of infection by single or multiple viruses (Fig. 4). To confirm and verify the sensitivity of the developed mRT-PCR, a 10-fold serial dilution test (100-10−7) was conducted to analyze the detection limits in this system. In simplex RT-PCR, showed highest dilution detected was 10−5 for CYVCV, followed by 10−4 for CLBV, 10−3 for CTV and CVEV (Fig. 5). The bands of CYVCV and CLBV were clear at a relatively higher dilution, while the CTV and CVEV band intensity dropped at 10−4. The result of mRT-PCR showed that the detection limit of four viruses was equally 10−3. As shown in above results, it was possible to effectively diagnose viruses at even a very low concentration (10−3) of total RNA by mRT-PCR. Five samples collected in Gongju, Chungnam province and one sample collected in Cheongju, Chungbuk province, were identified using a simultaneous detection system in this study to confirm the practicality of this method. CTV, CYVCV, CVEV, and CLBV were detected in all six samples, and amplified viral nucleotides were sequenced and identified. The result of mRT-PCR was identical to those of simplex RT-PCR using gene-specific primer sets. mRT-PCR is therefore a reliable method for the simultaneous detection of CTV, CYVCV, CVEV, and CLBV.
Viruses were identified in newly cultivated citrus species such as lemon, lime and calamansi, and the incidence pattern of the viruses was analyzed in Korea. Of the four viruses found in citrus leaves collected from nine regions of six provinces, CTV was the most prevalent virus as with previous studies (Hyun et al., 2017). Of the 118 Citrus spp. samples, 11 were found to be infected with CYVCV based on RT-PCR. A CYVCV incidence of 9.3% was observed in six host plants, including calamansi, kumquat, Persian lime, pummelo, and Eureka lemon. Lemon co-infected with CTV and CYVCV caused symptoms of leaf curling and yellow vein clearing, but lemon infected with CTV, CVEV, and CLBV did not show any symptoms. Only mild yellowing symptoms were detected in the leaves of infected hosts, except for one sample of Persian lime co-infection with CTV and CYVCV, which showed leaf curling and yellowing. This study was the first to confirm the occurrence of CYVCV in Korea, and the citrus plants infected with CYVCV have been discarded.
From May 2014 to April 2016, total 2,350 samples were collected to verify the viral infection of CYVCV in the 11 major citrus-growing provinces of China (Zhou et al., 2017). In RT-PCR assay, the virus infection rate was 19.5%, which higher than a survey identified in this study. Furthermore, the infection rate of CYVCV against lemon and mandarin in China were 43.8 and 23.5%, respectively, which were higher than 13.3 and 5.9% of this study (Zhou et al., 2017). Meanwhile, the infection rate of lemon and mandarin in Türkiye were 16.66 and 0%, respectively (Afloukou and Önelge, 2021), similar to this field research. However, the overall infection was 16.74% which is higher than 9.3% of this assay. CYVCV has not yet spread widely in Korea but considering that this study did not investigate Jeju Island, which produces the largest number of citrus fruits in Korea, and lack of the number of samples, an additional distribution study of CYVCV be needed. Additionally, CYVCV has been reported to naturally infect grapevine in Türkiye (Afloukou and Önelge, 2020), so it is necessary to confirm CYVCV infection in domestic grapevines. The virus can be transmitted by Aphids spiraecola, A. craccivora, and D. citri (Önelge et al., 2011; Zhang et al., 2018, 2019a), which are common in Korea, so there is possibility that it could spread nationwide. Moreover, if a virus is transmitted by insect vectors, symptoms may not appear for up to 12 months (Zhang et al., 2019a) and may not be identified by observation depending on the susceptibility of the host, making it difficult to confirm the spread of the virus. CYVCV, a phloem-limited virus, has been shown to be transmitted through sap (Alshami et al., 2003; Önelge et al., 2011; Zhang et al., 2019b), suggesting the possibility of rapid CYVCV spread. However, since pathological analysis of CYVCV isolates has not yet been conducted in Korea, additional investigations of symptoms, infection rates by mechanical, insect vector and major means of infection in domestic citrus hosts are needed to minimize the damage and prevent the spread of the virus caused by CYVCV in citrus-growing regions of Korea.
Pairwise identity and phylogenetic analysis of CYVCV suggested that genetic diversity of different hosts and geographical origin was low level as same of previous study (Meena et al., 2019; Zhou et al., 2017). Isolates from Korea were clustered into same clade and showed low level of genetic difference with the isolates from China, India, Pakistan, and Türkiye. In this study, genetic characteristics were analyzed through full genome sequencing of CYVCV that was first isolated in Korea, and this complete genome of CYVCV is expected to be utilized in further studies on the evolution or epidemiological investigation of the virus.
mRT-PCR is a time-saving, reliable, and effective detection method used to diagnose a variety of pathogens simultaneously. It is implemented using different sets of primers for two or more targets in one reaction (Pallás et al., 2018). Pathogens can also be diagnosed by conventional RT-PCR or serological methods, but those methods consume more time and have drawbacks in comparison with mRT-PCR in terms of cost and efficiency. Additionally, plant viruses often infect multiple viruses in a single sample, so it is more efficient to diagnose them using this mRT-PCR. Roy et al. (2005) developed a mRT-PCR assay to detect six RNA (citrus leaf rugose virus [CLRV], citrus psorosis virus [CPsV], CTLV, CTV, citrus variegation virus [CVV], and Indian citrus ringspot virus [ICRSV]) and one DNA (citrus yellow mosaic virus [CYMV]) virus from citrus plant, and Meena and Baranwal (2016) also construct the method for tree RNA (CYVCV, ICRSV, and CTV) and one DNA (CYMV) viruses along with HLB bacterium. However, these methods do not aim to detect CVEV and CLBV, which were recently isolated viruses in Korea. The simultaneous viral diagnosis method developed in this study can amplify four species of citrus viruses (CTV, CYVCV, CVEV, and CLBV) simultaneously using the designed primer sets. These four viruses were detected without any problem on mRT-PCR, and this result suggests that simultaneous detection could be used in agricultural institutions to diagnose viral disease in citrus trees. This rapid, efficient, and specific diagnostic method will help prevent the spread of these viruses and limit damage to citrus trees.
Acknowledgments
This work was supported by the research grant of the Chungbuk National University in 2019 and the Agenda Program (PJ014878, PJ016388) of the Rural Development Administration.
Fig. 2
Virus-like symptoms from collected citrus. (A-D) Yellow vein clearing, leaf distortion symptom in Persian lime (A-C) and Eureka lemon (D). (E) Yellowing across the vein on leaves of ‘Shiranui’. (F) Mild mottle on leaf of lemon.

Fig. 3
Phylogenetic tree based on CP (amino acids) of CYVCV. Constructed by maximum likelihood method using MEGA X software (version 10.1.8). Bold fonts represent the isolates that were discovered in Korea. Bootstrap replicates 1,000 and Bootstrap values >50% are indicated. Indian citrus ringspot virus (ICRSV, accession no. AF406744) is outgroup. CP, coat protein; CYVCV, citrus yellow vein clearing virus.

Fig. 4
Multiplexing of CYVCV (596 bp), CTV (738bp), CLBV (325 bp), and CVEV (472 bp) primers on polymerase chain reaction amplification. M, 100 bp ladder; lane 1, CYVCV; lane 2, CTV; lane 3, CYVCV + CLBV; lane 4, CTV + CLBV; lane 5, CTV + CVEV; lane 6, CTV + CYVCV; lane 7, CYVCV + CVEV + CLBV; lane 8, CTV + CVEV + CLBV; lane 9, CTV + CYVCV + CLBV; lane 10, CTV + CYVCV + CVEV; lane 11, CTV + CYVCV + CVEV + CLBV. CYVCV, citrus yellow vein clearing virus; CTV, citrus tristeza virus; CLBV, citrus leaf blotch virus; CVEV, citrus vein enation virus.
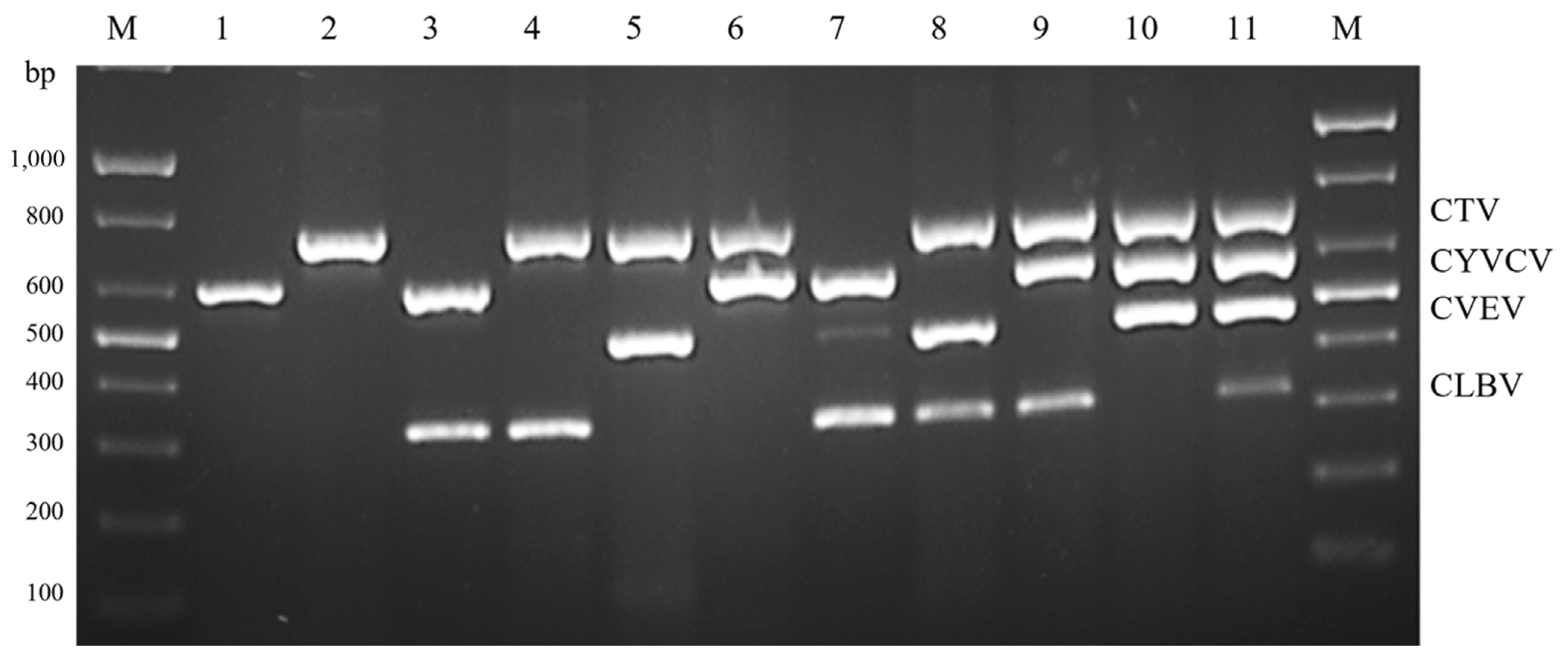
Fig. 5
Sensitivity assay of primer sets. (A) Detection of CTV. (B) Detection of CYVCV. (C) Detection of CVEV. (D) Detection of CLBV. (E) Detection of CTV + CYVCV + CVEV + CLBV. M, DNA size marker (100 bp DNA ladder); CTV, citrus tristeza virus; CYVCV, citrus yellow vein clearing virus; CVEV, citrus vein enation virus; CLBV, citrus leaf blotch virus. The total RNA is mixed with RNA extracted from infected citrus leaves.

Table 1
Citrus species collected in this study
Table 2
Gene-specific primer sets for RT-PCR
| Viruses | Name | Sequence (5′ → 3′) | Size (bp) | Reference |
|---|---|---|---|---|
| CTV | F | GARTTAATATCCGTTGACCC | 637 | This study |
| R | CTGATTTGATTACCCACCAC | |||
| CLBV | CP-for | ACTGCCAGTGACACTGGGA | 1,228 | Park et al. (2019) |
| CP-rev | GTGTAAAGTCCTGGCCCAC | |||
| CVEV | VE16F | AATTACGGCGTATCTATGGTGAGTCG | 413 | Vives et al. (2013) |
| VE17R | AATGAGATAGCCCGGTTGTCCAG | |||
| CYVCV | 1fw | GAAAAGCAAACAGTAACAAACACACCC | 921 | Loconsole et al. (2012) |
| 921rev | GGGCAAGAGCATTTGGGTATCT |
Table 3
Nucleotide sequence and positions of primers used for complete genome sequencing of CYVCV
| Primer namea | Sequence (5′ → 3′) | Locusb |
|---|---|---|
| Fragment 1 | ||
| 1F | GAAAAGCAAACAKTAWCAWACACACC | 1-26 |
| 2660R | GGCTTCGACATTGTWGCTTC | 2,660-2,641 |
| Fragment 2 | ||
| 2412F | GAACGCTTCCTCAACWCC | 2,412-2,429 |
| 5124R | CRGCAACAGCGTGTATGAC | 5,124-5,106 |
| Fragment 3 | ||
| 4893F | GCCTATCAACTTAAAGACGAG | 4,893-4,913 |
| 7531R | CAGAAAATGGAAACTGAAAGCC | 7,531-7,510 |
| Primers targeting the inner fragments for sequencing | ||
| 643F | CGCTACCCAGAGATATACCAG | 643-663 |
| 1495R | GGTGTTGAGTTGRTCGGAG | 1,495-1,477 |
| 1350F | CCAGACATCTCAGACCAAG | 1,350-1,368 |
| 2047R | GGTTAGTTGTGCAATCTCTAATG | 2,047-2,025 |
| 3207R | GCCTTAATCTCYACTCTTGACC | 3,207-3,186 |
| 4306R | CAGTTGATCATAATTTCGGGTG | 4,306-4,285 |
| 4127F | CCAACTCTTCCTGAAATCAC | 4,127-4,146 |
| 5662F | ACTCTAACCACCTTCTCATACTC | 5,662-5,684 |
| 6561R | CAACCTCAACCTAAGATACTCAC | 6,561-6,539 |
| 6361F | AACCTATCCCTGTACCCACTC | 6,361-6,381 |
Table 4
Primer sets designed for multiplex RT-PCR
Table 5
Assembled result of viral sequence by NGS
Table 6
The infection rate of viruses infecting different citrus tree
| Host | CTV | CLBV | CVEV | CYVCV | Infection ratio | Mixed- infection |
|---|---|---|---|---|---|---|
| Lemon | 2 | 1 | 1 | 2 | 3/15a | 2b |
| Lime | 11 | -c | 1 | 2 | 12/12 | 1 |
| Calamansi | 12 | 8 | - | 2 | 12/22 | 11 |
| Pummelo | - | - | - | 1 | 1/12 | - |
| Mandarin | ||||||
| Citrus reticulata × sinensis | 11 | 1 | - | 3 | 11/22 | 4 |
| ‘Shiranui’ | 12 | 1 | 3 | - | 12/12 | 4 |
| ‘Kanpei’ | 4 | 2 | 1 | - | 4/4 | 2 |
| ‘Setoka’ | 2 | - | - | - | 2/2 | - |
| ‘Winter Prince’ | - | - | - | - | 0/1 | - |
| ‘Ehime Kashi 28 gou’ | 3 | 2 | - | - | 3/3 | 2 |
| Satsuma mandarin | 6 | 3 | 2 | - | 6/6 | 4 |
| ‘YN-26’ | 1 | 1 | 1 | - | 1/1 | 1 |
| Natsudaidai | 1 | - | - | - | 1/4 | - |
| Kumquat | - | 1 | - | 1 | 1/1 | 1 |
| Citron | 1 | - | - | - | 1/1 | - |
| Total (%d) | 66 (55.9) | 20 (17.0) | 9 (7.6) | 11 (9.3) | 70/118 (59.3) | 32 (45.7) |
Table 7
Mixed-infection rate of viruses
| Infection case | No. of samples (%) | |
|---|---|---|
| Single infection (55.71%) | CTV | 36a (51.43)b |
| CYVCV | 3 (4.29) | |
| Duplex infection (37.14%) | CTV + CYVCV | 6 (8.57) |
| CTV + CVEV | 4 (5.71) | |
| CTV + CLBV | 15 (21.43) | |
| CYVCV + CLBV | 1 (1.43) | |
| Triplex infection (7.14%) | CTV + CYVCV + CVEV | 1 (1.43) |
| CTV + CVEV + CLBV | 4 (5.71) | |
| Total | 70 (100) 0 | |
Table 8
Pairwise identity of complete genome of CYVCV isolates
| No. | Isolates | 1a | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|---|
| 1 | Korea: calamansi GJ1 | |||||
| 2 | Korea: Persian lime GJ2 | 99.7b | ||||
| 3 | Korea: Persian lime GJ3 | 99.7 | 99.7 | |||
| 4 | Korea: Eureka lemon GJ4 | 99.7 | 99.7 | 99.6 | ||
| 5 | Korea: kumquat GJ5 | 99.5 | 99.5 | 99.5 | 99.4 | |
| KX156744.1 China: pummelo | 98.8 | 98.7 | 98.6 | 98.7 | 98.5 | |
| KX156747.1 China: sweet orange | 98.7 | 98.6 | 98.6 | 98.6 | 98.4 | |
| KX156743.1 China: pummelo | 98.6 | 98.5 | 98.5 | 98.6 | 98.4 | |
| NC_026592.1 China: lemon | 98.6 | 98.5 | 98.5 | 98.6 | 98.3 | |
| KX156752.1 China: sweet orange | 98.5 | 98.4 | 98.4 | 98.4 | 98.2 | |
| MK415923.1 China: satsuma | 98.5 | 98.4 | 98.4 | 98.4 | 98.2 | |
| MK415928.1 China: mandarin | 98.4 | 98.3 | 98.3 | 98.3 | 98.1 | |
| KP120977.1 China: lemon | 98.2 | 98.2 | 98.2 | 98.1 | 98.0 | |
| KY933797.1 China: tangor | 98.1 | 98.0 | 98.0 | 98.0 | 97.8 | |
| MG878869.1 China: lemon | 97.9 | 97.9 | 97.8 | 97.8 | 97.7 | |
| KT696512.1 India: sweet orange | 97.4 | 97.4 | 97.3 | 97.3 | 97.2 | |
| KP313241.1 Pakistan: lemon | 97.2 | 97.1 | 97.1 | 97.1 | 96.9 | |
| KT696513.1 India: kinnow | 97.1 | 97.0 | 97.0 | 97.0 | 96.8 | |
| KT345342.1 Pakistan: lemon | 96.9 | 96.9 | 96.9 | 96.9 | 96.8 | |
| KT696511.1 India: orange malta | 97.0 | 97.0 | 97.0 | 96.9 | 96.8 | |
| JX040635.1 Türkiye: lemon | 96.8 | 96.8 | 96.8 | 96.8 | 96.6 | |
| KT696510.1 India: etrog citron | 95.4 | 95.3 | 95.3 | 95.4 | 95.2 | |
| AF406744 ICRSVc | 71.6 | 71.6 | 71.6 | 71.6 | 71.5 |
References
Abobatta, W. F. 2019. Potential impacts of global climate change on citrus cultivation. MOJ Ecol. Environ. Sci. 4:308-312.

Afloukou, F. M. and Önelge, N. 2020. First report of natural infection of grapevine (Vitis vinifera) by Citrus yellow vein clearing virus. New Dis. Rep. 42:5.


Afloukou, F. and Önelge, N. 2021. Occurrence and molecular characterisation of citrus yellow vein clearing virus isolates in Adana province, Turkey. Physiol. Mol. Plant Pathol. 115:101655.

Alshami, A. A. A., Ahlawat, Y. S. and Pant, R. P. 2003. A hitherto unreported yellow vein clearing disease of citrus in India and its viral etiology. Indian Phytopathol. 56:422-427.
Catara, A., Azzaro, A., Davino, M. and Polizzi, G. 1993. Yellow vein clearing of lemon in Pakistan. In: International Organization of Citrus Virologists Conference Proceedings (1957-2010), eds. by P. Moreno, J. V. da Graça and L. W. Timmer, 12:pp. 364-367. International Organization of Citrus Virologists, Riverside, CA, USA.

Chen, H. M., Li, Z. A., Wang, X. F., Zhou, Y., Tang, K. Z., Zhou, C. Y., Zhao, X. Y. and Yue, J. Q. 2014. First report of Citrus yellow vein clearing virus on lemon in Yunnan, China. Plant Dis. 98:1747.

Hashmian, S. M. B. and Aghajanzadeh, S. 2017. Occurrence of citrus yellow vein clearing virus in citrus species in Iran. J. Plant Pathol. 99:290.
Hyun, J. W., Hwang, R. Y. and Jung, K. E. 2017. Development of multiplex PCR for simultaneous detection of citrus viruses and the incidence of citrus viral diseases in late-maturity citrus trees in Jeju Island. Plant Pathol. J. 33:307-317.



Kesanakurti, P., Belton, M., Saeed, H., Rast, H., Boyes, I. and Rott, M. 2016. Screening for plant viruses by next generation sequencing using a modified double strand RNA extraction protocol with an internal amplification control. J. Virol. Methods 236:35-40.


Kim, B.-S., Yang, H.-J., Lee, S.-H., Ko, S.-H., Park, K. N., Choi, E. J. and Lee, S.-J. 2019. Incidence of Citrus vein enation virus in Citrus spp. and Poncirus trifoliata in Korea. Res. Plant Dis. 25:233-236 (in Korean).


Kim, D.-H., Oh, D.-C., Hyun, C.-W., Kwon, H.-M., Kim, D.-H. and Lee, S.-C. 1999. Incidence of three major citrus viruses in Cheju Island. Plant Dis. Agric. 5:34-40.
Li, X.-T., Su, H.-N., Tan, K.-G., Tong, X.-N. and Zhong, B.-L. 2017. Fruit malformation of Satsuma mandarin (Citrus unshiu Marc.) infected by citrus yellow vein clearing virus. J. Phytopathol. 165:283-288.


Liu, C., Liu, H., Hurst, J., Timko, M. P. and Zhou, C. 2020. Recent advances on Citrus yellow vein clearing virus in citrus. Hortic. Plant J. 6:216-222.

Loconsole, G., Önelge, N., Potere, O., Giampetruzzi, A., Bozan, O., Satar, S., De Stradis, A., Savino, V., Yokomi, R. K. and Saponari, M. 2012. Identification and characterization of Citrus yellow vein clearing virus, a putative new member of the genus Mandarivirus. Phytopathology 102:1168-1175.


Meena, R. P. and Baranwal, V. K. 2016. Development of multiplex polymerase chain reaction assay for simultaneous detection of clostero-, badna- and mandari-viruses along with huanglongbing bacterium in citrus trees. J. Virol. Methods 235:58-64.


Meena, R. P., Prabha, K. and Baranwal, V. K. 2019. Genome characterization of citrus yellow vein-clearing virus: limited heterogeneity of viral genomes in Mandarivirus-infecting different citrus species. 3 Biotech 9:348.




Önelge, N., Bozan, O. and Gök-Güler, P. 2016. First report of Citrus yellow vein clearing virus infecting new natural host plants in Turkey. J. Plant Pathol. 98:373.
Önelge, N., Bozan, O., Gök, M. and Satar, S. 2010. Yellow vein clearing of lemons in Turkey. In: International Organization of Citrus Virologists Conference Proceedings (1957-2010), eds. by M. E. Hilf, R. G. Milne, L. W. Timmer and J. V. da Graça, 17:pp. 227-228. International Organization of Citrus Virologists, Riverside, CA, USA.
Önelge, N., Satar, S., Elibüyük, Ö, Bozan, O. and Kamberoğlu, M. 2011. Transmission studies on Citrus yellow vein clearing virus. International Organization of Citrus Virologists Conference Proceedings (1957-2010). 18:pp. 11-14. International Organization of Citrus Virologists, Riverside, CA, USA.
Pallás, V., Sánchez-Navarro, J. A. and James, D. 2018. Recent advances on the multiplex molecular detection of plant viruses and viroids. Front. Microbiol. 9:2087.


Park, C. Y., Park, J., Kim, H., Yi, S.-I. and Moon, J. S. 2019. First report of citrus leaf blotch virus in Satsuma mandarin in Korea. J. Plant Pathol. 101:1229.


Pecman, A., Kutnjak, D., Gutiérrez-Aguirre, I., Adams, I., Fox, A., Boonham, N. and Ravnikar, M. 2017. Next generation sequencing for detection and discovery of plant viruses and viroids: comparison of two approaches. Front. Microbiol. 8:1998.



Roossinck, M. J., Martin, D. P. and Roumagnac, P. 2015. Plant virus metagenomics: advances in virus discovery. Phytopathology 105:716-727.


Roy, A., Fayad, A., Barthe, G. and Brlansky, R. H. 2005. A multiplex polymerase chain reaction method for reliable, sensitive and simultaneous detection of multiple viruses in citrus trees. J. Virol. Methods 129:47-55.


Song, W. 2023. Climate change and tourism sustainability in Jeju Island landscape. Sustainability 15:88.

Umer, M., Liu, J., You, H., Xu, C., Dong, K., Luo, N., Kong, L., Li, X., Hong, N., Wang, G., Fan, X., Kotta-Loizou, I. and Xu, W. 2019. Genomic, morphological and biological traits of the viruses infecting major fruit trees. Viruses 11:515.



Vives, M. C., Velázquez, K., Pina, J. A., Moreno, P., Guerri, J. and Navarro, L. 2013. Identification of a new enamovirus associated with citrus vein enation disease by deep sequencing of small RNAs. Phytopathology 103:1077-1086.


Yang, H.-J., Oh, J., Lee, H.-K., Lee, D.-S., Kim, S.-Y., Kim, M.-H., Park, C.-Y., Kim, H., Lee, S.-I., Jeong, R.-D., Moon, J.-S. and Lee, S.-H. 2019. First report of citrus vein enation virus in Satsuma mandarin (Citrus unshiu) in Korea. Plant Dis. 103:2703.

Zhang, Y. H., Liu, C. H., Wang, Q., Wang, Y. L., Zhou, C. Y. and Zhou, Y. 2019a. Identification of Dialeurodes citri as a vector of Citrus yellow vein clearing virus in China. Plant Dis. 103:65-68.


Zhang, Y., Liu, Y., Wang, Y., Wang, Q., He, S., Li, X. and Zhou, Y. 2019b. Transmissibility of Citrus yellow vein clearing virus by contaminated tools. J. Plant Pathol. 101:169-171.


- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 687 View
- 61 Download
- ORCID iDs
-
Mikyeong Kim

https://orcid.org/0000-0003-3154-8178 - Related articles
-
Specific PCR Detection of Four Quarantine Fusarium Species in Korea2010 December;26(4)
Detection of Allexiviruses in the Garlic Plants in Korea2007 December;23(4)
Occurrence and Distribution of Viruses Infecting Pepper in Korea2005 September;21(3)
Occurrence and Detection of Rice black-streaked dwarf virus in Korea2005 June;21(2)



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print



