 |
 |
| Plant Pathol J > Volume 35(6); 2019 > Article |
Abstract
The genetic variability of ŌĆśCandidatus Liberibacter asiaticusŌĆÖ (CLas) population associated with huanglongbing (HLB) disease of citrus in North Eastern (NE) region of India, a geographically locked region, and home for the diversity of many citrus species was analyzed on the basis of tandem repeat numbers (TRN) in variable CLIBASIA_01645 genomic loci. Fifty-five CLas strains sampled from different groves of NE Hill (NEH) region of India were in single amplicon group, but there was remarkable genetic variability in TRNs. The TRN in HLB-associated CLas strains varied from 0-21 and two novel repeat motifs were also identified. Among the NE population of CLas, TRN5 and TRN9 were most frequent (total frequency of 36.36%) followed by TRN4 (14.55%) and TRN6, TNR7 with a frequency of 12.73% each. Class II type CLas genotypes (5 < TRN Ōēż 10) had highest prevalence (frequency of 60.00%) in the samples characterized in present study. Class I (TRN Ōēż 5) genotypes were second highest prevalent (29.09%) in the NEH region. Further analysis of genetic diversity parameters using NeiŌĆÖs measure (H value) indicated wide genetic diversity in the CLas strains of NE India (H value of 0.58-0.86). Manipur CLas strains had highest genetic variability (0.86) as compared to Eastern, Southern and Central India. The R10 values (TRN Ōēż 10/TRN > 10) of NE CLas population was 10.43 (73/7), higher from other regions of India. Present study conclusively reported the occurrence of high genetic variability in TRN of CLas population in North East Indian citrus groves which have evolved to adapt to the specific ecological niche.
Greening (ŌĆ£huanglongbingŌĆØ, HLB) is most destructive disease of citrus causing major economic losses to citrus industry worldwide including India (Ahlawat, 1997; Bov├®, 2006; Ghosh et al., 2015). From the citrus growing areas of India, the disease was confirmed by transmitting it through Asian citrus psyllid during 1967 (Capoor, 1963; Capoor et al., 1967). Association of ŌĆśCandidatus Liberibacter asiaticusŌĆÖ (CLas), a phloem limited ╬▒-proteobacteria was identified from symptomatic citrus tree in India (Capoor, 1963; Capoor et al., 1967; Jagoueix et al., 1994). Since then disease has been reported from almost all citrus growing regions of world including Taiwan, Japan, United States, South Africa, North and Central American countries etc. (Brown et al., 2011; Coletta-Filho et al., 2004; Halbert, 2005; Manjunath et al., 2010; Mart├Łnez et al., 2009; Matos et al., 2009; Miyakawa and Tsuno, 1989; Oberholzer et al., 1965) and has emerged as major bottleneck to citrus cultivation.
A phloem-limited, gram-negative fastidious, non-culturable bacteria belonging to the genus ŌĆśCandidatus LiberibacterŌĆÖ is associated with the HLB disease of citrus. So far three species of this fastidious bacteria, CLas found in Asian region, Brazil (Sao Paulo) and United States (Florida) (Bove, 2006); ŌĆśCa. Liberibacter africanusŌĆÖ (CLaf) found in African countries (Jagoueix et al., 1994); and ŌĆśCa. Liberibacter americanusŌĆÖ (CLam) found in Brazil and South America (Texeira et al., 2005) are reported. CLas and CLam are vectored by the Asian citrus psyllid (Diaphorina citri) and CLaf by the two-spotted citrus psyllid (Trioza erytreae) (Bov├®, 2006). The CLaf is heat sensitive and does not cause symptoms at temperatures greater than 25-30┬░C. The CLas primarily distributed in Asia, is heat tolerant and able to cause symptoms at temperatures greater than 35┬░C and considered the most destructive one. The CLam appears heat sensitive similar to that of the CLaf (Bov├®, 2006). CLas bacterium has been found most widespread and devastating due to its wide host range and is able to infect most rutaceae species and some solanaceae species (Halbert and Manjunath, 2004).
Characterization of bacterial strains or genotypes is essentially required for understanding the epidemiological patterns and for devising durable management strategies. Conventionally CLas strains have been characterized based on the most conserved genomic locus (16S rDNA). Due to low genetic variation in the 16S rDNA genomic region, it is not ideal target for adequate genetic resolution for categorization of strains or genotypes (Chen et al., 2010; Deng et al., 2008; Jagoueix et al., 1994). Other genomic regions like beta operon, ribosomal protein (rp) and outer membrane protein (omp) genes were alternatively used for characterizations of CLas strains but have limited resolution potential in genetic differentiation of CLas strains (Bastianel et al., 2005; Deng et al., 2008; Hocquellet et al., 1999). Single-nucleotide polymorphism based variations in the omp locus suggested the grouping of CLas population from China-Guangdong with those from Thailand and Nepal, whereas the strains from Philippines and China-Behai were in separate group (Deng et al., 2008). Genetic variations in the alternative loci like bacteriophage-type DNA polymerase revealed three distinct clusters of the CLas population from Southeast Asia (Tomimura et al., 2009). The clustering pattern was not in congruence with the geographical origin of the strains except for those from Indonesia.
Limited genetic variability in the most commonly characterized genetic loci of CLas has further demanded for targeting the variable and hypervariable loci with adequate genetic resolution. Simple sequence repeat or microsatellite markers with variable number of tandem repeats (VNTR) have been widely employed as robust marker in differentiating the bacterial species (Katoh et al., 2011; Lin et al., 2005; Ma et al., 2014). CLIBASIA_ 01645 loci in the CLas genome was identified and used as a sensitive indicator in differentiation of strains (Chen et al., 2010). This region putatively encodes for bacteriophage repressor protein (Duan et al., 2009), which regulates the phage/prophage activity. The variations in this genomic region could affect environmental adaptations and pathogenicity (Liu et al., 2011). Tandem repeat number (TRN) variations in the bacterial population are generated through DNA strand slippage (Bichara et al., 2006; Verstrepen et al., 2005) and have role in pathogenic or environmental fitness (Boles et al., 2004). TRN profiling has recently been used for fine genetic resolution of CLas population (Chen et al., 2010; Ghosh et al., 2015; Katoh et al., 2011; Ma et al., 2014).
Analysis of genetic diversity is important to understand the ecology and epidemiology of CLas bacterial population. The hilly terrains of North Eastern region of India, known as North Eastern Hill (NEH) region, a rich treasure of citrus diversity has not been surveyed for characterization of CLas strains. Large number of citrus species is grown in this region in wild, semi-wild and cultivated conditions. Unlike other parts of India, where citrus is generally grown as commercial crop and being propagated by budded plants, the citrus planted in North Eastern (NE) region is predominantly of seedling origin. Analysis of genetic diversity is important to understand the ecology and epidemiology of this otherwise genetically homogenous population of CLas bacteria. Hence systematic sampling of HLB infected citrus from this region was undertaken in the present study to unravel the genetic diversity in CLas population prevalent in the North East region of India based on the TRN in the CLIBASIA_01645 locus. The overall genetic profile of CLas population in India has been presented.
Samples were collected during the period June 2017 to July 2018 from the major citrus groves of NEH region of India, covering the states of Manipur (34 samples), Tripura (6 samples), Nagaland (6 samples), Meghalaya (5 samples), and Arunachal Pradesh (4 samples). Four to five twigs along with leaves were collected from the symptomatic citrus trees (exhibiting the symptoms of HLB) and one tree each from a citrus grove was considered as a strain. As in NEH region of India, the citrus is usually grown on un-terraced hill slopes, an aggregate of citrus plants in an isolated pocket was considered equivalent to one orchard/strain in the present study. The CLas strains were sampled from acid lime (Citrus aurantifolia), mandarin (Citrus reticulata), Assam lemon (Citrus limon), Kachai lemon (Citrus jambhiri), sweet orange (Citrus sinensis), pomelo (Citrus grandis), and Citrus macroptera grown in the surveyed groves (Table 1). Collected samples were brought to laboratory, mid ribs and petioles were excised and stored under deep freezing conditions (ŌłÆ80┬░C) till further analysis.
The midribs and petioles of the collected citrus leaves were used for total DNA extraction using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following manufacturesŌĆÖ protocol with 200 mg of the tissues. Initial testing for the CLas presence was done using primer pair CGB-F (5ŌĆ▓-TGGGTGGTTTACCATTCAGTG-3ŌĆ▓) and CGB-R (5ŌĆ▓-CAATGGGTTGCGAAGTCGCG-3ŌĆ▓) (Datar et al., 2014; Meena and Baranwal, 2016) targeting partial 16S rDNA gene of bacterial genome yielding an amplicon of ~450 bp.
The CLas positive samples were further used for the amplification of CLIBASIA_01645 locus employing the previously reported primer set, LapGP-1f (5ŌĆ▓-GACATTTCAACGGTATCGAC-3ŌĆ▓) and LapGP-1r (5ŌĆ▓-GCGACATAATCTCACTCCTT-3ŌĆ▓) (Chen et al., 2010). The PCR reaction was carried out in SimpliAmp Thermal Cycler (Applied Biosystems, Carlsbad, CA, USA) in 25 ╬╝l mixture containing 100 ng of template DNA, 2.5 ╬╝l of 10├Ś buffer, 1 ╬╝l of MgCl2 (25 mM), 1 ╬╝l of dNTP (10 mM), 1 ╬╝l each of primers (10 mM), 0.5 ╬╝l of Taq DNA polymerase (5 U/╬╝l) and nuclease free water. The thermal cycling conditions were: one cycle of 95┬░C for 4 min, followed by 35 cycles at 95┬░C for 30 s, 58┬░C for 45 s, 72┬░C for 1 min and final extension at 72┬░C for 10 min. The amplicons were electrophoresed in 1.5% agarose gel and visualized using UV light. Out of the 25 ╬╝l amplified product, 5 ╬╝l was used for gel electrophoresis based analysis and remaining 20 ╬╝l for purification of DNA using QIAquick PCR Purification Kit (Qiagen). The purified single amplicon DNAs were then sequenced directly bi-directionally (Eurofins Genomics India Pvt. Ltd., Bengaluru, India) using ABI BigDye terminator sequencing method. Each amplicon was sequenced twice and the resulting sequences of each amplicon which shared >99.6% identity were considered for further analysis.
The putative identification of the sequenced fragments was initially done using nucleotide BLAST programme of NCBI (https://blast.ncbi.nlm.nih.gov/). The VNTR, referred to as TRN in the CLIBASIA_ 01645 was calculated using Tandem Repeats Finder software (http://tandem.bu.edu) (Benson, 1999). The tandem repeats obtained in the software were further verified manually. Frequency of different TRN containing CLas genotypes was calculated and differences in the genetic variations were analyzed statistically using F test employing analysis of variance test (Gomez and Gomez, 1984), where if the computed F-value was greater than tabular F value at 5% level of significance, the differences in the TRN of CLas associated with specific citrus host cultivar was considered significant. In case if the computed F-value was lower than or equal to the tabular F-value at the 5% level of significance, the differences in the TRN of CLas associated with specific citrus host cultivar was considered non-significant. Based on the genetic variations in the CLI-BASIA_ 01645 locus, the CLas strains were divided into different classes as defined by Ghosh et al. (2015) with little modification as: class I, TRN Ōēż 5; class II, 5 < TRN Ōēż 10; class III, 10 < TRN Ōēż 15; class IV, 15 < TRN Ōēż 20; and class V, TRN > 20. The population genetic diversity among different populations of CLas were calculated using NeiŌĆÖs H value as H = 1 ŌłÆ Ōłæpi2, where ŌĆśpiŌĆÖ referred to as frequency of the allele ŌĆśiŌĆÖ at the locus (Nei, 1973). In order to describe the CLas population structure based on TRN10 threshold, the R10 values were calculated as ratio of TRN Ōēż 10 and TRN > 10 genotypes which were stabilized empirically (Chen et al., 2010; Ma et al., 2014).
PCR amplification using CLIBA-SIA_01645 specific primers LapGP-1f/LapGP-1r generated a single amplicon of ~650 bp in most of the 55 samples except for a few cases where a larger amplicons up to 700 bp were observed (data not shown). Sequencing of the amplified region and sequence analysis further indicated the variation in the targeted CLIBASIA_01645 loci of respective CLas strains. Analysis of 55 CLas strains characterized in the present study indicated that TRNs at this locus varied from 0 to 21 (Table 1). Interestingly, CLas strains sampled from acid lime in Manipur showed large genetic variations in the CLIBASIA_01645 locus ranging from 0 TRN (strain MnH-94) to maximum TRN of 21 (strain MnH-19). All the CLas strains had tandem repeats of ŌĆśAGACACAŌĆÖ sequences except for the strains CLas-NgH-7 (sampled from Assam lemon in Nagaland) and CLas-MnH-31 (sampled from acid lime in Manipur) which had ŌĆśTTGTTATCCTGTCGATACCGTTGAAATGTCŌĆÖ and ŌĆśACACAAGŌĆÖ as repeat sequence in the CLIBASIA_01645 locus.
Among the NEH population of CLas, TRN5 and TRN9 were the most frequent (total frequency of 36.36%) followed by TRN4 (14.55%) and TRN6, TRN7 with a frequency of 12.73% each (Table 2). Interestingly, CLas strains with TRN9 were found in all the NEH states sampled in the present study. Class II type CLas genotypes (5 < TRN Ōēż 10) had highest prevalence (frequency of 60.00%) in the samples characterized in the present study. Class I genotypes was second highest prevalent (29.09%) in the NEH region.
A demarcation of CLas genotypes of NEH region based on high altitude (> 500 meters above mean sea level) and low altitude (< 500 m above mean sea level) indicated that TRN5 genotypes were more prevalent in high altitude citrus groves (20.51% frequency) followed by TRN4 and TRN7 (each with a frequency of 17.94%). In contrast, in the low altitude citrus groves, CLas genotypes of TRN6 were most prevalent (42.85% frequencies) followed by TRN3 and TRN2 at a frequency of 21.42% and 14.28%, respectively (Table 2). This indicated the altitude based aggregation of CLas genotypes and their preferential accumulation in the citrus groves located at different altitudes.
The CLas population previously characterized from the other parts of India (Ghosh et al., 2015) were compared with the strains characterized in the present study in order to have overall picture of its genetic structure. In the NE states of India, where citrus is largely cultivated through seedling propagation but budded plants are less commonly planted, the CLas genotypes of TRN7 were more predominant (23.75% frequency) followed by TRN5 genotypes (15.00% frequency) (Table 2), which indicated that overall infection of class II genotypes of CLas were widely prevalent in the citrus groves of NE India. Whereas, in the other parts of India, CLas genotypes of TRN5 were most prevalent (28.57% frequency) followed by TRN6 and TRN9 (12.50% frequency for each).
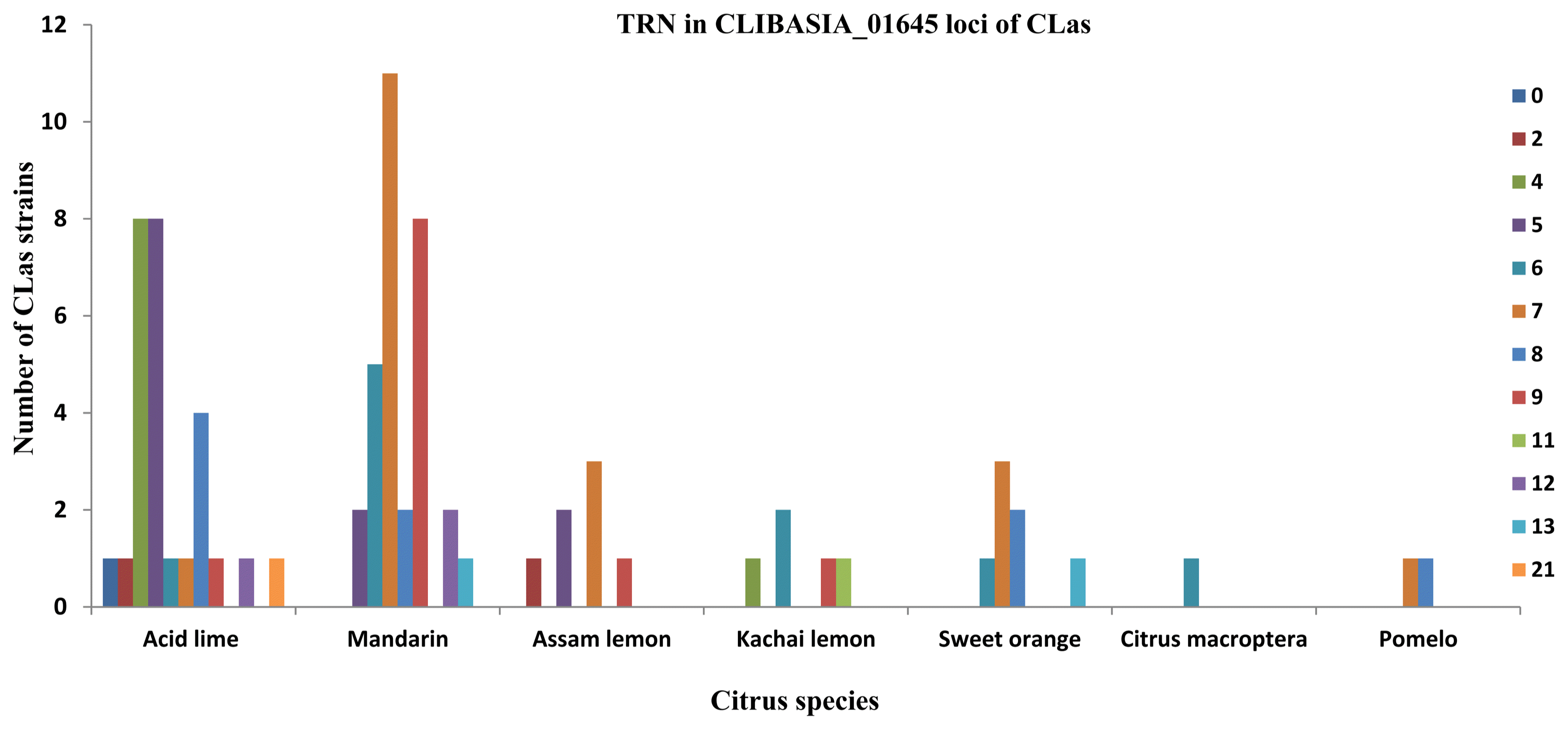
In the NE region, the mandarin was preferentially infected by CLas with TRN7 followed by TRN9 (Fig. 1), whereas in other part of India, they were infected predominantly by TRN5 followed by TRN9 genotypes. Acid lime trees in NE region were predominantly infected with TRN4 and TRN5 CLas (Fig. 1), whereas in rest parts of India, they were infected predominantly by TRN5 CLas genotypes. Assam lemons showed highest infection of CLas with TRN7 in the NE region. A strain of rough lemon (Citrus jambhiri) known as Kachai lemon, cultivated in selected pocket of Manipur state showed remarkable diversity of CLas genotypes (TRN4, TRN6, TRN9, and TRN11) although the trees were sampled from a small isolated groves of 4 km radius, the preferential accumulation of TRN6 CLas was detected (Fig. 1).
TRN typing of CLas strains in the present study along with the previously reported Indian CLas strains from different citrus growing regions was further analyzed for the genetic diversity parameters using NeiŌĆÖs measures (H values) (Table 3). Among the CLas strains sampled in the present study from NEH states, Manipur CLas strains had maximum H-values (0.86), followed by Nagaland (0.72), Meghalaya (0.72), and Mizoram (0.67) (Table 3), which indicates high genetic diversity in that order. The strains sampled from Tripura, Arunachal Pradesh and Sikkim has H values in a homogenous range (0.61-0.62), indicating similar patterns of genetic diversity of CLas in these states. The Assam CLas population showed lowest H-values among the NE states (0.58). The ratio of TRN Ōēż 10 and TRN > 10 genotypes (R10) for NE strains was highest (10.43) while for that of North, South and West Indian CLas strains were 5.00, 4.00, and 1.00, respectively (Table 3).
Variations in the TRN in CLIBASIA_01645 genomic loci of CLas have proven to be an important indicator of its genetic diversity and epidemiological studies. TRN in genomic loci like CLIBASIA_01645 is more precise in genetic differentiation of CLas isolates in comparison to single nucleotide polymorphisms (Katoh et al., 2011). The amplification of CLIBASIA_01645 loci of CLas strains from NEH region in present study revealed that all the strains belonged to single amplicon group (SAG) exhibiting identical electrophoretic profile as reported previously (Chen et al., 2010; Ma et al., 2014). Association of specific TRN class of CLas genotype with specific citrus cultivar has been observed e.g. mandarins (Khasi mandarin) were predominantly infected by TRN7 and TRN9 genotypes. In previous study preferential accumulation of TRN5 and TRN9 was reported form mandarins (Ghosh et al., 2015). Sweet oranges in NE region were more frequently infected with CLas of TRN7, whereas preferential accumulation of TRN6 was detected earlier from sweet oranges grown in other parts of India (Ghosh et al., 2015). Acid limes from NE region showed large diversity of CLas genotypes as TRN0 to TRN21 were detected in acid lime (classes I, II, III, and V) (Fig. 1), which to the best of our knowledge is the widest range of diversity for any single citrus species reported worldwide. Although the TRN specifically varied with the citrus cultivar sampled in the present study, the overall class (i.e., class II) remained same. The differences in the TRN of CLas associated with specific citrus host cultivar were non-significant (P > 0.05), as also indicated in the previous studies (Chen et al., 2010; Ghosh et al., 2015). Current study indicated that inter-population TRN distribution in the different citrus groves of India is different. The CLas strains from North East India belonged to SAG as like that of Guangdong and Florida (Chen et al., 2010) unlike that of existence of both SAG and multiple amplicon group reported from Southern China (Ma et al., 2014).
The uniqueness of the genetic profiles of CLas strains from NE region along with the high variability indicated that selection pressure and other evolutionary forces driving the evolution of CLas in this region are quite distinct compared to other parts of the India. Highest frequency of TRN7 and TRN5 CLas genotypes in entire NE region and TRN6 and TRN7 based on the earlier sampling (Ghosh et al., 2015), indicated that HLB-associated CLas population in NE India predominantly belonged to Class II (with TRNs of 7, 5, 6 and 9). CLas with TRN5 were the most commonly associated with the citrus grown in other parts of the country followed by TRN6 and TRN9. Class II (5 < TRN Ōēż 10) was found most common and widely distributed all over the India. However, TRN7 harbouring CLas strains were specifically aggregated in the citrus growing region of NE India. The class V (TRN21) CLas genotype was detected for the first time from citrus groves in India. Two novel TRN loci with new repeat motifs were also detected from the Assam lemon grown in Nagaland (NgH-7) and acid lime grown in Manipur (MnH-31). To the best of our knowledge, these are two novel repeat motifs in CLIBASIA_01645 locus of CLas genome, which might have role in distinct pathogenicity and ecological adaptation. The range of variation in TRN among CLas strains clearly demonstrates that the population of these otherwise genetically homogeneous bacteria is very diverse in India (as evident from the TRN0-TRN21 detected in the present study). A strain of rough lemon (Citrus jambhiri) grown in an isolated remote grove of Manipur for decades together (100% seedling origin) harboured CLas genotypes with TRN6, TRN9, and TRN11 (class II and class III).
CLas strain with TRN in the class IV has not been detected in the present study but it was reported in sweet orange earlier (Ghosh et al., 2015). The diversity of CLas genotypes indicated diverse evolutionary forces (ecological niche, environmental conditions, psyllid vector prevalence and host diversity niche), which has led to the evolution of diverse genotypes and unique environmental adaptation as evident from the diverse TRN in the CLIBASIA_01645 genomic region.
NeiŌĆÖs H value has been widely used as a genetic parameter for assessing the variability in genomic loci of bacterial population (Katoh et al., 2011; Ma et al., 2014). CLas strains from Eastern and Southern India had H-values in similar range (0.75-0.77), whereas that of North India was the lowest, indicating lowest genetic diversity of the CLas population in this region. Strains from Western India (Northern part of Maharashtra region) showed the high H-values (0.86). The range of variations in H-values was highest among NE states (0.58-0.86) as compared to other regions of India. The variations in the H-values among the characterized citrus groves were significant (P = 0.02). This could be possible because of the high variability of citrus species and prevalence of psyllid vectors along with the availability of alternate hosts in this region. Studies from other citrus growing parts of the world also reported the similar extent of genetic variation (H-value in the range of 0.83-0.88) for CLas population in the areas harbouring high genetic diversity in citrus (Central Japan, Southern Japan, Southern China) (Katoh et al., 2011; Ma et al., 2014).
As reported earlier by Ma et al. (2014), although NeiŌĆÖs H value represents population diversity of CLas, it does not necessarily reflect orderly distribution of corresponding TRN genotypes. Hence R10 values are used to describe the population structure and genotype aggregation based on TRN in CLIBASIA_01645 locus. Analysis based on R10 values (ratio of TRN Ōēż 10/TRN > 10) indicated that the NE CLas population had R10 values (10.48), significantly higher from other regions of India. The higher R10 values of NE population of CLas represent high TRN genotype aggregation. Earlier Ghosh et al. (2015) also indicated that there is possible aggregation in TRN genotype of CLas population in the unique niche of NE India as evident from comparatively high R10 values. The North East India, a geographically locked region with the high genetic diversity of citrus and its relatives, provides suitable niche and environment for evolution of CLas genotypes as reflected by the variations in TRN in CLIBASIA_01645 genomic loci. CLIBASIA_01645 genomic loci putatively encodes for a bacteriophage repressor, a protein which regulates the activity of phage/prophage (Duan et al., 2009). Variations in TRN in this locus were earlier reported to affect the gene translation thereby leading to truncated proteins, which potentially affect the pathogenicity (Chen et al., 2010). In many culturable bacterial species, TRN variations are predicted to have role in ecological adaptations (Zhou et al., 2014). These variations also lead to enhanced survival rate of bacterial populations (Moxon et al., 1994) particularly in case of pathogens like Haemophilus influenza and H. pylori (Razin et al., 1998). Through binary switching in transcription and translation, these changes substantially affect the bacterial adaptation under stress conditions (Zhou et al., 2014). The high genetic variation observed in the NE population of CLas as reflected by the TRN typing, suggests that these variation could lead to evolution of different regulatory proteins encoded by these genetic loci and affecting the pathogenic behaviour or citrus-CLas interactions (Ma et al., 2014; Zhang et al., 2011). Changes in the TRN are expected to alter the expressed proteins, their stability and enzyme activity (Lindstedt, 2005; van Belkum et al., 1998) thus putatively affect the biological functions (Chen et al., 2010). The sampling done in the present study was both from naturally grown (wild and semi-wild) citrus plantations and orchards, all of them were of seedling origin and hence it can be concluded that CLas population characterized here was predominantly native to the NE region and evolved here.
In summary, it can be concluded that CLas population infecting different citrus cultivars in NE India are genetically diverse and evolved differentially as compared to other citrus growing regions of India where citrus plants are propagated through budwood. The wide variations in the TRN in the variable CLIBASIA_01645 locus has implication in terms of epidemiology and pathogenicity of the bacterial populations. Indian CLas population was largely dominated by Class II TRN-type CLas population. We also hypothesize that CLas population of NE region is native and evolved over longer period of time.
Acknowledgements
Authors are grateful to the Department of Biotechnology (DBT), Govt. of India for financial support under DBT-twinning project Agri 2015/57 (Grant No. BT/PR16723/NER/95/264/2015) and Director, ICAR Research Complex for NEH Region, Umiam, Meghalaya for providing necessary lab facilities.
Fig.┬Ā1
Graphical illustration of tandem repeat numbers (TRN) distribution in ŌĆśCandidatus Liberibacter asiaticusŌĆÖ (CLas) strains sampled from different citrus hosts in North Eastern (NE) India (combined data of present study for hilly terrains of NE India and previous study of Ghosh et al., 2015). Color bars represent number of repeat motifs (different TRN).
Table┬Ā1
Analysis of tandem repeat sequence of HLB-associated CLas strains from North East Hill region of India
| No. | Sample namea | Host plant | Tandem repeat | GenBank accession No. | No. of repeat motifs present | Class |
|---|---|---|---|---|---|---|
| 1 | MnH-1 | Acid Lime | AGACACA | MH781492 | 4 | I |
| 2 | MnH-3 | Acid Lime | AGACACA | MH781493 | 4 | I |
| 3 | MnH-7 | Acid Lime | AGACACA | MH781494 | 4 | I |
| 4 | MnH-15 | Acid Lime | AGACACA | MH781495 | 12 | III |
| 5 | MnH -19 | Acid Lime | AGACACA | MH781496 | 21 | V |
| 6 | MnH -21 | Acid Lime | AGACACA | MH781497 | 4 | I |
| 7 | MnH -22 | Acid Lime | AGACACA | MH781498 | 8 | II |
| 8 | MnH -23 | Acid Lime | AGACACA | MH781499 | 2 | I |
| 9 | MnH -24 | Acid Lime | AGACACA | MH781500 | 5 | I |
| 10 | MnH -25 | Acid Lime | AGACACA | MH781501 | 5 | I |
| 11 | MnH -26 | Acid Lime | AGACACA | MH781502 | 5 | I |
| 12 | MnH -27 | Acid Lime | AGACACA | MH781503 | 5 | I |
| 13 | MnH -28 | Acid Lime | AGACACA | MH781504 | 5 | I |
| 14 | MnH -30 | Acid Lime | AGACACA | MH781505 | 5 | I |
| 15 | MnH -31 | Acid Lime | ACACAAG | MH781506 | 9 | II |
| 16 | MnH -35 | Acid Lime | AGACACA | MH781507 | 8 | II |
| 17 | MnH -36 | Mandarin | AGACACA | MH781508 | 12 | III |
| 18 | MnH -55 | Kachai lemon | AGACACA | MH781509 | 6 | II |
| 19 | MnH -56 | Kachai lemon | AGACACA | MH781510 | 6 | II |
| 20 | MnH -57 | Kachai lemon | AGACACA | MH781511 | 9 | II |
| 21 | MnH -62 | Kachai lemon | AGACACA | MH781512 | 11 | III |
| 22 | MnH -63 | C. macroptera | AGACACA | MH781513 | 6 | II |
| 23 | MnH -65 | Mandarin | AGACACA | MH781514 | 7 | II |
| 24 | MnH -66 | Acid lime | AGACACA | MH781515 | 5 | II |
| 25 | MnH -86 | Assam lemon | AGACACA | MH781516 | 7 | II |
| 26 | MnH -88 | Assam lemon | AGACACA | MH781517 | 5 | II |
| 27 | MnH -91 | Mandarin | AGACACA | MH781518 | 9 | II |
| 28 | MnH -93 | Acid Lime | AGACACA | MH781519 | 4 | I |
| 29 | MnH -94 | Acid Lime | AGACACA | MH781520 | 0 | I |
| 30 | MnH -99 | Acid Lime | AGACACA | MH781521 | 4 | II |
| 31 | MnH -101 | Acid lime | AGACACA | MH781522 | 6 | II |
| 32 | MnH -105 | Acid Lime | AGACACA | MH781523 | 7 | II |
| 33 | MnH -106 | Assam lemon | AGACACA | MH781524 | 7 | II |
| 34 | MnH -108 | Pumello | AGACACA | MH781525 | 7 | II |
| 35 | NgH-1 | Assam lemon | AGACACA | MH781526 | 5 | II |
| 36 | NgH-7 | Assam Lemon | ACTGTTGTTATCCTGTCGATACCGTTGAAATGTC | MH781527 | 2 | I |
| 37 | NgH-9 | Assam lemon | AGACACA | MH781528 | 9 | II |
| 38 | NgH-10 | Acid lime | AGACACA | MH781529 | 8 | II |
| 39 | NgH-11 | Sweet orange | AGACACA | MH781530 | 8 | II |
| 40 | NgH-12 | Acid lime | AGACACA | MH781531 | 5 | II |
| 41 | TrH-1 | Acid Lime | AGACACA | MH781532 | 4 | I |
| 42 | TrH-2 | Mandarin | AGACACA | MH781533 | 9 | II |
| 43 | TrH-3 | Mandarin | AGACACA | MH781534 | 9 | II |
| 44 | TrH-4 | Mandarin | AGACACA | MH781535 | 9 | II |
| 45 | TrH-9 | Mandarin | AGACACA | MH781536 | 6 | II |
| 46 | TrH-10 | Mandarin | AGACACA | MH781537 | 6 | II |
| 47 | MgH-1 | Acid lime | AGACACA | MH781538 | 4 | I |
| 48 | MgH-2 | Mandarin | AGACACA | MH781539 | 6 | II |
| 49 | MgH-4 | Mandarin | AGACACA | MH781540 | 9 | II |
| 50 | MgH-5 | Mandarin | AGACACA | MH781541 | 7 | II |
| 51 | MgH-6 | Mandarin | AGACACA | MH781542 | 7 | II |
| 52 | AnH-5 | Acid lime | AGACACA | MH781543 | 8 | III |
| 53 | AnH-8 | Mandarin | AGACACA | MH781544 | 12 | III |
| 54 | AnH-12 | Mandarin | AGACACA | MH781545 | 9 | II |
| 55 | AnH-13 | Mandarin | AGACACA | MH781546 | 9 | II |
Table┬Ā2
Distribution of TRNs in different states of NEH region of India TRN
| State | 0 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 16 | 17 | 21 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Manipur | 1 | 1 | - | 6 | 9 | 4 | 5 | 2 | 3 | - | 1 | 2 | - | - | - | 1 | 35 |
| Nagaland | - | 1 | - | - | 1 | - | - | 2 | 1 | - | - | - | - | - | - | - | 5 |
| Tripura | - | - | - | 1 | - | 2 | - | - | 3 | - | - | - | - | - | - | - | 6 |
| Meghalaya | - | - | - | 1 | - | 1 | 2 | - | 1 | - | - | - | - | - | - | - | 5 |
| Arunachal Pradesh | - | - | - | - | - | - | - | 1 | 2 | - | - | 1 | - | - | - | - | 4 |
| Total (NEH) | 1 | 2 | - | 8 | 10 | 7 | 7 | 5 | 10 | - | 1 | 3 | - | - | - | 1 | 55 |
| Frequency in NEH (%) | 1.82 | 3.64 | - | 14.55 | 18.18 | 12.73 | 12.73 | 9.09 | 18.18 | - | 1.82 | 5.45 | - | - | - | 1.82 | - |
| Total NE statesa | 1 | 2 | - | 9 | 12 | 10 | 19 | 9 | 11 | - | 1 | 3 | 2 | - | - | 1 | 80 |
| Frequency in NE (%) | 1.25 | 2.50 | - | 11.25 | 15.00 | 12.50 | 23.75 | 11.25 | 13.75 | - | 1.25 | 3.75 | 2.50 | - | - | 1.25 | - |
| Rest parts of Indiab | - | 1 | 1 | 4 | 16 | 7 | 3 | 5 | 7 | 2 | 2 | 2 | 4 | 1 | 1 | - | 56 |
| Frequency rest part of India (%) | - | 1.79 | 1.79 | 7.14 | 28.57 | 12.50 | 5.36 | 8.93 | 12.50 | 3.57 | 3.57 | 3.57 | 7.14 | 1.79 | 1.79 | - | - |
a North Eastern (NE) states including those reported by Ghosh et al. (2015).
b Rest parts of India (Eastern, Western, Central, and Southern parts of India) as reported by Ghosh et al. (2015).
Table┬Ā3
Population structures of CLas in different states of North East India and other regions described by NeiŌĆÖs H value and R10
The tandem repeat numbers of isolates characterized in the present study and earlier study (Ghosh et al., 2015) were used for analysis of genetic parameters.
References
Ahlawat, Y.S. 1997. Viruses, greening, bacterium and viroids associated with citrus (Citrus species) decline in India. Indian J Agric Sci. 67:51-57.
Bastianel, C., Garnier-Semancik, M., Renaudin, J., Bov├®, J.M. and Eveillard, S. 2005. Diversity of ŌĆ£Candidatus Liberibacter asiaticus,ŌĆØ based on the omp gene sequence. Appl Environ Microbiol. 71:6473-6478.



Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580.



Bichara, M., Wagner, J. and Lambert, I.B. 2006. Mechanisms of tandem repeat instability in bacteria. Mutat Res. 598:144-163.


Boles, B.R., Thoendel, M. and Singh, P.K. 2004. Self-generated diversity produces ŌĆ£insurance effectsŌĆØ in biofilm communities. Proc Natl Acad Sci U S A. 101:16630-16635.



Bov├®, J.M. 2006. Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol. 88:7-37.
Brown, S.E., Oberheim, A.P., Barret, A. and McLaughlin, W.A. 2011. First report of ŌĆśCandidatus Liberibacter asiaticusŌĆÖ associated with huanglongbing in the weeds Cleome rutidosperma, Pisonia aculeata and Trichostigma octandrum in Jamaica. New Dis Rep. 24:25

Capoor, S.P. 1963. Decline of citrus tree in India. Bull Natl Inst Sci India. 24:48-64.
Capoor, S.P., Rao, D.G. and Viswanath, S.M. 1967. Diaphorina citri Kuway: a vector of the greening disease of citrus in India. Indian J Agric Sci. 37:572-576.
Chen, J., Deng, X., Sun, X., Jones, D., Irey, M. and Civerolo, E. 2010. Guangdong and Florida populations of ŌĆśCandidatus Liberibacter asiaticusŌĆÖ distinguished by a genomic locus with short tandem repeats. Phytopathology. 100:567-572.


Coletta-Filho, H.D., Targon, M.L.P.N., Takita, M.A., De Negri, J.D., Pompeu, J. Jr., Marchado, M.A., do Amaral, A.M. and Muller, G.W. 2004. First report of the causal agent of Huanglongbing (ŌĆ£Candidatus Liberibacter asiaticusŌĆØ) in Brazil. Plant Dis. 88:1382

Datar, V.V., Chavan, V., Verma, R., Mungekar, D., Gaikwad, P. and Tripathi, S. 2014. Survey and identification of citrus greening bacterium from multiple locations in Maharashtra. J Plant Dis Sci. 9:60-63.
Deng, X., Chen, J. and Li, H. 2008. Sequestering from host and characterization of sequence of a ribosomal RNA operon (rrn) from ŌĆ£Candidatus Liberibacter asiaticusŌĆØ. Mol Cell Probes. 22:338-340.


Duan, Y., Zhou, L., Hall, D.G., Li, W., Doddapaneni, H., Lin, H., Liu, L., Vahling, C.M., Gabriel, D.W., Williams, K.P., Dickerman, A., Sun, Y. and Gottwald, T. 2009. Complete genome sequence of citrus huanglongbing bacterium, ŌĆśCandidatus Liberibacter asiaticusŌĆÖ obtained through metagenomics. Mol Plant-Microbe Interact. 22:1011-1020.


Ghosh, D.K., Bhose, S., Motghare, M., Warghane, A., Mukherjee, K., Ghosh, D.K. Sr., Sharma, A.K., Ladaniya, M.S. and Gowda, S. 2015. Genetic diversity of the Indian populations of ŌĆśCandidatus Liberibacter asiaticusŌĆÖ based on the tandem repeat variability in a genomic locus. Phytopathology. 105:1043-1049.


Gomez, K.A. and Gomez, A.A. 1984. Statistical procedures for agricultural research. 2nd ed. John Wiley and Sons, New York, NY, USA. 680.
Halbert, S.E. 2005. In: The discovery of huanglongbing in Florida, In: Proceedings of the 2nd International Citrus Canker and Huanglongbing Research Workshop; eds. by In : T.R. Gottwald, W.N. Dixon, J.H. Graham and P. Berger, 1-3. Florida Citrus Mutual, Orlando FL, USA.
Halbert, S.E. and Manjunath, K.L. 2004. Asian citrus psyllids (Sternorrhyncha: Psyllidae) and greening disease of citrus: a literature review and assessment of risk in Florida. Fla Entomol. 87:330-353.

Hocquellet, A., Toorawa, P., Bov├®, J.M. and Garnier, M. 1999. Detection and identification of the two Candidatus Liberobacter species associated with citrus huanglongbing by PCR amplification of ribosomal protein genes of the ╬▓ operon. Mol Cell Probes. 13:373-379.


Jagoueix, S., Bove, J.M. and Garnier, M. 1994. The phloem-limited bacterium of greening disease of citrus is a member of the ╬▒ subdivision of the Proteobacteria. Int J Syst Bacteriol. 44:379-386.


Katoh, H., Subandiyah, S., Tomimura, K., Okuda, M., Su, H.-J. and Iwanami, T. 2011. Differentiation of ŌĆ£Candidatus Liberibacter asiaticusŌĆØ isolates by variable-number tandem-repeat analysis. Appl Environ Microbiol. 77:1910-1917.



Lin, H., Civerolo, E.L., Hu, R., Barros, S., Francis, M. and Walker, M.A. 2005. Multilocus simple sequence repeat markers for differentiating strains and evaluating genetic diversity of Xylella fastidiosa. Appl Environ Microbiol. 71:4888-4892.



Lindstedt, B.A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis. 26:2567-x2582.


Liu, R., Zhang, P., Pu, X., Xing, X., Chen, J. and Deng, X. 2011. Analysis of a prophage gene frequency revealed population variation of ŌĆśCandidatus Liberibacter asiaticusŌĆÖ from two citrus-growing provinces in China. Plant Dis. 95:431-435.


Ma, W., Liang, M., Guan, L., Xu, M., Wen, X., Deng, X. and Chen, J. 2014. Population structures of ŌĆśCandidatus Liberibacter asiaticusŌĆÖ in southern China. Phytopathology. 104:158-162.


Manjunath, K.L., Ramadugu, C., Majil, V.M., Williams, S., Irey, M. and Lee, R.F. 2010. First report of the citrus Huanglongbing associated bacterium ŌĆśCandidatus Liberibacter asiaticusŌĆÖ from sweet orange, Mexican lime, and Asian citrus psyllid in Belize. Plant Dis. 94:781

Mart├Łnez, Y., Llauger, R., Batista, L., Luis, M., Iglesia, A., Collazo, C., Pe├▒a, I., Cas├Łn, J.C., Cueto, J. and Tablada, L.M. 2009. First report of ŌĆśCandidatus Liberibacter asiaticusŌĆÖ associated with Huanglongbing in Cuba. Plant Pathol. 58:389

Matos, L., Hilf, M.E. and Camejo, J. 2009. First report of ŌĆśCandidatus Liberibacter asiaticusŌĆÖ associated with citrus huanglongbing in the Dominican Republic. Plant Dis. 93:668

Meena, R.P. and Baranwal, V.K. 2016. Development of multiplex polymerase chain reaction assay for simultaneous detection of clostero-, badna- and mandari-viruses along with huanglongbing bacterium in citrus trees. J Virol Methods. 235:58-64.


Miyakawa, T. and Tsuno, K. 1989 Occurrence of citrus greening disease in the southern islands of Japan. Ann Phytopathol Soc Jpn. 55:667-670 (In Japanese).

Moxon, E.R., Rainey, P.B., Nowak, M.A. and Lenski, R.E. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 4:24-33.


Nei, M. 1973. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A. 70:3321-3323.



Oberholzer, P.C.J., von Standen, D.F.A. and Basson, W.J. 1965. In: Greening disease of sweet orange in South Africa, In: Proceedings of the 3rd Conference of the International Organization of Citrus Virologists; eds. by In : W.C. Price, 213-219. University of Florida Press, Gainesville, FL, USA.
Razin, S., Yogev, D. and Naot, Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 62:1094-1156.



Texeira, D.C., Ayres, J., Kitajima, E.W., Danet, L., Jagoueix-Eveillard, S., Saillard, C. and Bov├®, J.M. 2005. First report of a huanglongbing like disease of citrus in S├Żo Paulo State, Brazil, and association of a new Liberibacter species, ŌĆśCandidatus Liberibacter americanusŌĆÖ, with the disease. Plant Dis. 89:107

Tomimura, K., Miyata, S.-I., Furuya, N., Kubota, K., Okuda, M., Subandiyah, S., Hung, T.-H., Su, H.-J. and Iwanami, T. 2009. Evaluation of genetic diversity among ŌĆśCandidatus Liberibacter asiaticusŌĆÖ isolates collected in Southeast Asia. Phytopathology. 99:1062-1069.


van Belkum, A., Scherer, S., van Alphen, L. and Verbrugh, H. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 62:275-293.



Verstrepen, K.J., Jansen, A., Lewitter, F. and Fink, G.R. 2005. Intragenic tandem repeats generate functional variability. Nat Genet. 37:986-990.



Zhang, S., Flores-Cruz, Z., Zhou, L., Kang, B.-H., Fleites, L.A., Gooch, M.D., Wulff, N.A., Davis, M.J., Duan, Y.-P. and Gabriel, D.W. 2011. ŌĆ£Ca. Liberibacter asiaticusŌĆÖ carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant infections. Mol Plant Microbe Interact. 24:458-468.


- TOOLS
-
METRICS

-
- 6 Crossref
- 0 Scopus
- 2,541 View
- 15 Download
- Related articles



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print



