Plant immunity is activated by several triggers, which may consist of not only biotic factors, such as beneficial microbes, arthropods, and pathogens (Boller and Felix, 2009; Engelberth et al., 2004; Koo et al., 2020; Pieterse et al., 2014; Yoo and Sang, 2017), but also abiotic stresses including chemical inducers and salinity (Beckers and Conrath, 2007). The local immune system can recognize these key stimuli and respond by inducing local and systemic immune responses. Certain substances, namely salicylate, azelaic acid, and jasmonate, are used for this systemic induction of plant immunity (Dempsey and Klessig, 2012).
The bacterium Pseudomonas fluorescens Q2-87, a well-known biocontrol agent, produces 2,4-diacetylphloroglucinol (DAPG) (Raaijmakers and Weller, 2001). DAPG is a phenolic compound exhibiting a wide spectrum of antibiotic activity against bacteria, fungi, and nematodes (Haas and Keel, 2003; Weller et al., 2007). Genetic analyses using different DAPG-producing or -nonproducing P. fluorescens strains have revealed that DAPG is indeed a key antibiotic agent which is effective against different soilborne pathogens (Keel et al., 1992). In the host plant, DAPG may either directly inhibit soil-borne pathogens or indirectly do so by activating induced systemic resistance (ISR) (Kwak and Weller, 2013). ISR is phenotypically and biochemically related to systemic acquired resistance (SAR) in plants, which is usually induced via infection with avirulent pathogens. SAR is mediated by the salicylic acid (SA)-signaling pathway in plants acquiring systemic resistance and is regulated by the redox-regulated protein NONEX-PRESSER OF PR GNENS 1 (NPR1) (Cao et al., 1997). It was recently shown that SAR contributes to the expression of PATHOGENESIS-RELATED (PR-1), PR-2, and PR-5, which are SA signal-responsive genes (Nie et al., 2017). By way of comparison, ISR is mediated by jasmonic acid (JA)/ethylene (ET)-signaling pathways, and NPR1 also plays important roles in how this defensive response is executed by plants (Pieterse et al., 1998). It is known that ISR can switch on JA- and ET-responsive genes’ expression, namely of ChiB and PDF1.2, LOX1, LOX2, PAL1, and PIN2, as well as VSP (van Wees et al., 1999).
DAPG-induced ISR has been shown to confer plant resistance to a variety of plant pathogens. In Arabidopsis, root-colonizing P. fluorescens CHA0 prevented sporulation of the oomycete Peronospora parasitica, whereas DAPG-deficient strains, such as CHA 89 (HCN, DAPG-, and pyoluteorin-deficient) and CHA631 (DAPG-deficient strain) did not protect the plant from the pathogen’s attack (Iavicoli et al., 2003). Importantly, the P. fluorescens- or DAPG-mediated ISR responses did not occur in the Arabidopsis mutants npr1-1 (non-expressing NPR1 protein), jar1-1 (JA-insensitive), and eir1-1 (ET-insensitive) (Iavicoli et al., 2003). Similarly, ISR mediated by DAPG was critical for enacting plant resistance via the JA/ET-signaling pathway against Pseudomonas syringae (Weller et al., 2012). The DAPG-producing P. fluorescens strain Q2-87 was capable of induced resistance against the pathogen P. syringae pv. tomato, which causes bacterial speck disease, and this protection occurred in transgenic Arabidopsis plants carrying the SA degrading gene (NahG) but not in Arabidopsis mutants npr1-1, jar1, and etr1 (Weller et al., 2012). Collectively, these findings indicate that DAPG-mediated ISR relies on the JA/ET-signaling pathway.
Nevertheless, using mutants that lack a specific hormonal signaling pathway can produce misleading results and a crucial misunderstanding of the role of specific hormones in the evolutionary development of disease resistance (Seif El-Yazal et al., 2015). Ethylene and auxin, for example, can govern a number of common processes in plants, including their primary root elongation, root hair formation, hook formation, epinasty of leaves, and leaf abscission (Stepanova et al., 2005). Additionally, the hormones underpinning ISR figure prominently in shaping the resistance of plants to pathogens. SA is involved in plants’ resistance to biotrophic pathogens, whereas JA/ET is involved in their resistance to necrotrophic pathogens/insects, respectively (Lievens et al., 2017). Therefore, studying the ISR system with only single gene knockout Arabidopsis mutant may provide only partial or indirect evidence for the mechanisms of plant defense.
In this study, DAPG-tolerance Arabidopsis lines were developed by transforming the activity of DAPG hydrolase, via the phlG gene. The phlG encodes an enzyme that catalyzes the functioning of acetyltransferase to convert antibiotic DAPG to its non-toxic monoacetylphloroglucinol (MAPG) form (Bottiglieri and Keel, 2006). This work further dissects the role and function of DAPG in ISR by using a transgenic plant that degrades DAPG.
Materials and Methods
Plant, bacterial and fungal materials
Arabidopsis thaliana Col-0 was used as the experimental model plant. The plants were cultivated in bed soil or media. Seed was sown in the bed soil (a mixture of cocopeat, peat moss, zeolite, and vermiculite), and each pot held three or four seeds. After 7 days of cultivation, when the plant had grown large enough to be identified, one plant was transferred into another pot to grow. For plant growth on media, seed sterilization was first performed with 1% NaOCl. Vernalization was achieved with 1 ml of distilled water, at 4°C, for 7 days under a dark condition. Murashige and Skoog (MS) agar media (2.1 g of MS salt, 30 g of sucrose, 0.7 g per 1 liter of phytoagar) was adjusted to pH 5.7-5.8 with 1 M of KOH and autoclaving. Arabidopsis was grown in a growth chamber (Seyoung Scientific, Bucheon, Korea) under the following conditions: 12-h/12-h, light/dark cycle, 22°C, 60% relative humidity and 1,390 lux (light intensity).
The bacterium Pseudomonas fluorescens Q2-87 was incubated on 1/5 TSA (tryptic soy broth [6 g/l] with agar [20 g/l]) at 27°C for 2 days. The challenging pathogen Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) was incubated on 1/5 TSA at 27°C for 2 days. A single colony was cultured in 5 ml of TSB liquid medium (tryptic soy broth, 30 g/l) at 27°C for 48 h, and then inoculated into 200 ml of TSB. The cultured cells were collected by centrifugation and re-suspended in 10-mM MgCl2, then 0.01% (v/v) of the Silwet L-77 (PhytotechLABS, Shawnee Mission, KS, USA) was added and cell density adjusted to 1 × 107 colony-forming unit (cfu)/ml.
The fungal pathogen Botrytis cinerea was cultivated on PDA (potato dextrose broth [24 g/l] with agar [20 g/l]) at 27°C for 7 days. Its mycelium and spores were collected in sterilized water with a cotton swab, which was filtered through three layers of sterilized cheesecloth to collect the individual spores. The spore concentration was adjusted to 1 × 105 cfu/ml by using a hemocytometer (SUPERIOR, Lauda, Germany).
DAPG sensitivity test of A. thaliana Col-0
HPLC-grade methanol was used as a solvent for DAPG (Santa Cruz Biotechnology, Dallas, TX, USA), which had a stock concentration of 100 mM. This stock was stored at -20°C under dark conditions. DAPG stock was added to MS media to obtain final concentrations of 20, 40, 60, 80, and 100 μM, and each respectively poured into a (90 mm × 15 mm) petri dish and dried on a clean bench. The vernalized seeds were sown at a density of 100-200 seeds per dish and cultivated in the growth chamber for 10 days. This experiment was conducted using three replications. Statistical analysis was performed by Tukey’s honestly significant difference (HSD) test (P < 0.05).
Detection and purification of phlG gene
The phlG cloning primers (Supplementary Table 1) were designed with P. fluorescens Q2-87 reference sequences currently registered in NCBI [Locus_tag: PflQ2_2239]. The primers were synthesized by Cosmo Genetech (Seoul, Korea). The PCR mixture was 50 μl, comprising 2 mM of dNTPs, 2× PCR Buffer of KOD FX Neo, 1 μg of template DNA, 1.0 U KOD FX Neo, and 0.2 μM of each primer. The PCR reaction was performed as touchdown PCR, whose cycling began with a denaturation and annealing temperature of 83°C for 5 cycles, then reducing the annealing temperature 1°C every 5 cycles to 73°C, followed by 10 more cycles run at 73°C. Each primer was designed to amplify an 882-bp product, and the PCR product identified by gel electrophoresis in 1% agarose. Gel purification was done using Expin Gel SV (GeneAll, Seoul, Korea). To ligate with a donor vector, the purified PCR product underwent an A-tailing reaction (20 μl) as follows; dATP (NEB, Ipswich, MA, USA), 10× Reaction Buffer, and 5 units of TOP DNA Polymerase (Bioneer, Daejeon, Korea); the mixture was treated at 70°C for 30 min. DNA quantification was measured on a NanoDrop 2000C (Thermo Fisher Scientific, Waltham, MA, USA) to confirm its concentration and quality, after which it was stored at -20°C.
Binary vector construction
The PCR8/GW/TOPO (Invitrogen, Waltham, MA, USA) plasmid served as the entry vector. The reaction mixture (2 μl of the A-tailed PCR product, 0.5 μl of salt solution, and 0.5 μl of the PCR8/GW/TOPO vector) was incubated at room temperature for 5 min. The plasmid was transformed into Escherichia coli DH5α using the heat shock protocol. The selection of transformed donor vector E. coli was performed in Luria-Bertani (LB) media containing the antibiotic spectinomycin (50 μg/ml). For plasmid isolation, using a single colony inoculation, cells were cultured in 5 ml of LB broth at 37°C for 16 h in a shaking incubator. The plasmid was extracted with the DokDo-Prep plasmid mini-prep kit (ELPIS Biotech, Daejeon, Korea). The gateway destination vector (pMDC43) containing a hygromycin resistance gene (hpt) was used for transgenic Arabidopsis selection; pMDC43 was introduced into transformed E. coli DB3.1. The selection was performed on LB media containing kanamycin (50 μg/ml). The expression clone was constructed through the LR reaction of the entry clone and destination vector. This reaction solution contained 1.5 μl of the destination vector mixed with 0.5 μl of PCR8 clone and 0.5 μl of LR Clonase (Invitrogen), and it was incubated at room temperature for 24 h. E. coli transformation and plasmid isolation proceeded as described above. The restriction enzymes EcoRI and EcoRV (NEB) were used to confirm the correct insert in the entry clone and the expression clone. The reaction volume (20 μl) comprised 2 μl of EcoRI, 2 μl of NEBuffer 2.1, 1 μg of the plasmid, and distilled water. After incubation at 37°C for 3 h, gel electrophoresis was used to examine the product’s size whose DNA sequencing was then done by Cosmo Genetech.
Agrobacterium tumefaciens-mediated transformation
Agrobacterium tumefaciens GV3101 was used for plant transformation, via the electroporation methods. The binary vector was added into 100 μl of GV3101 competent cells, and this mixture homogenized by gently stirring it with a pipette several times. The mixture was placed in a pre-chilled cuvette in which electroporation proceeded at a voltage (v) 2.5 kV/cm. The cuvette was removed and 1 ml of LB broth was immediately added; the solution was then transferred into a new Eppendorf-tube and incubated for 2 h in a 30°C-incubator. The cells were selected in LB media containing gentamycin (10 μg/ml), rifampicin (100 μg/ml), and kanamycin (50 μg/ml). The floral-dip method was used for the Arabidopsis transformation (Clough and Bent, 1998). The GV3101 cells were cultured in 500 ml of LB broth for 12 h and then harvested by centrifugation (4,000 rpm) for 20 min. The harvested cells were suspended using an infiltration buffer (5% sucrose, 0.01% Silwet), after which Arabidopsis flowers were dipped in Agrobacterium mixture for 5 min. Transformed plants were incubated for 2 days, at room temperature, under cover of the plastic pot, and grown in the chamber.
Selection of the phlG transgenic homozygous line (T3)
Arabidopsis T1 selection was performed in media containing DAPG and hygromycin. Arabidopsis was screened using a concentration of 80/100 μM of DAPG that completely inhibited germination of the wild type (Col-0) (Supplementary Fig. 1); hygromycin was used in a final concentration of 57 μM. On each plate 100-150 seeds were sown in MS agar medium. After their 15-day cultivation in a growth chamber, resistant plants were transferred to bed soil. Each selected T1 plant was grown in the chamber for 2 months and harvested to obtain T2 generation seeds. From these, seedlings were sown on MS agar medium containing hygromycin (57 μM) or DAPG (100 μM). Segregation analysis was performed for the selection of the homozygous line. For this, 100-200 seeds were sown per plate and cultivated in the growth chamber for 7 days. Resistant and non-resistant populations were counted, for which a segregation analysis was based on significant differences assessed by the chi-square test in SAS software (v9.4, SAS Institute Inc., Cary, NC, USA).
Phenotype observations
Phenotypes of A. thaliana Col-0 and phlG transgenic lines (D16, H2) were observed. MS medium (1.2% agar) was used for their initial growth on plates grown vertically for 15 days in a growth chamber, after which the root length and weight of each plant were measured. The former was obtained from photographs analyzed using the ImageJ program (64-bit java v1.8.0_112). Fresh weight and leaf area were determined from 3-week-old plants in bed soil. The area per leaf was measured in ImageJ software. The data were statistically analyzed by Tukey’s HSD test (P < 0.05), based on five biological replications.
Plant DNA extraction and inserted phlG identification
Genomic DNA from plant tissue was obtained using the Edward extraction protocol (Edwards et al., 1991). To identify the phlG transgenic line (T3) PCR was used according to the gDNA and cDNA conditions. PCR volume (50 μl) consisted of 2 mM of dNTPs, 2× the PCR buffer for KOD FX Neo, 1 μg of template DNA, KOD FX Neo (1.0 U/μl) and 0.2 μM of each primer (same as before). PCR cycling using gDNA started with a denaturation and annealing temperature of 83°C for 5 cycles, then the annealing temperature was reduced by 1°C every 5 cycles to 73°C (same as above). The PCR conditions using the cDNA sample was carried out at 95°C for 30 s, 30 cycles of the denaturation at 95°C for 30 s, annealing at 56°C for 30 s, followed by an extension at 72°C for 30 s, with the final extension done at 72°C for 10 min.
Plant RNA extraction and qRT-PCR
The A. thaliana Col-0 and phlG transgenic lines (D16, H2) were grown on MS medium for 10 days, snap frozen with liquid nitrogen and homogenized using a mortar and pestle. Their total RNA was extracted with the TRIzol reagent (Thermo Fisher Scientific), following the manufacturer’s protocol. After quantification, cDNA was synthesized with 2 μg of total RNA by using ReverTra Ace-α-® (Toyobo, Osaka, Japan). Reaction volumes (20 μl) were obtained by adding 4 μl of 5× RT buffer, 2 μl of dNTP Mixture (10 mM each), 1 μl of RNase inhibitor (10 U/μl), 1 μl of Oligo (dT) 20 (10 pmol/μl), and 1 μl of ReverTra Ace, volume-topped by RNase-free H2O. Incubation proceeded at 42°C for 20 min in a PCR machine and heat applied at 99°C for 5 min. All synthesized cDNA samples were stored at -20°C. For the quantitative real time polymerase chain reaction (qRT-PCR), the SYBR Green Realtime PCR Master Mix (Toyobo) was used; the sample reaction volume (20 μl) consisted of 1 μl of 50 dilutions of cDNA, 10 μl of SYBR Green, and 1 μl of phlG_RT forward primer, and 1 μl of phlG_RT reverse primer (Supplementary Table 1). The internal control for this experiment used ubc, sand house-keeping genes, for which the reaction volume was the same as above. Reverse transcription was performed by a thermocycler (Bio-Rad, Hercules, CA, USA) with these conditions: initial denaturation step at 95°C for 1 min, 44 cycles at 95°C for 15 s, 56°C for 15 s, for 72°C for 30 s, 95°C for 10 s, and 55°C for 5 s, followed by a dissociation curve analysis. This experiment had three replicates per treatment. Relative gene expression levels were determined by the averaged fold-change (Pfaffl, 2004), calculated by dividing the ΔΔCt of the selected plants (D16, H2) by the ΔΔCt of the wild-type Col-0. The phlG gene-relative expression was quantified for the internal control genes.
DAPG treatment and ISR analysis
Three-week-old plants were used to identify ISR activity by DAPG. Before a pathogen’s inoculation, the plants were treated with water, methanol (DAPG solvent), and various concentrations of DAPG (10, 100, or 200 μM). The DAPG stocks (10, 100, 200 mM) were dissolved in methanol to their final concentration, and then further diluted with sterile water (200 ml) so the same volume (i.e., 1,000-fold dilution, 200 μl) of methanol could be added to each solution individually. For the DAPG-priming treatment, DAPG was dissolved in water and 13 ml of it applied onto the plant root via a needleless syringe. The same procedure was applied but with methanol (0.1%), as the experimental control. After 2 days, two challenging pathogens (B. cinerea and Pst DC3000) were introduced. For B. cinerea, its spores (5 μl, 1 × 105 cfu/ml) were placed on A. thaliana leaves in three treatment groups: untreated control, DAPG non-priming plus B. cinerea inoculation, and DAPG priming plus Botrytis inoculation. For Pst DC3000, its suspension (2 ml, 1 × 107 cfu/ml) was sprayed onto the leaves, likewise in three treatment groups: untreated control plant, DAPG non-priming plus Pst DC3000 inoculation, and DAPG priming plus Pst DC3000 inoculation. All treated plants were incubated for 4 days, covered on a tray, at 22°C and 95% relative humidity under 12-h/12-h light/dark conditions in the chamber. The extent of B. cinerea infection was gauged by measuring leaf necrosis diameters (mm) in ImageJ. For Pst DC3000 infection, experimental Arabidopsis leaf samples were collected from 0 to 4 days, from 0.1-g leaf material (0.1 g) were sterilized by 70% ethanol and washed thrice with distilled water. Each sample was homogenized using a mortar and pestle with 10 mM MgCl2, and a suitable dilution then plated onto King’s medium B agar containing 100 μg/ml of cycloheximide and 50 μg/ml of rifampicin (Pieterse et al., 1998). These plates were incubated at 27°C for 2 days after formed colonies were counted for bacterial cell density (cfu/ml). This value was compared among treatment groups by Duncan’s new multiple range test (MRT) (P < 0.05). To confirm the differential disease symptoms in the wild-type, D16, and H2 plants, their harvested plant leaves were stained with trypan blue (Fernández-Bautista et al., 2016) by putting them directly onto plates containing this staining solution (10 ml of lactic acid, 10 ml of phenol, 10 ml of glycerol, 10 ml of distilled water, with 40 mg of trypan blue). After 1-h incubation, each solution was removed and refilled with 100% ethanol. The solution was removed after 24 h and refilled with a 60%-glycerol solution, and the staining then visualized.
Relative expression of SA- and JA/ET-related defense genes
To identify the difference in ISR-related genes’ expression between A. thaliana Col-0 and phlG transgenic lines (D16, H2), qRT-PCR was performed with known ISR marker genes (i.e., for SA/JA/ET signaling). 100 μM of DAPG was applied to roots for 2 days prior to the challenge from pathogens (2 ddt, day-post-DAPG-treatment), after which the pathogens were introduced and allowed to incubate for 4 days. RNA was extracted from Arabidopsis leaves and cDNA was synthesized from 2 μg of total RNA. Five genes in all were examined for the expression of JA/ET signaling (i.e., WRKY11, PDF1.2) and SA signaling (i.e., EDS5, PR1, PR2) (Journot-Catalino et al., 2006; Wang et al., 2010). The same reference genes (SAND, UBC) were used to quantify gene expression patterns (Czechowski et al., 2005). All primers’ information can be found in Supplementary Table 1.
Results
DAPG inhibits A. thaliana Col-0 seed germination and growth
DAPG’s phytotoxicity to A. thaliana Col-0 was verified and its concentration that inhibits seed germination also determined. For the untreated control, 100% of its seeds had germinated at 2 days. Germination, however, decreased markedly depending on the DAPG concentration (Supplementary Fig. 1B). At 80 or 100 μM DAPG concentrations, all seeds did not germinate until 10 days later. This delay was statistically distinguishable from the other treatments (control, 20 μM, 40 μM, 60 μM) (Supplementary Table 2). Therefore, the final DAPG concentration for the selection of phlG transgenic Arabidopsis was set at 80 and 100 μM for 10 days.
phlG-transgenic Arabidopsis shows tolerance against DAPG
The phlG gene was amplified using the phlGF and phlGR primers from the genomic DNA of P. fluorescens Q2-87 (Supplementary Table 1). Their PCR products were identified as an amplicon of 879-bp spanning the entire length of the phlG gene in P. fluorescens Q2-87 (Supplementary Fig. 2). The phlG fragment was successfully cloned into a donor vector (PCR8/GW/TOPO), and this clone validated by EcoRV and 365-bp and 3,331-bp restriction enzyme reactions (Supplementary Fig. 2). EcoRI confirmed the expression vector by the LR reaction for the entry vector and destination vector (pMDC43). The binary vector containing the phlG gene (Supplementary Fig. 1A) was transformed into A. thaliana Col-0 and selection of the phlG transgenic Arabidopsis lines (T1) were achieved using 80 or 100 μM of DAPG or 57 μM hygromycin in MS media (Supplementary Fig. 1C and D). From the DAPG and hygromycin media, ca. 4,000 T1 lines were screened, resulting in total of 26 and 7 lines selected from each, respectively ( Table 3). The T1 selected plants were labeled D1-D26 (DAPG) and H1-H7 (hygromycin) and grown to obtain the next generation. These T2 generation lines were tested by segregation analysis, which validated the presence of a single copy of the chromosome-flanking vector (Supplementary Table 4). Both DAPG and hygromycin selections yielded similar results. The D16 line has been described as DAPG-tolerant for 144 plants and DAPG-sensitive for 45 plants. By way of comparison, 178 plants flourished in the hygromycin medium, and 58 plants were bleached out as sensitive lines. The DAPG-tolerant plants in the H2 line had 132 progenies of which 50 were prone to DAPG. In the hygromycin media, 90 plants were selected from the H2 line of which 31 plants were not resistant to hygromycin. Thus phenotype segregation ratio was 3:1, which was confirmed statistically (chi-square test; P < 0.05) (Supplementary Table 4). This experiment was done three times, all producing similar results.
Detection of the phlG inserted gene in the D16 and H2 lines
Genomic DNA (gDNA) and complementary DNA (cDNA) from the selected phlG transgenic lines, D16 and H2 were prepared. Both no DNA (blank) and non-transgenic A. thaliana Col-0 DNA served as the negative controls. Genomic DNA of P. fluorescens Q2-87 was a positive control. The blank sample, and DNA of the wild-type A. thaliana Col-0 were not amplified in phlG, in either the gDNA or cDNA, by PCR. Samples of P. fluorescens Q2-87, D16, and H2 demonstrated amplification of the phlG gene in both their gDNA (879-bp) and cDNA (157-bp) (Supplementary Fig. 3A and B). However, in A. thaliana Col-0 Cq value of the phlG gene was undetectable. The D16 line showed that phlG expression was 256.3-fold (UBC) and 270.9-fold (SAND) greater when compared when A. thaliana Col-0. For H2 line, phlG was expressed at 74.8-fold (UBC) and 49.6-fold (SAND) greater levels (Supplementary Fig. 3C). These results confirmed the D16 line had higher phlG expression than did the H2 line, a difference that was significant according to Duncan’s new MRT (P < 0.05).
Phenotype of A. thaliana Col-0, D16 and H2
To assess whether the phenotypes of A. thaliana Col-0, D16, and H2 differed, their growth was observed between 15 and 21 days. At 15 days, the fresh weight of A. thaliana Col-0 was 29.5 mg, 31.0 mg for D16, and 30.4 mg for H2. For A. thaliana Col-0, its primary root’s length was 40.6 mm, 41.3 mm for D16, and 40.5 mm for H2 (Supplementary Fig. 4A, C and D). Fresh weight recorded at 21 days (rosette stage) was 111.8 mg for A. thaliana Col-0, 115.9 mg for D16, and 116.3 mg for H2, with corresponding leaf areas of 4.1 cm2, 3.8 cm2, and 4.2 cm2 (Supplementary Fig. 4B, E and F). There was no evidence of any distinction between plant phenotypes after repeated this experiment five times (Tukey HSD test, P < 0.05). These finding indicated that the D16 and H2 are sufficient for further ISR research.
DAPG-primed A. thaliana Col-0 is resistant against B. cinerea and Pst DC3000
For this experiment, a necrotrophic fungal pathogen (B. cinerea) and a biotrophic bacterial pathogen (Pst DC3000) were used to evaluate ISR development in DAPG-primed A. thaliana Col-0 plants. Upon DAPG-priming, the B. cinerea-inoculated plants with disease symptoms had lesion diameters that were 0 mm at 0 day post inoculation (dpi), 0.2 mm at 1 dpi, 1.1 mm at dpi, 2 mm at 3 dpi, and 2.5 mm at 4 dpi. By contrast, in non-primed plants, their necrosis lesion diameters were 0 mm at 0 dpi, 1.2 mm at 1 dpi, 2.2 mm at 2 dpi, 4.9 mm at dpi, and 7.9 mm at 4 dpi (Fig. 1). After DAPG-priming, the Pst DC3000-challenged plants could recognize bacterial cell densities (log cfu/0.1 g of leaves) of 0 at 0 dpi, 1.8 at 1 dpi, 3.4 at 2 dpi, 4.4 at 3 dpi, and 5.4 at 4 dpi. The Pst DC3000 density in DAPG non-primed plants were 0 at 0 dpi, 1.2 at 1 dpi, 2.2 at 2 dpi, 4.9 at 3 dpi, and 7.9 at 4 dpi, respectively (Fig. 2). Together, these findings indicated that DAPG-priming of A. thaliana increased its ISR activity against both fungal and bacterial pathogens.
phlG transgenic D16 and H2 failed DAPG mediated ISR
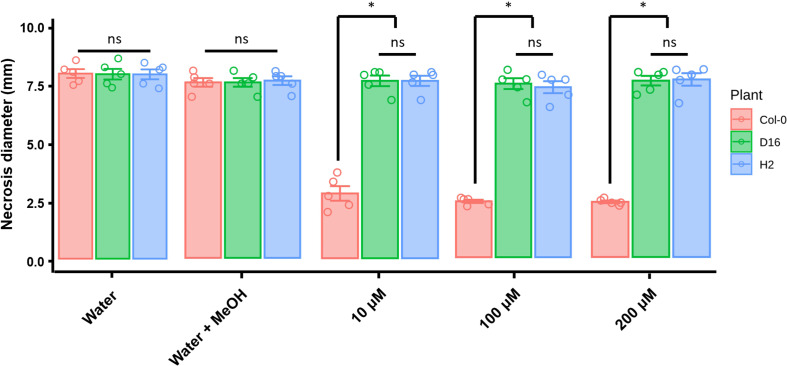
DAPG functioning for ISR was investigated in the A. thaliana Col-0, D16, and H2 plants by apply different concentrations of DAPG (10, 100, and 200 μM). In the absence of DAPG treatment, all transgenic and non-transgenic plants showed similar disease symptoms and lesion diameters (Fig. 3). The mock treatment (0.1% methanol, DAPG solvent) slightly enhanced disease resistance of both non-transgenic (Col-0) and transgenic (D16 and H2) Arabidopsis against B. cinerea infection. Importantly, the DAPG-primed A. thaliana Col-0 showed significantly reduced the disease symptoms when compared with those of non-primed plants. The lesion diameters were 2.5, 2.4, and 2.4 mm in 10, 100, and 200 μM DAPG-treated plants, respectively. Conversely, DAPG-treated D16 and H2 plants experienced similar effects to those from water and methanol treatments. Under all three concentrations of DAPG, the lesion diameters were 7 mm in D16 plants, very similar to the 7.1 mm measured in DAPG-primed H2 plants.
The non-primed A. thaliana Col-0, D16, and H2 plants harbored similar cell densities of Pst DC3000 bacterial pathogen (>7 log cfu/0.1 g of leaves) (Fig. 4). In DAPG-primed A. thaliana Col-0, the pathogen density was 6.4 (Col-0), 6.8 (D16), and 6.5 (H2) log cfu/0.1 g of leaves per 10, 100, and 200 μM of DAPG, respectively. However, the DAPG-primed D16 and H2 failed to trigger their ISR and were unable to suppress the Pst DC3000 pathogen densities: these were 6.4 (10 μM DAPG), 6.3 (100 μM DAPG), and 6.2 (200 μM DAPG) in D16 and 6.5 (10 μM DAPG), 6.1 (100 μM DAPG), and 6.8 (200 μM DAPG) in H2.
These results were confirmed by analyzing the infected cells with trypan blue staining, which let cell deaths be visualized (Keogh et al., 1980). The D16 and H2 leaves presented more stained areas than did wild-type A. thaliana Col-0 after treatment with 100 μM of DAPG (Supplementary Fig. 5). In sum, these results showed phlG-transgenic plants were unable to activate the DAPG-mediated defense system against both necrotrophic and biotrophic pathogens.
DAPG triggers the JA/ET-mediated defense system
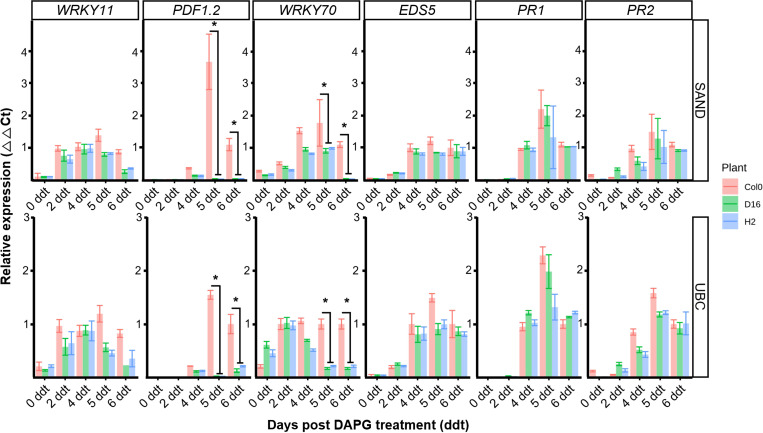
SA-mediated defense (EDS5, PR1, PR2), JA/ET-mediated defense (WRKY11, PDF1.2) or SA, JA integrated signaling (WRKY70) were assessed to establish the induced defense signal pathway (Jiang et al., 2016; Li et al., 2004). The respective expression levels of these five genes were assessed from 0 to 6 ddt by qRT-PCR (Figs. 5, 6). The B. cinerea inoculation of the DAPG-primed Col-0 led to upregulation of PDF1.2 and WRKY70 compared with the D16, H2 plants, at both 5 and 6 ddt (Student t-test, P < 0.05) (Fig. 5). By contrast, the wild-type A. thaliana Col-0 and phlG transgenic D16 and H2 plants all showed a similar expression of the SA-mediated defense genes.
The A. thaliana Col-0 plants inoculated with Pst DC3000 after treatment with DAPG showed no difference in their gene expression levels of WRKY11, EDS5, PR1, and PR2 when compared to those of D16 and H2, but there was a difference in the expression of WRKY70 and PDF1.2 genes (Fig. 6). At 4 ddt, different gene expression activity was observed and PDF1.2 was not expressed in D16 and H2 lines (Student t-test, P < 0.05). Similarly, at 4 ddt, WRKY70 gene expression was statistically higher in the A. thaliana Col-0 than either D16 or H2 (Student t-test, P < 0.05). Interestingly, expression levels of the SA-mediated defense genes (EDS5, PR1, PR2) were similar between Col-0, D16, and H2 plants.
Discussion
DAPG has a wide range of antibiotic activities capable of killing bacteria, fungi, nematodes, and even plants. It is known that DAPG prevents seed germination and plants’ growth (Keel et al., 1992), and that it can alter root architecture by interacting with the auxin-dependent signaling pathway (Brazelton et al., 2008). The phlG gene encodes DAPG hydrolase, which can catalyze non-toxic MAPG in the DAPG biosynthetic loci of P. fluorescens (Bottiglieri and Keel, 2006). DAPG-producing P. fluorescens harbor antibiotic resistance depending on the action of this hydrolase. In our study, the phlG gene was transformed into A. thaliana Col-0 and we selected the DAPG-degrading plant lines to comprehensively investigate the roles of DAPG in the plant ISR mechanism.
Root-colonizing P. fluorescens CHA0, a biological control agent producing DAPG, successfully protected A. thaliana from infection with Hyaloperonospora arabidopsidis by altering its ISR defense system (Iavicoli et al., 2003). The same CHA0 strain also defended this plant against Meloidogyne javanica, a root-knot nematode (Siddiqui et al., 2007). To better understand the mechanism and participation of DAPG in ISR, some studies have relied on testing responses of pathway-related mutants with a single or double mutant Arabidopsis line, mainly with respect to SA or JA/ET hormone signaling. SA is usually involved in processes confering resistance to biotrophic pathogens while JA/ET activity contributed to resistance against necrotrophic pathogens (Lievens et al., 2017). Both SA, JA signaling-deficient mutants of Arabidopsis were reportedly more vulnerable to B. cinerea and Pst DC3000 than wild-type plants (Jiang et al., 2016; Nie et al., 2017). Nevertheless, other physiological functions in plant growth involve these hormones. For example, SA shapes phenotypic features, such as the numbers of cotyledons and rumpled leaves, while the JA/ET hormones influence root phenotypic development, namely primary root, lateral root, and root hair characteristics, as well as also hypocotyl weight, apical hook, and plant height (Peng et al., 2009). Such reports point to many phenotypic imbalances arising in mutant Arabidopsis lines, so it may be inappropriate to use a hormone related mutant for studying the plant’s ISR function against pathogens. A way around this problem is to use insensitive transgenic plant lines of DAPG (D16, H2), which as confirmed here have no phenotypic distinctions from wild-type counterparts. Furthermore, the Col-0, D16, and H2 treated with water showed no difference in their responses to B. cinerea and Pst DC3000. However, DAPG-primed D16 and H2 showed significantly greater disease severity and higher pathogen cell density than did the DAPG-primed wild-type plant. This result indicates the phlG transgenic plants were not able to induce a defense system because of DAPG catalysis.
In DAPG-primed plants, the expression levels of PDF1.2, and WRKY70 were lower in the D16 and H2 than Col-0 following infection with either B. cinerea or Pst DC3000. But levels of expression for SA’s EDS5, PR1, and PR2 genes were not specific to D16, H2, or wild-type plants. These results suggest the JA/ET signaling pathway plays a more critical role in plants’ ISR activated by DAPG than does the SA-based pathway. In other work, P. fluorescens CHA0 could not mount mediated resistance to Peronospora parasitica in the Arabidopsis mutants npr1-1, jar1-1, and eir1-1 (Lavicoli et al., 2007). In that study, synthetic DAPG, for comparison, also failed to trigger induced resistance response on the Arabidopsis mutants eir1 and jar1. Similarly, P. fluorescens Q2-87 failed to defend Pst DC3000 against the npr1-1, jar1, and etr1 mutants (Weller et al., 2012). Those findings suggested that DAPG-mediated ISR relied on the signal pathway based on JA/ET, which is consistent with our results.
In conclusion, DAPG-hydrolyzing plants showed tolerance against toxicity to DAPG and JA/ET defense system plays a critical role in the DAPG-mediated mechanism of ISR. These observations made from using phlG-transgenic plants in experimental investigations of the ISR system provide more clear proof of how plant-microbe interactions can operate.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).








 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print






