 |
 |
| Plant Pathol J > Volume 37(1); 2021 > Article |
|
Abstract
Acidovorax citrulli (Ac) is the causal agent of bacterial fruit blotch (BFB) in watermelon, a disease that poses a serious threat to watermelon production. Because of the lack of resistant cultivars against BFB, virulence factors or mechanisms need to be elucidated to control the disease. Glycerol-3-phosphate dehydrogenase is the enzyme involved in glycerol production from glucose during glycolysis. In this study, we report the functions of a putative glycerol-3-phosphate dehydrogenase in Ac (GlpdAc) using comparative proteomic analysis and phenotypic observation. A glpdAc knockout mutant, AcΔglpdAc(EV), lost virulence against watermelon in two pathogenicity tests. The putative 3D structure and amino acid sequence of GlpdAc showed high similarity with glycerol-3-phosphate dehydrogenases from other bacteria. Comparative proteomic analysis revealed that many proteins related to various metabolic pathways, including carbohydrate metabolism, were affected by GlpdAc. Although AcΔglpdAc(EV) could not use glucose as a sole carbon source, it showed growth in the presence of glycerol, indicating that GlpdAc is involved in glycolysis. AcΔglpdAc(EV) also displayed higher cell-to-cell aggregation than the wild-type bacteria, and tolerance to osmotic stress and ciprofloxacin was reduced and enhanced in the mutant, respectively. These results indicate that GlpdAc is involved in glycerol metabolism and other mechanisms, including virulence, demonstrating that the protein has pleiotropic effects. Our study expands the understanding of the functions of proteins associated with virulence in Ac.
Acidovorax citrulli (Ac), previously known as Pseudomonas pseudoalcaligenes subsp. citrulli, causes bacterial fruit blotch (BFB) disease in cucurbit crops such as watermelon (Latin and Rane, 1990; Sowell and Schaad, 1979). The disease was first observed in 1989 in Florida, USA, and has since spread throughout the US and other countries, including Korea (Burdman and Walcott, 2012). Under favorable conditions, the crop losses caused by BFB have been greater than 90% on marketable crops. The disease generally begins with contaminated seeds because Ac is known as a seed-borne pathogen (Latin and Hopkins, 1995). The initial symptoms are water-soaked lesions on cotyledons and wilt development in seedlings. In fruit, the infected surfaces also show water-soaked lesions, which expand, leading to necrosis of the infected fruit (Latin and Hopkins, 1995). Thus, BFB is a major threat to the production of watermelon. Despite its negative impact on the industry, resistant cultivars of watermelon have not been developed. Therefore, virulence factors or mechanisms in Ac have to be elucidated to control the disease.
There was a focus on controlling BFB spread in the 1990s, and various studies regarding virulence factors and mechanisms in Ac were subsequently carried out. It was elucidated that twitching motility and biofilm formation, which are mediated by type IV pili, are indispensable for Ac virulence (Bahar et al., 2009). It was also reported that quorum sensing and ferric uptake systems are involved in the regulation of its virulence (Johnson and Walcott, 2013; Liu et al., 2019). Additionally, research focusing on the type III secretion system is actively ongoing (Jim├®nez-Guerrero et al., 2019). Recently, Kim et al. (2020) reported that CmpAc, which is a bifunctional chorismate mutase and prephenate dehydratase, is involved in virulence and other mechanisms in Ac. However, other genes and mechanisms that are related to virulence in Ac have not been definitively identified.
Glycerol-3-phosphate (G3P) dehydrogenase is a cytosolic enzyme involved in carbohydrate and lipid metabolism, and it controls the conversion process between dihydroxyacetone phosphate and G3P (Guindalini et al., 2010). This G3P dehydrogenase has been shown to be the key enzyme that produces glycerol from glucose during glycolysis (Hao et al., 2015). Further, G3P dehydrogenase plays an important role in the virulence of Pseudomonas aeruginosa (Daniels et al., 2014). In Magnaporthe oryzae, the deletion of G3P dehydrogenase had negative effects on fungal development and virulence (Shi et al., 2018). The function of G3P dehydrogenase in Colletotrichum gloeosporioides is also related to conidiation (Wei et al., 2004). Thus, G3P and glycerol assimilation and carbon utilization through G3P dehydrogenase play important roles in pathogenicity.
In this study, we report the functions of a putative G3P dehydrogenase in Ac (GlpdAc) in virulence and other biological mechanisms using a G3P dehydrogenase knockout mutant (AcΔglpdAc). To predict and characterize the functions of GlpdAc, the label-free shotgun proteomics following clusters of orthologous groups (COGs) was carried out. In addition to comparative proteomic analysis, diverse phenotypic assays were also conducted to elucidate the roles of GlpaAc in Ac.
For this study, the Ac group II strain, KACC17005, whose genome information has been fully sequenced (Park et al., 2017), was used as the wild-type strain. Ac strains were grown in TSB (tryptic soy broth, 30 g/l) with appropriate antibiotics at 28┬░C. For generating the plasmid constructs, Escherichia coli strain DH5╬▒ was used, and for cloning Tn5-inserted DNA fragments, the E. coli strain EC100D was used (Epicentre, Madison, WI, USA). E. coli strains were grown in LB (Luria-Bertani; 1% tryptone, 0.5% yeast extract, and 1% NaCl) with appropriate antibiotics. The final antibiotic concentrations in the media were as follows: kanamycin, 50 ╬╝g/ml; rifampicin, 50 ╬╝g/ml; gentamicin, 10 ╬╝g/ml; and ampicillin, 100 ╬╝g/ml.
To generate the knockout of glpdAc, the EZ-Tn5 <R6K╬│ori/KAN-2> Insertion Kit (Lucigen, Middleton, WI, USA) was used. Following the kit procedure, Tn5 was randomly inserted on one site of the Ac genome, and this insertion was verified using Tn5-specific primers 5ŌĆ▓-GGATCGCGATGGATATGTTCT-3ŌĆ▓ and 5ŌĆ▓-AACAGGAATCGAATGCAACC-3ŌĆ▓. Genomic DNA was extracted, and the Tn5-disrupted DNA fragment was cloned following the manufacturer's protocol (Lucigen). After cloning the DNA fragment, its DNA sequence was verified by Sanger sequencing with the following primers: forward (KAN-2F), 5ŌĆ▓-ACCTACAACAAAGCTCTCATCAACC-3ŌĆ▓ and reverse (R6KAN-2R), 5ŌĆ▓-CTACCCTGTTGGAACACCTACATCT-3ŌĆ▓ (Lucigen). The confirmed strain was named Ac╬öglpdAc. To create the construct for the complemented strain, the open reading frame of glpdAc was amplified using the following gene-specific primers: 5ŌĆ▓-CTCGAGATGAAGATCATCGT-3ŌĆ▓ and 5ŌĆ▓-AAGCTTCAGTGGTGGTGGTGGTGGTGGCAGGAGCGCAA-3ŌĆ▓. The amplicon was cloned into the pGem-T Easy vector (Promega, Madison, WI, USA), generating pGem-GlpdAc, and the cloned DNA was confirmed by Sanger sequencing. The construct was digested with restriction enzymes XhoI and HindIII, and the digested DNA fragment was subcloned into the pBBR1-MCS5 vector, which is a broad host range vector containing the lacZ promoter (Kovach et al., 1995), generating pBBR1-GlpdAc. The generated plasmid was introduced into Ac╬öglpdAc and selected on tryptic soy agar (TSA) with appropriate antibiotics. The selected transformant was confirmed using PCR with GlpdAc-specific primers and called Ac╬öglpdAc(GlpdAc). To control for vector side-effects, an empty pBBR1-MCS5 vector was also introduced into Ac and Ac╬öglpdAc, generating Ac(EV) and Ac╬öglpdAc(EV) strains, respectively. The transformants were selected on TSA with appropriate antibiotics and then confirmed by PCR with the following pBBR1-MCS5-specific primers: 5ŌĆ▓-CAGGGTTTTCCCAGTCACGA-3ŌĆ▓ and 5ŌĆ▓-ATGCTTCCGGCTCGTATGTT-3ŌĆ▓. The generated plasmids and strains are described in Supplementary Table 1.
To predict the putative 3D structure of GlpdAc, the deduced amino acid sequence of GlpdAc was evaluated through the I-TASSER server (Roy et al., 2010), and the PDB file was obtained. This file was imported into the PyMOL program (Molecular Graphics System, San Carlos, CA, USA), generating the putative 3D structure. For comparison, the 3D structure of GpsA, a G3P dehydrogenase in Coxiella burnetii, was also generated using the same method.
Watermelon (Citrullus lanatus var. vulgaris) line SBA, which was kindly provided by Partner Seed Company (Gimje, Korea), was used for virulence testing. A germinated-seed inoculation was used according to a previously established method with slight modifications (Bahar et al., 2009). For effective inoculation, a pre-germination step was added. To prepare the germinated seeds for inoculation, they were placed on sterilized filter papers with 4 ml sterilized water and incubated at 28┬░C for 2 days; germination was confirmed by visual observation. The Ac strains were grown on TSA with antibiotics at 28┬░C. The bacteria were collected and suspended in 10 mM MgCl2 to an optical density at 600 nm (OD600) of 0.3 (approximately 108 colony forming units [cfu]/ml) and then diluted to 106 cfu/ml with 10 mM MgCl2. Ten pre-germinated seeds were incubated in 20 ml of bacterial suspension and gently agitated for 1 h. After incubation, the inoculated seeds were sown in a 50-pot tray with sterilized soil and sealed with a tray cover for 2 days. The planted seeds were then grown in a growth chamber and checked for disease severity for 7 days. Disease severity was evaluated using a 0-2 scale (0, no symptoms; 1, water-soaked region; 2, seedling wilt) as described previously (Kim et al., 2020; Song et al., 2020). A disease index was then calculated as follows:
The infiltration of watermelon leaves was performed according to a previous protocol (Bahar et al., 2011). The pre-germinated watermelon seeds were sown and grown for 2 weeks, at which time the seedlings had acquired four true leaves. The Ac strains were suspended in 10 mM MgCl2 to an OD600 of 0.3 and diluted to 105 cfu/ml with 10 mM MgCl2. The undersides of first and second leaves were inoculated with bacterial suspension using a 3 ml needleless syringe. For counting CFUs, the inoculated leaves were punched using cork-borers (0.4 cm diameter), and two punches were ground in 200 ╬╝l sterilized water using a tissue grinder. The leaf extracts were serially diluted (10-fold). The diluted suspensions were spotted onto TSA with antibiotics and incubated at 28┬░C for 2 days. Cfus were determined 0, 2, 4 6, and 8 days after inoculation. Data from three biological replicates were collected.
The proteomics protocols and conditions, including protein extraction, peptide generation, liquid chromatography-tandem mass spectrometry (LCMS/MS), peptide quantification/identification, comparison of identified proteins, and COG classification, were as reported previously (Park et al., 2020). Briefly, three biological replicates of the Ac strains (a total of six samples) were grown in TSB to an OD600 of 0.5 and collected by centrifugation. Total soluble proteins were then extracted from the bacteria, and peptides were generated. A split-free nano-LC system (EASY-nLC II, Thermo Fisher Scientific, Waltham, MA, USA) combined with an LTQ Velos Pro instrument (Thermo Fisher Scientific) was used for the proteomics analysis. Mass spectra from the LC-MS/MS analysis were identified using the Ac strain KACC17005 database from the National Center for Biotechnology Information, and the identified peptides were quantified using Thermo Proteome Discoverer 1.3 (ver. 1.3.0.399) and the SEQUEST program. After peptide quantification, protein levels in Ac and AcΔglpdAc were compared. Finally, differentially expressed proteins were categorized by COG analysis. Student's t-tests (P < 0.05) were used for statistical comparisons.
For bacterial growth assay in TSB medium, Ac(EV), Ac╬öglpdAc(EV), and Ac╬öglpdAc(GlpdAc) were harvested, washed twice with sterilized water, and suspended in TSB to an OD600 of 0.3 (108 cfu/ml). The bacterial suspensions were then diluted to 105 cfu/ml with TSB and measured for 96 h at intervals of 12 h using a spectrophotometer. To determine the effect of carbon sources on the Ac strains, bacterial growth was assayed in M9 minimal medium containing glucose or glycerol (Hames et al., 2009). For the assay, M9 minimal medium (5├Ś M9 salts, 200 ml; 1 M MgSO4, 2 ml; 1 M CaCl2, 0.1 ml in 1 l) with 20 mL of 20% glycerol or 20% glucose was used. The bacteria were grown on TSA, harvested, washed twice, and suspended in 10 mM MgCl2 to an OD600 of 0.05. The bacterial growth was measured for 10 days at 24 h intervals using a spectrophotometer. Three biological replicates were used in this assay.
The assay for cell-cell aggregation was carried out as previously described (Bahar et al., 2009). Ac strains were grown in TSA, washed, and suspended in TSB. The bacterial suspension was adjusted to an OD600 of 0.1 (4 ml in 15 ml tube), incubated for 12 h at 28┬░C, and kept without shaking at 22┬░C for 2 h. After the bacterial cells had settled at the bottom of the tube, the absorbance at 600 nm (ODs) of the upper 1 ml of liquid was measured with a spectrophotometer. The residual bacteria were completely dispersed by vortexing and the absorbance at 600 nm was measured (ODt). The cell-cell aggregation rate was calculated as [(ODt ŌĆÆ ODs) ├Ś 100]/ODt. For this assay, six biological replicates were collected.
To estimate bacterial tolerance to stress conditions, treatments with 2.5% NaCl for osmotic stress, and 0.1 ╬╝g/ml ciprofloxacin for antibiotic stress were used. For osmotic stress, the bacterial strains were grown, harvested, and adjusted to an OD600 of 0.3. The bacteria were then treated with 2.5% NaCl for 10 min. For antibiotic stress, the bacteria strains were harvested, suspended at an OD600 of 0.1, and exposed to 0.1 ╬╝g/ml ciprofloxacin for 2 h. The treated and untreated bacterial suspensions were serially diluted and plated on TSA with appropriate antibiotics. After incubation for 2 days at 28┬░C, the numbers of colonies were counted. The ratio of stress-treated bacterial cell count to untreated bacterial cell count was calculated for each strain and stress treatment. For this study, at least three biological replicates were used for each strain.
By screening a Tn5 insertion library, we identified one mutant with reduced virulence in watermelon and confirmed that one gene (accession no. ATG93866), identified as G3P dehydrogenase, was disrupted by Tn5. The predicted 3D structure of ATG93866 is similar to that of G3P dehydrogenase (GpsA) from C. burnetii, which is the causal agent of Q fever in humans (Fig. 1A and B). As shown in Fig. 1C, the deduced amino acid sequence of ATG93866 has high similarity with the NAD(P)H-dependent GlpdAc strain AAC00-1, G3P dehydrogenase in Nocardioides sp. SLBN-35, and G3P dehydrogenase in Burkholderiales bacterium 64-34. Therefore, ATG93866 was named GlpdAc (glycerol-3-phosphate dehydrogenase in Ac).
To investigate the role of GlpdAc in virulence, germinatedseed inoculation and leaf infiltration were performed. Ac(EV) (wild-type strain carrying an empty vector), AcΔglpdAc(EV) (glpdAc knockout mutant carrying an emphajeoty vector), and the complemented strain, AcΔglpdAc(GlpdAc) (AcΔglpdAc carrying glpdAc on pBBR1MCS5) were tested. In the germinated-seed inoculation, the disease index of Ac(EV) continuously increased and reached a value of 2 at 7 days after inoculation (DAI). In contrast, AcΔglpdAc(EV) did not cause disease, with a disease index was 0 at 7 DAI (Fig. 2A), demonstrating that GlpdAc is indispensable for virulence in Ac. The virulence of AcΔglpdAc(GlpdAc) was similar to that of Ac(EV), indicating that there were no positional effects by the insertion of Tn5 in GlpdAc. As shown in Fig. 2B, watermelon infected by AcΔglpdAc(EV) did not show disease symptoms. The results of the leaf-infiltration experiments were similar to those of the seed-germinated inoculation. Although the AcΔglpdAc(EV) population slightly increased during the observation period, it was significantly lower than that of Ac(EV) (Fig. 2C). However, the growth of the complemented strain was not different from that of Ac(EV). Similar to the seed-germinated inoculation, AcΔglpdAc(EV) did not induce symptoms on the leaves of watermelon (Fig. 2D). However, leaves infiltrated with Ac(EV) and AcΔglpdAc(GlpdAc) showed necrosis near the site of inoculation.
It is clear that GlpdAc is involved in virulence. In addition, it has been shown that one protein related to biochemical pathways can produce pleiotropic effects in bacteria, including the alteration of virulence and other physiological mechanisms (Felgner et al., 2016; Kim et al., 2020). Therefore, we carried out comparative proteomics to postulate biological and cellular mechanisms related to GlpdAc using the wild-type and AcΔglpdAc strains. In three biological replicates of the Ac strain, 751, 757, and 752 proteins were found from 59,405, 58,831, and 59,569 peptide spectral matches (PSMs), respectively; 726, 695, and 722 proteins from 59,367, 58960, and 59,261 PSMs, respectively, were identified from the AcΔglpdAc strain (Supplementary Table 2). Among them, 696 and 658 proteins were commonly detected in the three biological replicates of Ac and AcΔglpdAc, respectively, and these proteins were used for the comparative analysis. It was found that 51 and 27 proteins were more abundant (>2-fold) in Ac and AcΔglpdAc, respectively (Fig. 3A). Seventy-eight proteins were subjected to COG classification. The COG analysis revealed that proteins classified in groups F (nucleotide metabolism and transport), G (carbohydrate metabolism and transport), N (cell motility), and U (intracellular trafficking and secretion) were uniquely found in the Ac strain, and proteins categorized in groups I (lipid metabolism), L (replication and repair), and Q (secondary structure) were detected only in AcΔglpdAc (Fig. 3B, Supplementary Tables 3 and 4). Proteins belonging to other categories were mostly abundant in the Ac strain. Interestingly, fructose 1/6-bisphosphatase, C4-dicarboxylate ABC transporter substrate-binding protein, twitching motility protein PilT, and universal stress protein were only detected in Ac. These results indicate that GlpdAc is involved in diverse cellular processes in Ac, including carbohydrate metabolism and cell wall/membrane biogenesis.
Sequence comparison and proteomics analysis of GlpdAc reveal that the protein may be associated with glucose/glycerol metabolism. Therefore, bacterial growth in the presence of glucose or glycerol as a sole carbon source was investigated. First, we examined the growth of Ac(EV), AcΔglpdAc(EV), and AcΔglpdAc(GlpdAc) in a rich medium to test the effect of GlpdAc on bacterial multiplication (Fig. 4A). During the measurement period, the OD600 values of the three strains were not significantly different, indicating that GlpdAc is not associated with bacterial reproduction. As shown in Fig. 4B, the growth of AcΔglpdAc(EV) barely increased in M9 medium with glucose, but the complemented strain showed similar growth to that of Ac(EV). Notably, the growth of AcΔglpdAc(EV) in M9 medium supplemented with glycerol was not different from that of Ac(EV) and AcΔglpdAc(GlpdAc) (Fig. 4C). These data suggest that the function of GlpdAc is related to glycolysis.
In the comparative proteomics analysis, the abundance of diverse proteins related to cell wall/membrane/envelope, including PilT, which is associated with cell aggregation (Bahar et al., 2009), was altered (Supplementary Tables 3 and 4). To examine the effect of GlpdAc on cell aggregation, a bacterial cell sedimentation assay was conducted using fully grown Ac(EV), AcΔglpdAc(EV), and AcΔglpdAc(GlpdAc). As shown in Fig. 5, the bacteria that remained in suspension after allowing the medium to stand for 2 h and those that had pelleted were quantified, and the cell-cell aggregation ratio was calculated. Ac(EV) showed a cell-cell aggregation ratio of 7.76%. The ratio for AcΔglpdAc(EV) was 16%, which was approximately 2-fold higher than that for Ac(EV). The cell-cell aggregation in AcΔglpdAc(GlpdAc) was restored to the level in Ac(EV).
In the proteomics analysis, a universal stress protein (ATG96448) was found to be more abundant in Ac than in AcΔglpdAc. Proteins associated with cell membrane/wall/envelope biogenesis were also abundantly detected in the comparative proteomics analysis. Thus, the tolerance to osmotic stress and antibiotics in Ac(EV), AcΔglpdAc(EV), and AcΔGglpdAc(GlpdAc) was investigated. After treatment with 2.5% NaCl, 6.61% of AcΔglpdAc(EV) had survived, compared to 16.7% of Ac(EV) (Fig. 6A), indicating that the mutant was less tolerant to osmotic stress. Survival was restored to wild-type levels in the complemented strain. Notably, with 0.1 μg/ml ciprofloxacin treatment, AcΔglpdAc(EV) displayed higher survival (7.79%) than both Ac(EV) (5.93%) and AcΔglpdAc(GlpdAc) (4.22%) (Fig. 6B).
In many pathogens, glucose and glycerol metabolism has been implicated in virulence. For example, Mycoplasma glycerol metabolism is essential to its cell cytotoxicity and is involved in its virulence (Bl├Čtz and St├╝lke, 2017; Schmidl et al., 2011). This glycerol metabolism is mediated by G3P dehydrogenase, which is known as a key enzyme for glycerol production from glucose during glycolysis (Andr├® et al., 1991). In P. aeruginosa, the analogous G3P dehydrogenase plays a role in producing virulence factors (Daniels et al., 2014). In addition, in the rice blast fungus, M. oryzae, conidia formation and disease development were affected by G3P dehydrogenase, and the G3P dehydrogenase knockout mutant was less virulent than the wild-type strain (Shi et al., 2018). Thus, the G3P dehydrogenase that is indispensable for glycerol production from glucose is an important component of virulence. In agreement with these previous studies, Ac╬öglpdAc(EV) characterized in our study also showed significantly reduced disease symptom development and bacterial populations on watermelon, indicating that GlpdAc contributes to virulence in Ac. Nevertheless, Ac╬öglpdAc(EV) showed little difference compared to Ac(EV) and Ac╬öglpdAc(GlpdAc) in bacterial growth assays in TSB. These results suggest that the reduction in virulence of Ac╬öglpdAc(EV) is not related to bacterial multiplication.
Comparative proteomics indicated that GlpdAc is associated with diverse metabolic processes, including a carbohydrate metabolic pathway. Especially, fructose 1/6-bisphosphatase (ATG94560), which is a glycolytic enzyme (Spoering et al., 2006), was detected only in Ac. Furthermore, proteins categorized in groups E (amino acid metabolism and transport) and H (coenzyme metabolism) were found to be abundant in Ac. In addition, proteins involved in other metabolic processes were more abundant in Ac than in AcΔglpdAc. Thus, GlpdAc is likely associated with carbohydrate metabolism as well as other metabolic processes. In the growth assay, AcΔglpdAc(EV) did not grow in the presence of glucose but did grow in the presence of glycerol. These data indicate that GlpdAc is a key enzyme involved in the glucose-dependent pathway of carbohydrate metabolism, rather than the glycerol-dependent pathway. The sequence analysis of GlpdAc also supported this observation. Because GlpdAc encodes a putative G3P dehydrogenase, GlpdAc likely catalyzes the conversion of glucose to glycerol during glycolysis. In addition, the twitching motility protein PilT, which belongs to group N (cell motility), was detected in Ac but not in AcΔglpdAc. In a previous study, the group I Ac strain knockout mutant of PilT showed high cell-cell aggregation (Bahar et al., 2009). Similarly, AcΔglpdAc(EV) displayed higher cell-cell aggregation than Ac(EV) or AcΔglpdAc(GlpdAc) in our study. PilT is involved in the retraction of type IV pili, and the loss of this function could negatively affect virulence. Thus, our proteomics analysis and phenotypic observations reveal that impaired mechanisms caused by the loss of GlpdAc function trigger abnormal phenotypes, which may contribute to Ac virulence.
In E. coli, G3P dehydrogenase is a monotopic enzyme that is bound to the inner membrane and is involved in energy regulation (Yeh et al., 2008). According to a previous study, G3P dehydrogenase also participates in sustainable cell formation in E. coli (Spoering et al., 2006). Therefore, it can be predicted that the altered mechanisms in Ac╬öglpdAc(EV) could reduce tolerance to external stress. For example, the deletion mutant of G3P dehydrogenase in Saccharomyces cerevisiae showed enhanced sensitivity to osmotic stress conditions (Albertyn et al., 1994). Similar to this observation, Ac╬öglpdAc(EV) was less tolerant to growth in 2.5% NaCl than both Ac(EV) and Ac╬öglpdAc(GlpdAc). In contrast, the impaired function of GlpdAc enhanced tolerance to ciprofloxacin. It was also reported that the loss of G3P dehydrogenase function in P. aeruginosa increased cell formation persistence, resulting in enhanced tolerance to antibiotic stress (Shuman et al., 2018). Our proteomics analysis also supports these phenotypic observations because the abundance of many proteins involved in cell membrane or wall processes was changed. These results suggest that impaired GlpdAc alters metabolic processes and the expression of various proteins, resulting in varying persistence to different stressors. In both plants and humans, glycerol-3-phosphate dehydrogenase (GAPDH) is reported as a target of plant defense-related hormone salicylic acid (SA) (Choi et al., 2015; Tian et al., 2015). In humans, SAŌĆÖs interaction with GAPDH suppressed its nuclear translocation and subsequent cell death. On the other hands, SA inhibited Tomato bushy stunt virus (TBSV), a single-stranded RNA virus, replication in tobacco plants by inhibiting the binding of cytosolic GAPDH to the negative (ŌĆÆ)RNA strand of TBSV. Our study suggests that GlpdAc, which encodes GAPDH, is an important virulence factor in Ac. Thus it will be very interesting whether GlpdAc can be a target of SA, and SAŌĆÖs binding to GlpdAc can reduce virulence in Ac.
Taken together, the functions of GlpdAc were characterized using sequence analysis, comparative proteomics, and diverse phenotypic assays in this study. Consequently, GlpdAc was shown to be involved in pathogenicity, glucose metabolism, cell-cell aggregation, and tolerance to osmotic stress and ciprofloxacin antibiotics in Ac. But, specific molecular mechanisms of GlpdAc regarding various phenotypes are still elusive, and it has to be elucidated in further study. The lack of availability of resistant cultivars against BFB highlights the need to characterize virulence mechanisms and factors in Ac to control the disease. Our study findings indicate that GlpdAc is a potential target to develop antivirulence agents for suppressing the virulence of Ac.
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. NRF-2020R1A2C1013040) (to S.W. Han).
Fig.┬Ā1
Predicted 3D structures and amino acid sequence alignments for GlpdAc. Predicted 3D structures of GlpdAc (A) and glycerol-3-phosphate dehydrogenase (GpsA) (B) from Coxiella burnetii were generated using the I-TASSER sever and PyMol program. (C) Deduced amino acid sequence alignments of GlpdAc (ATG93866), NAD(P)H-dependent glycerol-3-phosphate dehydrogenase from Ac strain AAC00-1 (ABM34399), glycerol-3-phosphate dehydrogenase (NAD(P)+) from Nocardioides sp. SLBN-35 (TGK64598), and glycerol-3-phosphate dehydrogenase from Burkholderiales bacterium 64-34 (OJV61281). These results were obtained using the Clustal Omega Program. The ŌĆś*ŌĆÖ, ŌĆś:ŌĆÖ, and ŌĆś.ŌĆÖ indicate identical residues, conserved substitutions, and semi-conserved substitutions, respectively.
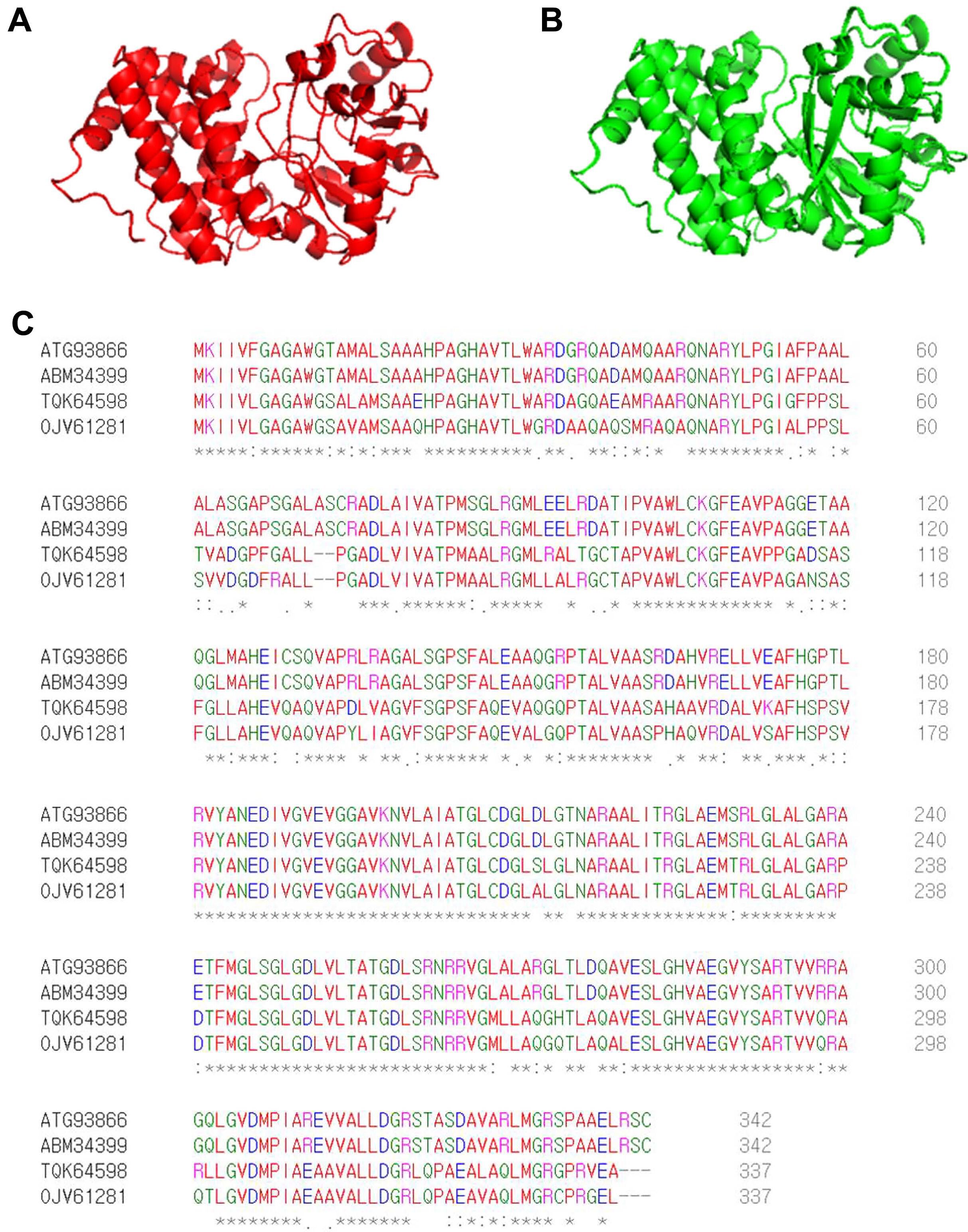
Fig.┬Ā2
Pathogenicity test for Ac(EV), Ac╬öglpdAc(EV), and Ac╬öglpdAc(GlpdAc) on watermelon. (A) Disease index from the seedgerminated method. Disease index = [(Number of normal plants) ├Ś 0 + (Number of plants with water-soaked regions) ├Ś 1 + (Number of wilted plants) ├Ś 2]/(Total number of plants). (B) Symptoms of infected seedlings in the seed-germinated method. The photograph was taken 7 days after inoculation (DAI). The index scale of watermelon infected by Ac(EV) and Ac╬öglpdAc(GlpdAc) was 2 and this of Ac╬öglpdAc(GlpdAc) was 0 at 7 DAI. (C) Bacterial multiplication in watermelon after leaf infiltration. Two-week-old watermelon plants at the 4-true-leaf stage were inoculated in their first and second leaves via needleless syringe. In the inoculated area, leaf discs were punched. Through serial dilution, log10 cfus were measured. Cfu counting was conducted for 8 days, in 2-day intervals. The different letters indicated significant differences (P < 0.05) using ANOVA with TukeyŌĆÖs HSD post-hoc analysis. (D) Images from the leaf infiltration. The punched areas indicate where the bacterial suspension infiltrated. The photographs were taken 8 days after inoculation.
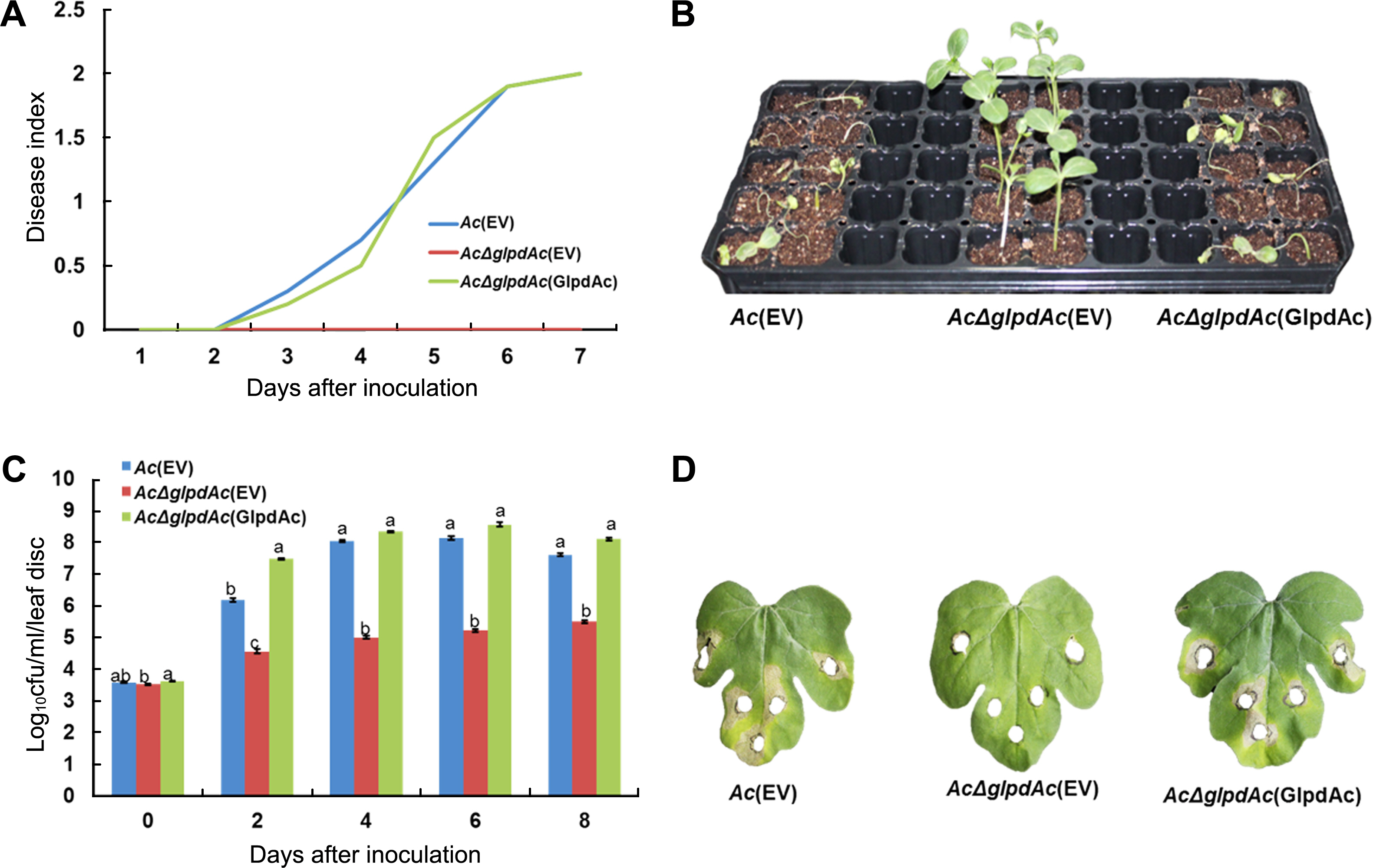
Fig.┬Ā3
Comparison of proteome analysis of Acidovorax citrulli (Ac) and AcΔglpdAc. (A) Venn diagram reveals that abundance of 51 and 27 proteins were changed (>2-fold) in comparative proteomic analysis. Fifty and 26 proteins were uniquely detected in Ac and AcΔglpdAc, respectively. One protein was more abundant in Ac and in AcΔglpdAc. (B) Classification of clusters of orthologous groups of differentially abundant proteins (>2-fold) by proteomics analysis.

Fig.┬Ā4
Growth of Acidovorax citrulli (Ac) strains in rich medium and M9 supplemented with glucose or glycerol. (A) Growth of three strains, Ac(EV), Ac╬öglpdAc(EV), and Ac╬öglpdAc(GlpdAc), in tryptic soy broth medium was measured for 4 days at 12-h intervals using a spectrophotometer (OD600). There was no statistical difference among strains at each time point. Growth of the three strains was examined in M9 medium supplemented with glucose (B) or glycerol (C) for 10 days at 24-h intervals using a spectrophotometer (OD600). Three biological replicates were analyzed. The different letters indicate significantly different values (P < 0.05) using ANOVA with TukeyŌĆÖs HSD post-hoc analysis. Error bars indicate standard deviations.
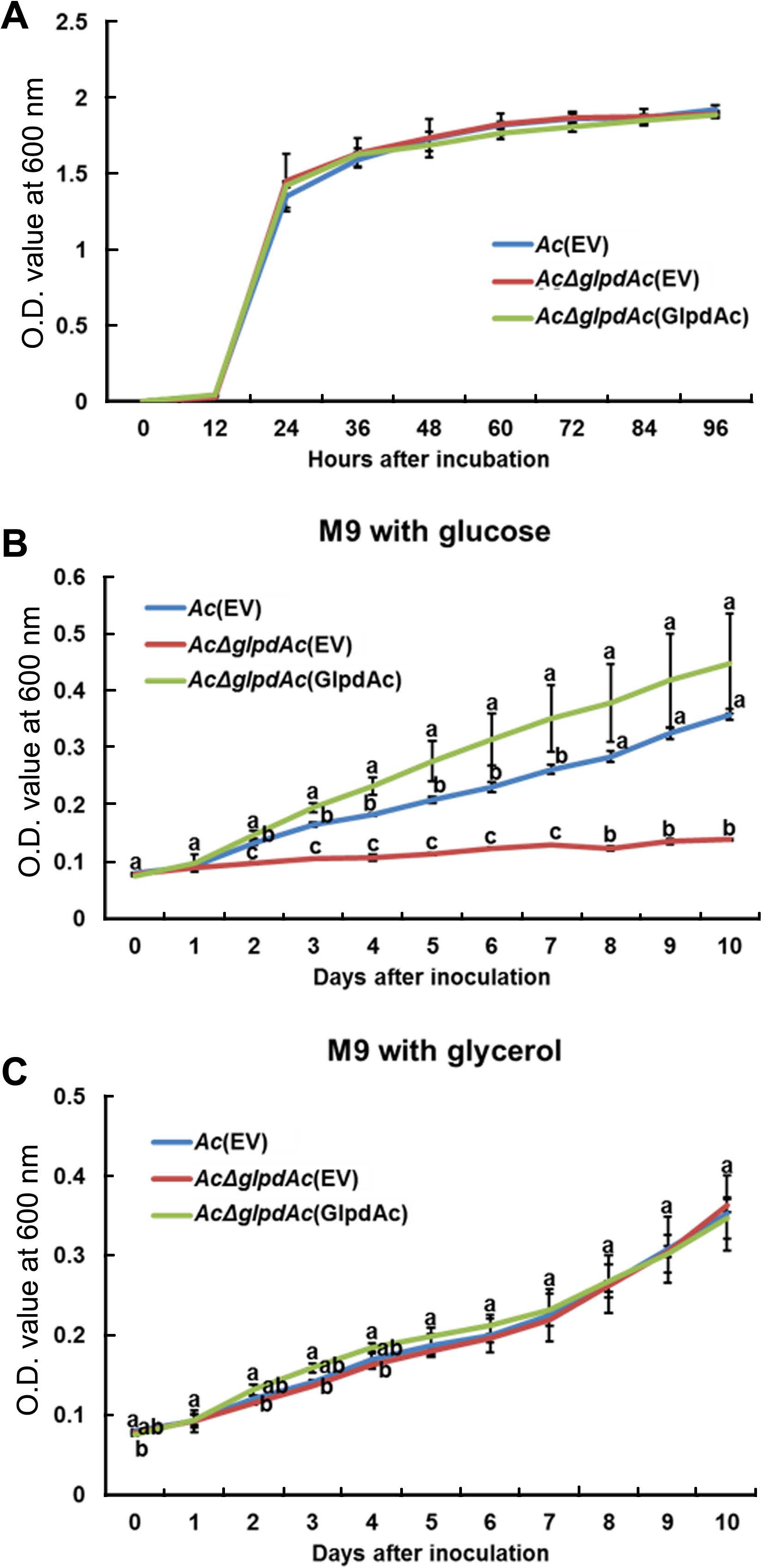
Fig.┬Ā5
Cell-cell aggregation assay. The bacterial suspension was adjusted to an OD600 of 0.1 and incubated without mixing at 22┬░C for 2 h. The supernatant was carefully removed and its turbidity measured by a spectrophotometer (ODs). The remaining pellet was completely resuspended by vortexing and its turbidity was measured by a spectrophotometer (ODt). The cell-cell aggregation percentage was calculated as: [(ODt ŌĆÆ ODs) ├Ś 100]/ODt. The different letters represent significant differences using ANOVA (P < 0.05) with TukeyŌĆÖs HSD post-hoc test. Error bars indicate standard deviations. Six biological replicates were used for this assay.
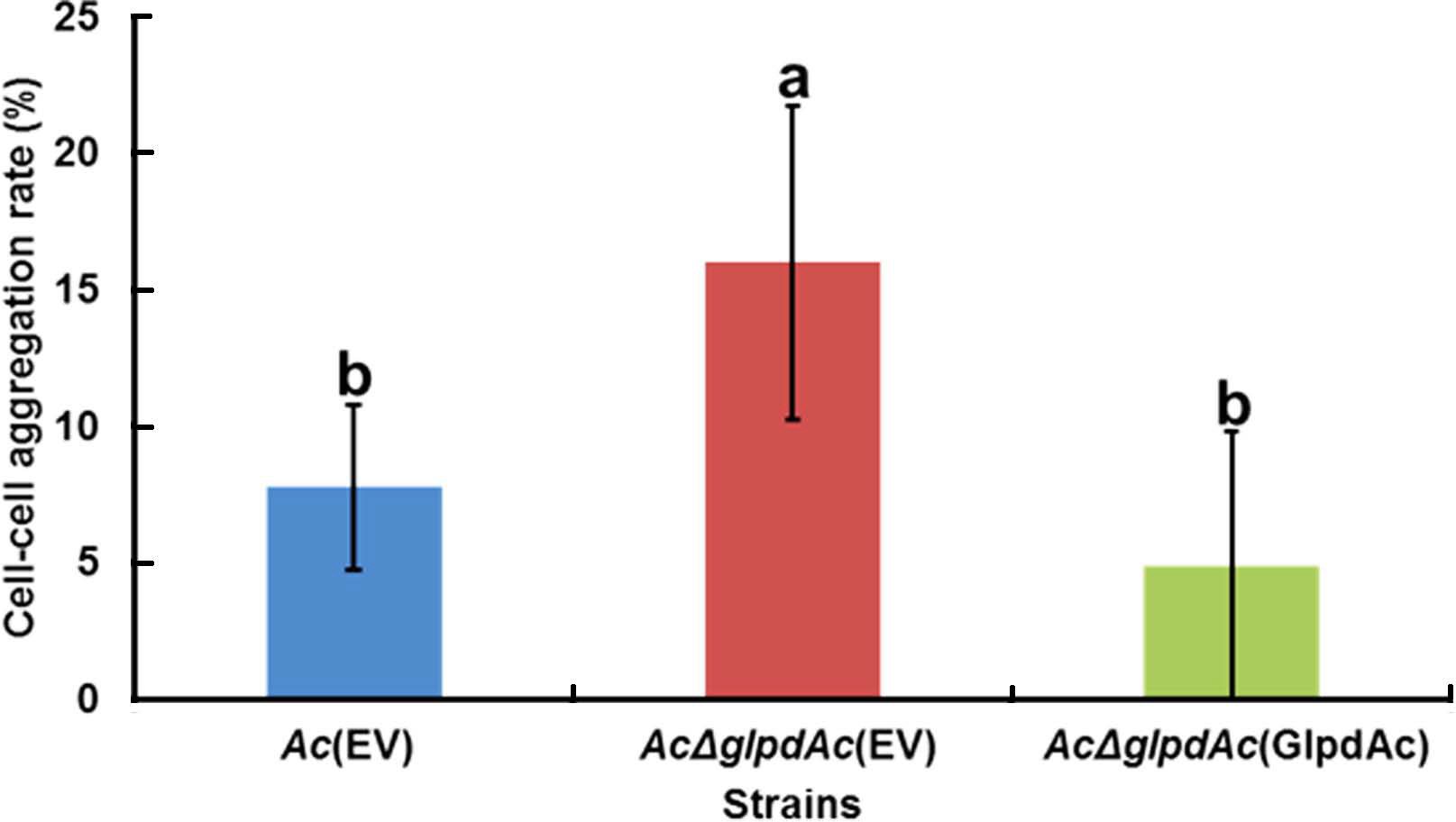
Fig.┬Ā6
Tolerance test against osmotic stress and ciprofloxacin. Tolerance against two stress conditions, 2.5% NaCl for 10 min (A) and 0.1 ╬╝g/ml ciprofloxacin for 2 h (B), was investigated. Treated bacterial suspensions were serially diluted and plated on tryptic soy agar containing appropriate antibiotics. Water was used as a control. Survivability was established from the numbers of bacteria after treatment to those in the water control using the colony counting method. Different letters indicate significantly different means as determined by ANOVA (P < 0.05), followed by TukeyŌĆÖs HSD post-hoc analysis. Error bars indicate standard errors. Three biological replicates were used for each strain.

REFERENCES
Albertyn, J., Hohmann, S., Thevelein, J. M. and Prior, B. A. 1994. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14:4135-4144.



Andr├®, L., Hemming, A. and Adler, L. 1991. Osmoregulation in Saccharomyces cerevisiae: studies on the osmotic induction of glycerol production and glycerol-3-phosphate dehydrogenase (NAD+). FEBS Lett. 286:13-17.


Bahar, O., Goffer, T. and Burdman, S. 2009. Type IV pili are required for virulence, twitching motility, and biofilm formation of Acidovorax avenae subsp. citrulli. Mol. Plant-Microbe Interact. 22:909-920.


Bahar, O., Levi, N. and Burdman, S. 2011. The cucurbit pathogenic bacterium Acidovorax citrulli requires a polar flagellum for full virulence before and after host-tissue penetration. Mol. Plant-Microbe Interact. 24:1040-1050.


Bl├Čtz, C. and St├╝lke, J. 2017. Glycerol metabolism and its implication in virulence in Mycoplasma. FEMS Microbiol. Rev. 41:640-652.


Burdman, S. and Walcott, R. 2012. Acidovorax citrulli: generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol. Plant Pathol. 13:805-815.



Choi, H. W., Tian, M., Manohar, M., Harraz, M. M., Park, S.-W., Schroeder, F. C., Snyder, S. H. and Klessig, D. F. 2015. Human GAPDH is a target of aspirin's primary metabolite salicylic acid and its derivatives. PLoS ONE. 10:e0143447



Daniels, J. B., Scoffield, J., Woolnough, J. L. and Silo-Suh, L. 2014. Impact of glycerol-3-phosphate dehydrogenase on virulence factor production by Pseudomonas aeruginosa. Can. J. Microbiol. 60:857-863.


Felgner, S., Frahm, M., Kocijancic, D., Rohde, M., Eckweiler, D., Bielecka, A., Bueno, E., Cava, F., Abraham, W.-R., Curtiss, R., rd, H├ż, ussler, S., Erhardt, M. and Weiss, S. 2016. aroAdeficient Salmonella enterica serovar Typhimurium is more than a metabolically attenuated mutant. mBio. 7:e01220-16.




Guindalini, C., Lee, K. S., Andersen, M. L., Santos-Silva, R., Bittencourt, L. R. A. and Tufik, S. 2010. The influence of obstructive sleep apnea on the expression of glycerol-3-phosphate dehydrogenase I gene. Exp. Biol. Med. 235:52-56.

Hames, C., Halbedel, S., Hoppert, M., Frey, J. and St├╝lke, J. 2009. Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J. Bacteriol. 191:747-753.


Hao, G., Chen, H., Gu, Z., Zhang, H., Chen, W. and Chen, Y. Q. 2015. Metabolic engineering of Mortierella alpina for arachidonic acid production with glycerol as carbon source. Microb. Cell Fact. 14:205



Jim├®nez-Guerrero, I., P├®rez-Monta├▒o, F., Da Silva, G. M., Wagner, N., Shkedy, D., Zhao, M., Pizarro, L., Bar, M., Walcott, R., Sessa, G., Pupko, T. and Burdman, S. 2019. Show me your secret(ed) weapons: a multifaceted approach reveals a wide arsenal of type III-secreted effectors in the cucurbit pathogenic bacterium Acidovorax citrulli and novel effectors in the Acidovorax genus. Mol. Plant Pathol. 21:17-37.




Johnson, K. L. and Walcott, R. R. 2013. Quorum sensing contributes to seed-to-seedling transmission of Acidovorax citrulli on watermelon. J. Phytopathol. 161:562-573.

Kim, M., Lee, J., Heo, L. and Han, S.-W. 2020. Putative bifunctional chorismate mutase/prephenate dehydratase contributes to the virulence of Acidovorax citrulli. Front. Plant Sci. 11:569552



Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M. and Peterson, K. M. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 166:175-176.


Latin, R. X. and Hopkins, D. L. 1995. Bacterial fruit blotch on watermelon: the hypothetical exam question becomes reality. Plant Dis. 79:761-765.

Latin, R. X. and Rane, K. K. 1990. Bacterial fruit blotch of watermelon in Indiana. Plant Dis. 74:331

Liu, J., Tian, Y., Zhao, Y., Zeng, R., Chen, B., Hu, B. and Walcott, R. R. 2019. Ferric uptake regulator (FurA) is required for Acidovorax citrulli virulence on watermelon. Phytopathology. 109:1997-2008.


Park, H.-J., Lee, J., Kim, M. and Han, S.-W. 2020. Profiling differentially abundant proteins by overexpression of three putative methyltransferases in Xanthomonas axonopodis pv. glycines. Proteomics. 20:e1900125



Park, H.-J., Seong, H. J., Sul, W. J., Oh, C.-S. and Han, S.-W. 2017. Complete genome sequence of Acidovorax citrulli strain KACC17005, a causal agent for bacterial fruit blotch on watermelon. Korean J. Microbiol. 53:340-341.
Roy, A., Kucukural, A. and Zhang, Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725-738.



Schmidl, S. R., Otto, A., Lluch-Senar, M., Pi├▒ol, J., Busse, J., Becher, D. and St├╝lke, J. 2011. A trigger enzyme in Mycoplasma pneumoniae: impact of the glycerophosphodiesterase GlpQ on virulence and gene expression. PLoS Pathog. 7:e1002263



Shi, Y., Wang, H., Yan, Y., Cao, H., Liu, X., Lin, F. and Lu, J. 2018. Glycerol-3-phosphate shuttle is involved in development and virulence in the rice blast fungus Pyricularia oryzae. Front. Plant Sci. 9:687



Shuman, J., Giles, T. X., Carroll, L., Tabata, K., Powers, A., Suh, S.-J. and Silo-Suh, L. 2018. Transcriptome analysis of a Pseudomonas aeruginosa sn-glycerol-3-phosphate dehydrogenase mutant reveals a disruption in bioenergetics. Microbiology. 164:551-562.


Song, Y.-R., Hwang, I. S. and Oh, C.-S. 2020. Natural variation in virulence of Acidovorax citrulli isolates that cause bacterial fruit blotch in watermelon, depending on infection routes. Plant Pathol. J. 36:29-42.



Sowell, G. J and Schaad, N. W. 1979. Pseudomonas pseudoalcaligenes subsp. citrulli on watermelon: seed transmission and resistance of plant introductions. Plant Dis. Rep. 63:437-441.
Spoering, A. L., Vuli─ć, M. and Lewis, K. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188:5136-5144.



Tian, M., Sasvari, Z., Gonzalez, P. A., Friso, G., Rowland, E., Liu, X. M., van Wijk, K. J., Nagy, P. D. and Klessig, D. F. 2015. Salicylic acid inhibits the replication of tomato bushy stunt virus by directly targeting a host component in the replication complex. Mol. Plant-Microbe Interact. 28:379-386.






 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




