 |
 |
| Plant Pathol J > Volume 36(2); 2020 > Article |
|
Abstract
Pseudomonas syringae pv. tabaci 6605 has two multidrug resistance (MDR) efflux pump transporters, MexAB-OprM and MexEF-OprN. To understand the role of these MDR efflux pumps in virulence, we generated deletion mutants, ŌłåmexB, ŌłåmexF, and ŌłåmexBŌłåmexF, and investigated their sensitivity to plant-derived antimicrobial compounds, antibiotics, and virulence. Growth inhibition assays with KB soft agar plate showed that growth of the wild-type (WT) was inhibited by 5 ┬Ąl of 1 M catechol and 1 M coumarin but not by other plant-derived potential antimicrobial compounds tested including phytoalexins. The sensitivity to these compounds tended to increase in ŌłåmexB and ŌłåmexBŌłåmexF mutants. The ŌłåmexBŌłåmexF mutant was also sensitive to 2 M acetovanillone. The mexAB-oprM was constitutively expressed, and activated in the ŌłåmexF and ŌłåmexBŌłåmexF mutant strains. The swarming and swimming motilities were impaired in ŌłåmexF and ŌłåmexBŌłåmexF mutants. The flood inoculation test indicated that bacterial populations in all mutant strains were significantly lower than that of WT, although all mutants and WT caused similar disease symptoms. These results indicate that MexAB-OprM extrudes plant-derived catechol, acetovanillone, or coumarin, and contributes to bacterial virulence. Furthermore, MexAB-OprM and MexEF-OprN complemented each otherŌĆÖs functions to some extent.
Plants respond to infection by microbial pathogens, and express defense responses, including preexisting antimicrobial compounds, phytoanticipins, and de novo production of antimicrobial compounds, phytoalexins (Morrissey and Osbourn, 1999; VanEtten et al., 2001). In order to establish infection, phytopathogens need to eliminate the effects of these antimicrobial compounds to convert the antimicrobials to ineffective structures by enzymes, to convert the structure of target proteins to those not affected by antimicrobial substances, and to extrude antimicrobials outside the cell. To extrude antimicrobials, bacteria have five structural groups of multidrug resistance (MDR) efflux pump transporters: resistance-nodulation-cell division (RND), small multidrug resistance, multi-antimicrobial extrusion, the major facilitator superfamily, and ATP-binding cassette superfamilies (Hernando-Amado et al., 2016; Li et al., 2015).
Some of the most relevant roles of RND efflux pumps so far identified include bacterial virulence, plant-bacteria interactions, transport of quorum sensing molecules, and extrusion of various kinds of toxic compounds (Alcalde-Rico et al., 2016; Alvarez-Ortega et al., 2013). The RND efflux pump is a tripartite complex composed of an inner membrane protein (IMP), an outer membrane protein (OMP), and a periplasmic membrane-fusion protein (MFP). The genes encoding each subunit are usually found in a single operon. In Pseudomonas syringae, there are different operons for the RND efflux pump transporter, mexAB-oprM and mexEF-oprN. The mexA and mexE encode MFP, whereas mexB and mexF encode IMP, and oprM and oprN encode OMP. We previously investigated the gene expression profiles in P. syringae pv. tabaci 6605 (Pta 6605) wild-type (WT) and its mutant strains by microarray analysis (Taguchi et al., 2015), and found that mexAB-oprM is constitutively expressed, whereas mexEF-oprN is hardly expressed in the WT strain. However, these expressions were sometimes dramatically changed in the mutant strains. A ’¼éagellin-defective mutant Ōłå’¼éiC, a flagellin-glycosylation-defective mutant Ōłåfgt1, and an N-acyl-homoserine lactone (AHL) synthase-defective mutant ŌłåpsyI were incapable of synthesizing AHL and upregulating mexEF-oprN expression (Sawada et al., 2018; Taguchi et al., 2015). On the other hand, Ōłåvfr, a mutant of the virulence factor regulator, and ŌłåpilA, a mutant of pilin protein in type 4 pili upregulated the expression of mexAB-oprM (Taguchi and Ichinose, 2011, 2013). These results provided an insight: flagella-mediated and pili-mediated motility-defective mutants have to adapt to a given condition and change their gene expression profile, including mexAB-oprM and mexEF-oprN, for survival (Sawada et al., 2018; Taguchi and Ichinose, 2011; Taguchi et al., 2015). Thus, the expressions of MDR efflux pump-related genes are responsive to environmental and genetic changes.
The mutant strains of ŌłåmexAB-oprM of P. syringae pv. phaseolicola 1448A, pv. syringae B728a, and pv. tomato DC3000 had reduced populations after inoculation into their own host leaves, indicating that MexAB-OprM contributes to virulence (Stoitsova et al., 2008). The susceptibilities of these mutants to aminoglycosides, ╬▓-lactams, chloramphenicol, ethidium bromide, fluoroquinolones, macrolides, nitrofurantoin, quinolones, rifampin, and tetracycline were increased, indicating that these compounds are potential substrates of MexAB-OprM (Fernando and Kumar, 2013; Stoitsova et al., 2008). Characterization of an RND transporter system in P. syringae pv. syringae B301D, the PseABC efflux system, revealed that PseABC has an important role in secretion of syringomycin and syringopeptin, phytotoxins of this pathogen (Kang and Gross, 2005). However, the role of RND efflux pumps in the secretion of the plant antimicrobial metabolites is not clear yet. In this study, we generated two single mutants for two major RND efflux pumps, ŌłåmexB and ŌłåmexF, and a double mutant, ŌłåmexBŌłåmexF, and investigated the growth inhibition effect of plant-derived antimicrobials, such as phytoanticipins and phytoalexins. Furthermore, the effects of the mutation on the expression of these RND pump genes, motility, and virulence were characterized. The roles of MexAB-OprM and MexEF-OprN in bacterial virulence are discussed.
All bacterial strains used in this study are listed in Table 1. Pta 6605 strains, Escherichia coli strains, and Chromobacterium violaceum CV026 were grown as described previously (Ichinose et al., 2020).
An approximately 5.8 kb DNA fragment encoding mexAB-oprM was amplified with a set of primers, mexA5 and oprM3, using KOD FX DNA polymerase (Toyobo, Osaka, Japan) with the genomic DNA of Pta 6605 as a template, then introduced into a pCR-Blunt II-TOPO plasmid vector (Invitrogen, Carlsbad, CA, USA) to obtain pCR-mexAB-oprM (Fig. 1A). To delete the mexB region from the plasmid, inverse PCR was carried out with a set of primers, mexB-d5NheI and mexB-d3NheI, and self-ligated by a Ligation-Convenience kit (Nippon Gene, Tokyo, Japan) after NheI and DpnI digestions. The mutated genetic region was introduced into pK18mobsacB (Sch├żfer et al., 1994) via an EcoRI site to generate pK18-ŌłåmexB, then introduced into E. coli S17-1 for conjugation with Pta 6605 strains. Deletion mutants were obtained by conjugation and homologous recombination according to the previously reported methods (Taguchi et al., 2006). The DNA sequence of the mutated region in the bacterium was confirmed by DNA sequencing with a Big Dye terminator cycle sequencing kit and an ABI PRISM 3100 sequencer (Thermo Fisher Scientific, Waltham, MA, USA). Primer sequences were designed based on the sequence of Pta 6605 provided by Dr. Studholme, University of Exeter, UK.
Using a previously generated ŌłåmexF mutant (Sawada et al., 2018) as a recipient, mexB was deleted by a conjugation and homologous recombination as described above, and the ŌłåmexBŌłåmexF double mutant was generated.
An overnight culture was centrifuged at 1,500 ├Śg for 5 min at 4┬░C, the bacterial pellet was resuspended in LB medium with 10 mM MgCl2, and the OD660 was adjusted to 0.02. Three ml of the bacterial culture was incubated at 27┬░C with shaking, and the bacterial OD660 was measured with an OD meter at each time point.
Potential antibiotics, trans-cinnamic acid, naringenin, acetovanillone, (+)-catechin, catechol, scopoletin, and coumarin were obtained from Tokyo Chemical Industry Co. Ltd. (Tokyo, Japan), phloretin, chlorogenic acid, and p-coumaric acid were obtained from Sigma-Aldrich (St. Louis, MO, USA), and acetosyringone was obtained from Cayman Chemical (Ann Arbor, MI, USA). Phytoalexins, capsidiol, debneyol, and rishitin were purified in the plant pathology laboratory of Nagoya University.
Potential antibiotics and antibiotics were dissolved in sterilized and distilled water or dimethyl sulfoxide. Bacteria (3 ml) cultured overnight in KingŌĆÖs B (KB) medium were mixed with 50 ml of melted 0.6% agar KB medium, then poured into three Petri dishes.
Compound-absorbed paper discs were put on the Petri dish, and the dishes were incubated at 27┬░C. After 24 h incubation, the growth inhibition zone was photographed and measured using Adobe Photoshop CC version 19.1.9 (Adobe, San Jose, CA, USA). The photographic data was gray-scaled, then noise was removed with a high-pass filter to a middle value of brightness, the image was converted into two gradations, and the growth inhibition zone was measured.
Overnight cultured bacteria were harvested by centrifugation and further incubated in MMMF medium (50 mM potassium phosphate buffer, 7.6 mM (NH4)2SO4, 1.7 mM MgCl2, and 1.7 mM NaCl, pH 5.7, supplemented with 10 mM each of mannitol and fructose) for an additional 1 h.
Total RNA was extracted from the cells collected by centrifugation using Trizol (Thermo Fisher Scientific). For qRT-PCR, the cDNA was synthesized using 0.5 ┬Ąg of total RNA and ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo) according to the manufacturerŌĆÖs instructions. The speci’¼üc primer sets for mexA (mexA-F1 and mexA-R1), mexE (mexE-F1 and mexE-R1), and pcaG (pcaG-F and pcaG-R) were designed according to the registered sequences of Pta 6605 (Table 2). Real-time qRT-PCR was carried out using a GVP-9600 (Shimadzu, Kyoto, Japan) and KAPA SYBR FAST qPCR Master Mix (2├Ś) Kit (Sigma-Aldrich), with primers at a final concentration of 0.4 ┬ĄM each and 1 ┬Ąl of cDNA product as a template in a 20 ┬Ąl reaction mixture. PCR was performed with one denaturation cycle of 20 s at 95┬░C and 40 cycles of 15 s at 95┬░C and 30 s at 62┬░C. The expression of each gene was normalized by pcaG, which encodes protocatechuate 3,4-dioxygenase subunit alpha, as a stably expressed gene (Taguchi et al., 2015). Melting curve analysis confirmed that each PCR was a single product. The PCRs were performed with a high linearity. The data were analyzed using the ŌłåŌłåCT (cycle threshold) method.
An inoculation test of Pta 6605 was conducted with a flood assay (Ishiga et al., 2011) optimized for tobacco plants (Nicotiana tabacum L. cv. Xanthi). Tobacco seeds were sterilized and sown on 0.8% Murashige-Skoog (MS) agar plates containing 1% sucrose and vitamin stock solution (thiamin hydrochloride 3 mg/l, nicotinic acid 5 mg/l, pyridoxine hydrochloride 0.5 mg/l), and grown at 28┬░C under 16 h light:8 h dark conditions for 2 weeks. Tobacco seedlings were transplanted to 0.8% MS agar plates containing 0.1% sucrose and vitamin stock solution as above and grown for 2 days under the same conditions. Bacteria were grown overnight at 27┬░C in KB medium with 50 ╬╝g/ml of nalidixic acid (Nal). The bacterial inoculum was adjusted to OD600 = 0.002 with sterilized water containing 0.025% Silwet L-77 (Biomedical Sciences, Tokyo, Japan). Sterilized water containing 0.025% Silwet L-77 was used as a mock treatment. The inoculation test was carried out on a clean bench. The bacterial suspension (approximately 30 ml) was poured into the plate of tobacco seedlings. After about 10 s incubation, the bacterial suspension was decanted, and the plate was air-dried on a clean bench for 15 min. The plants were incubated under 16 h light:8 h dark conditions at 22┬░C, and disease symptoms were observed for 4 days post-inoculation (dpi) and 10 dpi. To determine the bacterial population at 0 dpi and 4 dpi, leaf disks were punched out using a disposable biopsy hole punch and then ground with mortar and pestle. The homogenates were serially diluted in sterile distilled water and then spread on KB plates containing Nal. The plates were dried and incubated at 27┬░C for 2 days, after which the bacterial population
was measured by counting the number of colonies (colony forming unit).
Swimming and swarming assays were performed as described previously (Taguchi and Ichinose, 2011). An overnight culture with 50 ┬Ąg/ml of Nal was centrifuged at 1,500 ├Śg for 10 min, the bacterial pellet was resuspended in 10 mM MgSO4, and the OD660 was adjusted to 1.0. For the swimming assay, 3 ╬╝l of bacterial suspension was inoculated on the center of 0.25% Bactoagar (Becton Dickinson & Company, Glencoe, MD, USA) (w/v) MMMF plates. For the swarming assay, 3 ╬╝l of the bacterial suspension was spotted on the center of a 0.45% Bactoagar (w/v) SWM (0.5% peptone and 0.3% yeast extract) plate and allowed to dry. Plates were incubated overnight at 23┬░C. Swimming and swarming motilities were observed after 24 h incubations.
Bacterial strains were grown in KB medium for 24 h at 27┬░C. AHLs were extracted with an equal volume of ethyl acetate and detected using C18 reversed-phase thin layer chromatography (TLC Silica gel 60, Merck, Darmstadt, Germany) and the biosensor C. violaceum CV026 (Taguchi et al., 2006) as described in Ichinose et al. (2020).
Statistical analysis was performed using One-way ANOVA DunnettŌĆÖs test and Tukey honestly significant difference test with R software version 3.6.1 (https://www.r-project.org/).
The specific deletion of mexB and/or mexF was confirmed by genomic PCR (data not shown). The growth of each bacterial strain in in vitro culture was compared. There was no significant difference in growth speed between the WT strain and the mutant strains (Fig. 1B).
Growth inhibition assays of the plant-derived potential antimicrobial compounds in KB soft agar plates showed that 5 ┬Ąl of 0.9 M trans-cinnamic acid, 0.5 M p-coumaric acid, 1 M acetosyringone, 1 M scopoletin, 1 M chlorogenic acid, 0.3 M phloretin, 1 M naringenin, and 200 mg/ml (+)-catechin did not produce any clear growth inhibition zone in all tested strains (data not shown). Furthermore, 5 ┬Ąl of 500
┬ĄM capsidiol, debneyol, and rishitin, phytoalexins of Nicotiana tabacum, Nicotiana debneyi, and Solanum tuberosum, respectively, also did not inhibit any bacterial growth (Supplementary Fig. 1). When the concentration of capsidiol was increased to 10 mM, no growth inhibition was observed (Supplementary Fig. 1). This concentration of phytoalexins completely inhibits the growth of oomycetes and fungal pathogens (Egea et al., 1996). However, 5 ┬Ąl of 1 M catechol and 1 M coumarin inhibited the growth of the Pta 6605 WT strain (Fig. 2A and B). Furthermore, the size of the growth inhibition zone by 1 M catechol increased in ŌłåmexF, and further expanded in ŌłåmexB and ŌłåmexBŌłåmexF. The ŌłåmexB and ŌłåmexBŌłåmexF mutant strains showed a slightly higher sensitivity than the WT strain to 1 M coumarin; however, the ŌłåmexF mutant produced a smaller inhibition zone than the WT strain. Although 5 ┬Ąl of 2 M acetovanillone did not inhibit growth of the WT strain, it inhibited those of ŌłåmexB and ŌłåmexF, and an additive growth inhibition effect was observed in the ŌłåmexBŌłåmexF mutant (Fig. 2C).
The sensitivity of bacterial strains to some bacteria-derived antibiotics was also investigated. The sensitivities to most of the antibiotics used in this study, such as kanamycin, spectinomycin, tetracycline, streptomycin, and hygromycin B, did not differ among the bacterial strains (data not shown). However, only ŌłåmexB and ŌłåmexBŌłåmexF were sensitive to 5 ┬Ąl of 50 mg/ml ampicillin, while the growths of the WT strain and ŌłåmexF were not inhibited by it (Fig. 2D). On the contrary, although the WT and ŌłåmexB mutant strains showed only weak growth inhibition zone to 5 ┬Ąl of 50 mg/ml chloramphenicol, the growth of ŌłåmexF and ŌłåmexBŌłåmexF mutant strains was remarkably inhibited (Fig. 2E). These results indicate that MexAB-OprM and MexEF-OprN specifically extrude ampicillin and chloramphenicol, respectively.
The effect of the deletion of mexB, mexF, or both mexB/mexF on mexA and mexE gene expression was analyzed by real-time qRT-PCR. As shown in Fig. 3A, the expression of mexA was enhanced in the ŌłåmexF and ŌłåmexBŌłåmexF strains. In addition, the expression of mexE was very weak, and seemed to be slightly increased in the ŌłåmexB strain (Fig. 3B).
Surface swimming and swarming motilities of the ŌłåmexB mutant were comparable to WT strain, whereas those of ŌłåmexF and ŌłåmexBŌłåmexF double mutant strains were abolished (Fig. 4A).
Pta 6605 utilizes AHLs as quorum-sensing molecules. AHLs production in ŌłåmexB and ŌłåmexBŌłåmexF mutants was reduced. However, the ŌłåmexF mutant retained AHL production ability as good as that of the WT strain (Fig. 4B).
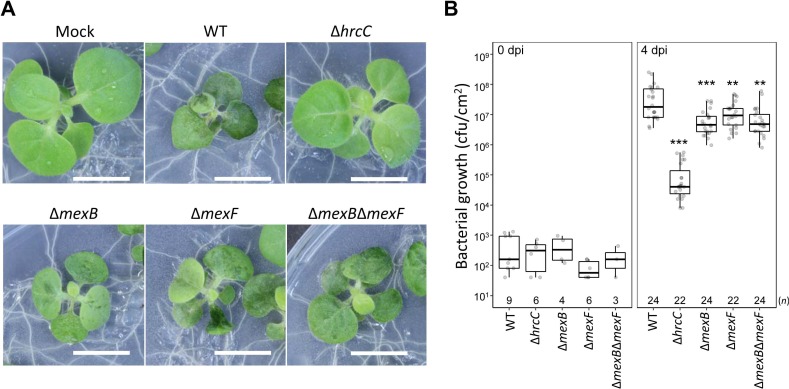
To investigate whether MexAB-OprM and/or MexEF-OprN of Pta 6605 is involved in the virulence, a flood inoculation test of the WT, ŌłåmexB, ŌłåmexF, ŌłåmexBŌłåmexF, and ŌłåhrcC mutant strains (Marutani et al., 2005) on host tobacco seedlings was performed. The ŌłåhrcC mutant did not show any disease symptoms, and bacterial growth was limited (Fig. 5). On the other hand, all mutants except ŌłåhrcC showed disease symptoms similar to those of the WT strain at 4 dpi (Fig. 5A) and 10 dpi (Supplementary Fig. 2). However, the bacterial populations in the leaves of ŌłåmexB, ŌłåmexF, and ŌłåmexBŌłåmexF mutant strains were significantly lower than that of WT at 4 dpi (Fig. 5B), indicating that MexAB-OprM and MexEF-OprN might partially contribute to virulence at the early stage of infection.
Stoitsova et al. (2008) reported that deletion mutants of mexAB-oprM of P. syringae pv. tomato DC3000, pv. phaseolicola 1448A, and pv. syringae B728a exhibited increased susceptibility to bacterial and plant-derived antimicrobials, detergents, and dyes. In this study, we focused on the functions of MexAB-OprM and MexEF-OprN on the plant-derived antimicrobials, phytoanticipins and phytoalexins in P. syringae pv. tabaci. The increased growth inhibition in ŌłåmexB and ŌłåmexBŌłåmexF of Pta 6605 by acetovanillone, catechol, and coumarin indicates that MexAB-OprM may extrude these compounds. Baker et al. (2015) reported that tobacco leaves accumulated acetovanillone, acetosyringone, and chlorogenic acid in response to the inoculation of P. fluorescens, P. syringae pv. tabaci, and P. syringae pv. syringae. Therefore, Pta 6605 may extrude these phytoanticipins during infection. Analysis of growth inhibition by formation of growth inhibition zones has the advantage that even hydrophobic substances can be absorbed in a paper disc and their antimicrobial activity examined. However, we did not observe antimicrobial activity in many plant-derived compounds with this method. Therefore, it is possible that some of these compounds might show an inhibitory activity with other methods.
It was reported that the expression of mexAB-oprM of P. syringae pv. tomato DC3000 increases in the presence of flavonoids such as phloretin and naringenin (Vargas et al., 2011). The mexA mutant exhibits higher susceptibility to these compounds and caused fewer symptoms compared to the WT strain in the inoculation test on tomato leaves (Vargas et al., 2011). Furthermore, the mexA mutant bore fewer flagella, and the presence of phloretin reduced the ratio of flagellated cells and swarming motility (Vargas et al., 2013). These results indicate that the MexAB-OprM transporter contributes to the colonization on host tomato plants. Although we observed similar swimming and swarming motilities in WT and ŌłåmexB mutant, the mutation of mexF resulted in the loss of these motilities, indicating relationship between MexEF-OprN and motility.
An extract of Arabidopsis thaliana contains 4-methylsulfinylbutyl isothiocyanate, a natural product derived from aliphatic glucosinolates, which inhibited the growth of non-host Pseudomonas bacteria (Fan et al., 2011). In contrast, P. syringae pathovars like P. syringae pv. tomato DC3000 that are pathogenic to A. thaliana can grow with the same extract from Arabidopsis because these pathovars have the sax (survival in Arabidopsis extract) genes. Parts of the sax genes encode MexAB-OprM and MexEF-OprN and confer the ability to extrude a wide range of substrates including antibiotics and host-derived molecules (Fan et al., 2011). The major phytoanticipin varies from plant to plant. Because acetovanillone, catechol, and coumarin inhibit the growth of Pta 6605, these compounds may be major phytoanticipins of tobacco, and both MexAB-OprM and MexEF-OprN contribute to the virulence of Pta 6605.
As Gnanamanickam and Mansfield (1981) already reported that isoflavonoid (kievitone and phaseollin), flavonoid (hydroxyflavans), furanoacetylenic (wyerone), and sesquiterpenoid (capsidiol and rishitin) phytoalexins do not inhibit the growth of a wide range of bacteria, none of the phytoalexins tested also showed an inhibitory effect on Pta 6605 or even ŌłåmexB, ŌłåmexF, and the ŌłåmexBŌłåmexF double mutant (Supplementary Fig. 1). Fifty ┬Ąg of capsidiol and rishitin did not inhibit the growth of six Gram-positive and eight Gram-negative bacteria including saprophytes and plant and animal pathogens. Camalexin, a phytoalexin in A. thaliana, was accumulated in response to the inoculation of P. syringae pv. syringae (Tsuji et al., 1992). However, a defective mutant that cannot synthesize the camalexin of A. thaliana still exhibits resistance to avirulent P. syringae pathogens, indicating that the accumulation of camalexin is not major defense response to bacterial pathogens (Glazebrook and Ausubel, 1994). These mutants displayed enhanced sensitivity to virulent P. syringae strains, suggesting that camalexin limits the growth of virulent bacteria (Glazebrook and Ausubel, 1994).
The mexEF-oprN of the WT strain is hardly expressed in the general culture conditions. However, it was weakly activated in the ŌłåmexB mutant. MexEF-OprN might compensate for the lost functionality of MexAB-OprM. MexAB-OprM and MexEF-OprN were expected to extrude acetovanillone, catechol, and coumarin. We will now investigate whether these compounds induce expressions of mexAB-oprM and mexEF-oprN genes.
The virulence of ŌłåmexB, ŌłåmexF, and ŌłåmexBŌłåmexF was reduced (Fig. 5), and swimming and swarming motilities of ŌłåmexF and ŌłåmexBŌłåmexF was impaired (Fig. 4A). Furthermore, ŌłåmexB and ŌłåmexBŌłåmexF mutants showed reduced production of AHL (Fig. 4B), suggesting that an RND-type multidrug efflux pump transporter not only directly extrudes antimicrobials but also indirectly controls other types of virulence, such as surface motility and the quorum sensing system. This may be the result of environmental adaptation of the mutant strains in the absence of MexAB-OprM or MexEF-OprN.
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
We would like to thank the Leaf Tobacco Research Labo- ratory of Japan Tobacco Inc. for providing Pta 6605. This work was supported in part by JSPS KAKENHI Grant Numbers 16K14861 and 19H02956.
References
Alcalde-Rico, M., Hernando-Amado, S., Blanco, P. and Mart├Łnez, J. L. 2016. Multidrug ef’¼éux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front. Microbiol. 7:1483


Alvarez-Ortega, C., Olivares, J. and Mart├Łnez, J. L. 2013. RND multidrug ef’¼éux pumps: what are they good for? Front. Microbiol. 4:7


Baker, C. J., Mock, N. M., Smith, J. M. and Aver'yanov, A. A. 2015. The dynamics of apoplast phenolics in tobacco leaves following inoculation with bacteria. Front. Plant Sci. 6:649



Egea, C., Alc├Īzar, M. D. and Candela, M. E. 1996. Capsidiol: its role in the resistance of Capsicum annuum to Phytophthora capsici. Physiol. Plant. 98:737-742.

Fan, J., Crooks, C., Creissen, G., Hill, L., Fairhurst, S., Doerner, P. and Lamb, C. 2011. Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Arabidopsis. Science. 331:1185-1188.


Fernando, D. M. and Kumar, A. 2013. Resistance-nodulation-division multidrug ef’¼éux pumps in Gram-negative bacteria: role in virulence. Antibiotics. 2:163-181.



Glazebrook, J. and Ausubel, F. M. 1994. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 91:8955-8959.



Gnanamanickam, S. S. and Mansfield, J. W. 1981. Selective toxicity of wyerone and other phytoalexins to Gram-positive bacteria. Phytochemistry. 20:997-1000.

Hernando-Amado, S., Blanco, P., Alcalde-Rico, M., Corona, F., Reales-Calder├│n, J. A., S├Īnchez, M. B. and Mart├Łnez, J. L. 2016. Multidrug ef’¼éux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resist. Updat. 28:13-27.


Ichinose, Y., Tasaka, Y., Yamamoto, S., Inoue, Y., Takata, M., Nakatsu, Y., Taguchi, F., Yamamoto, M., Toyoda, K., Noutoshi, Y. and Matsui, H. 2020. PsyR, a transcriptional regulator in quorum sensing system, binds lux box-like sequence in psyI promoter without AHL quorum sensing molecule and activates psyI transcription with AHL in Pseudomonas syringae pv. tabaci 6605. J. Gen. Plant Pathol. 86:124-133.

Ishiga, Y., Ishiga, T., Uppalapati, S. R. and Mysore, K. S. 2011. Arabidopsis seedling ’¼éood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions. Plant Methods. 7:32



Kang, H. and Gross, D. C. 2005. Characterization of a resistancenodulation-cell division transporter system associated with the syr-syp genomic island of Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 71:5056-5065.




Li, X.-Z., Pl├®siat, P. and Nikaido, H. 2015. The challenge of ef’¼éux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28:337-418.



Marutani, M., Taguchi, F., Shimizu, R., Inagaki, Y., Toyoda, K., Shiraishi, T. and Ichinose, Y. 2005. Flagellin from Pseudomonas syringae pv. tabaci induced hrp-independent HR in tomato. J. Gen. Plant Pathol. 71:289-295.

McClean, K. H., Winson, M. K., Fish, L., Taylor, A., Chhabra, S. R., Camara, M., Daykin, M., Lamb, J. H., Swift, S., Bycroft, B. W., Stewart, G. S. A. B. and Williams, P. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 143:3703-3711.


Morrissey, J. P. and Osbourn, A. E. 1999. Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 63:708-724.



Sawada, T., Eguchi, M., Asaki, S., Kashiwagi, R., Shimomura, K., Taguchi, F., Matsui, H., Yamamoto, M., Noutoshi, Y., Toyoda, K. and Ichinose, Y. 2018. MexEF-OprN multidrug efflux pump transporter negatively controls N-acyl-homoserine lactone accumulation in Pseudomonas syringae pv. tabaci 6605. Mol. Genet. Genomics. 293:907-917.


Sch├żfer, A., Tauch, A., J├żeger, W., Kalinowski, J., Thierbach, G. and P├╝hler, A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of de’¼üned deletions in the chromosome of Corynebacterium glutamicum. Gene. 145:69-73.


Stoitsova, S. O., Braun, Y., Ullrich, M. S. and Weingart, H. 2008. Characterization of the RND-type multidrug efflux pump MexAB-OprM of the plant pathogen Pseudomonas syringae. Appl. Environ. Microbiol. 74:3387-3393.



Taguchi, F. and Ichinose, Y. 2011. Role of type IV pili in virulence of Pseudomonas syringae pv. tabaci 6605: correlation of motility, multidrug resistance, and HR-inducing activity on a nonhost plant. Mol. Plant-Microbe Interact. 24:1001-1011.


Taguchi, F. and Ichinose, Y. 2013. Virulence factor regulator (Vfr) controls virulence-associated phenotypes in Pseudomonas syringae pv. tabaci 6605 by a quorum sensing-independent mechanism. Mol. Plant Pathol. 14:279-292.


Taguchi, F., Inoue, Y., Suzuki, T., Inagaki, Y., Yamamoto, M., Toyoda, K., Noutoshi, Y., Shiraishi, T. and Ichinose, Y. 2015. Characterization of quorum sensing-controlled transcriptional regulator MarR and Rieske (2Fe-2S) cluster-containing protein (Orf5), which are involved in resistance to environmental stresses in Pseudomonas syringae pv. tabaci 6605. Mol. Plant Pathol. 16:376-387.


Taguchi, F., Ogawa, Y., Takeuchi, K., Suzuki, T., Toyoda, K., Shiraishi, T. and Ichinose, Y. 2006. A homologue of the 3-oxoacyl-(acyl carrier protein) synthase III gene located in the glycosylation island of Pseudomonas syringae pv. tabaci regulates virulence factors via N-acyl homoserine lactone and fatty acid synthesis. J. Bacteriol. 188:8376-8384.



Tsuji, J., Jackson, E. P., Gage, D. A., Hammerschmidt, R. and Somerville, S. C. 1992. Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv syringae. Plant Physiol. 98:1304-1309.



VanEtten, H., Temporini, E. and Wasmann, C. 2001. Phytoalexin (and phytoanticipin) tolerance as a virulence trait: why is it not required by all pathogens? Physiol. Mol. Plant Pathol. 59:83-93.

Fig.┬Ā1
Generation of ŌłåmexB mutant stain and in vitro bacterial growth. (A) Schematic organization of mexAB-oprM in Pseudomonas syringae pv. tabaci 6605. PCR was carried out to isolate and clone the entire region of mexAB-oprM. Using pCR-mexAB-oprM as a template and mexB-d5NheI and mexB-d3NheI as primers, inverse PCR was carried out to obtain pCR-ŌłåmexB. (B) In vitro growth of each bacterium. The same density of bacterium (OD660 = 0.02) was used to incubate at 27┬░C with shaking, and bacterial OD660 was measured. The bars represent standard deviations for three independent experiments.

Fig.┬Ā2
Sensitivity of Pseudomonas syringae pv. tabaci 6605 wild-type (WT) and its mutant strains to several antimicrobial compounds. Growth inhibition on plates with 5 ┬Ąl of 1 M catechol (A), 1 M coumarin (B), 2 M acetovanillone (C), 50 ┬Ąg/ml ampicillin (D), and 50 ┬Ąg/ml chloramphenicol (E). N.D., not detected. Photographs were taken at 24 h after incubation. Data shown below photographs are the averages of three independent replicates of growth-inhibited areas (mm2). Statistical groups were determined using TukeyŌĆÖs honestly significant difference test (P < 0.05), and significant differences are indicated by different letters below right of each photograph.
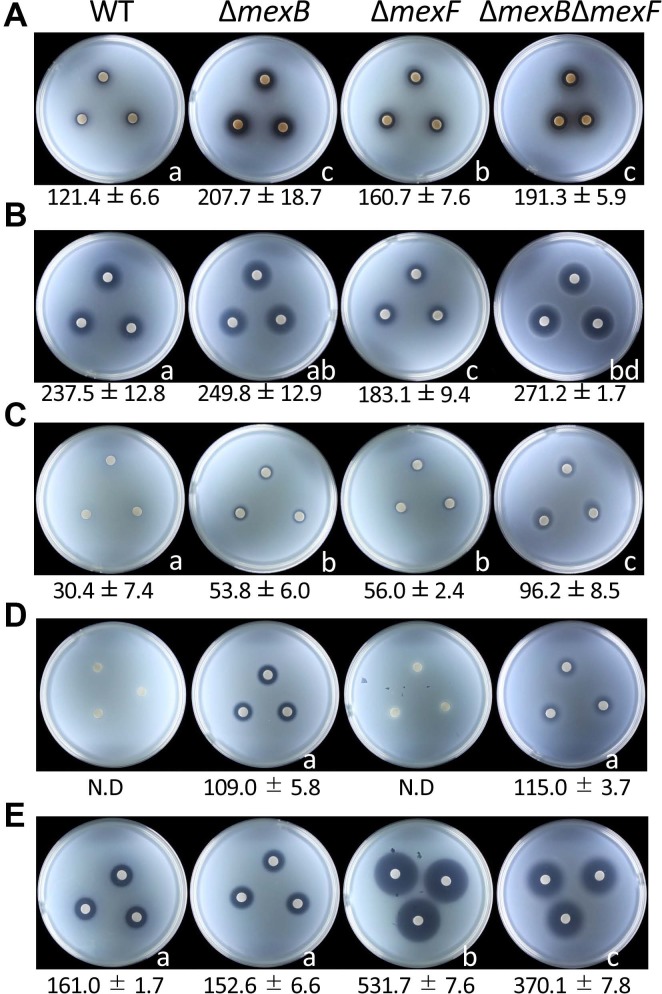
Fig.┬Ā3
Quantitative reverse transcription PCR analysis of gene expression of mexA (A) and mexE (B) in wild-type (WT) and mutant strains. Asterisk indicates significant difference from the WT (***P < 0.001) determined by one-way ANOVA DunnettŌĆÖs test.
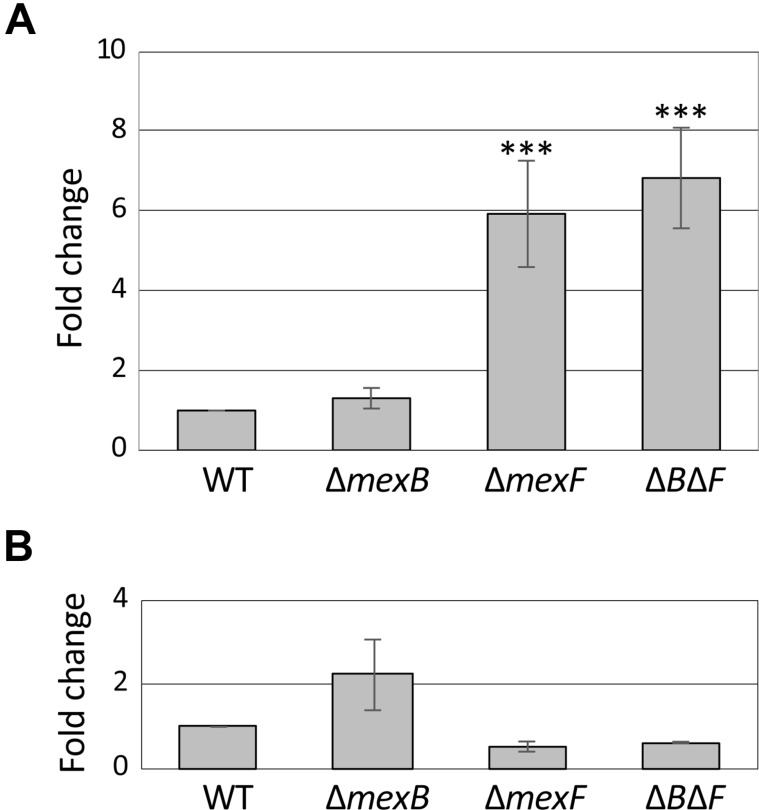
Fig.┬Ā4
Swarming and swimming motilities and N-acyl-homoserine lactone (AHL) production. (A) Surface swarming and swimming motilities after 24 h incubation. (B) AHL production. Ethyl acetate extract from 0.2 ml of each bacterial culture was spotted on the thin layer chromatography plate. AHL was visualized as described in ŌĆ£Materials and MethodsŌĆØ.
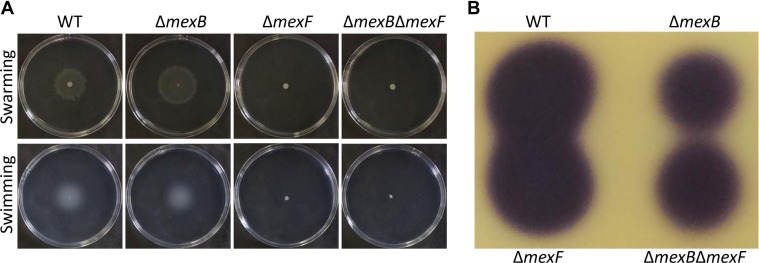
Fig.┬Ā5
Inoculation test. (A) Tobacco seedlings were inoculated with wild-type (WT) or ŌłåhrcC, ŌłåmexB, ŌłåmexF, ŌłåmexBŌłåmexF, and ŌłåhrcC strains and incubated at 23┬░C. Photographs taken 4 days post-infection (dpi) shows representative results. Scale bars = 1 cm. (B) Bacterial populations were determined at 0 or 4 dpi. Box plots and Jitter plots representing the bacterial growth in inoculated plants from three biological replicates. Boxes show the upper and lower quartiles of the data, and horizontal black lines represent the medians. Jitter plots indicate raw data. Statistical significance was determined using one-way ANOVA DunnettŌĆÖs test where each group was compared with the WT. (**P < 0.01, ***P < 0.001). n is shown above the stain names.
Table┬Ā1
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Escherichia coli strain | ||
| DH5╬▒ | F-, ╬╗-, ├Ė80dLacZ╬öM15, ╬ö(lacZYA-argF)U169, recA1, endA1, hsdR17(rK-mK+), supE44, thi-1, gyrA, relA1 | Takara Bio |
| S17-1 | Thi, pro, hsdR-, hsdM+, recA [chr::RP4-2-Tc::Mu-Km::Tn7] | Sch├żfer et al. (1994) |
| Chromobacterium violaceum CV026 | Double mini-Tn5 mutant from C. violaceum ATCC 31532; AHL biosensor, Kmr | McClean et al. (1997) |
| P. syringae pv. tabaci | ||
| Isolate 6605 | Wild-type, Nalr | Taguchi et al. (2006) |
| ŌłåhrcC | Isolate 6605 ŌłåhrcC, Nalr | Marutani et al. (2005) |
| ŌłåmexB | Isolate 6605 ŌłåmexB, Nalr | This study |
| ŌłåmexF | Isolate 6605 ŌłåmexF, Nalr | Sawada et al. (2018) |
| ŌłåmexBŌłåmexF | Isolate 6605 ŌłåmexBŌłåmexF, Nalr | This study |
| Plasmid | ||
| pCR Blunt II TOPO | Cloning vector for blunt ended PCR products, Kmr | Thermo Fisher Scientific |
| pCR-mexAB-oprM | pCR Blunt II TOPO with 5,789-bp of mexAB-oprM, Kmr | This study |
| pCR-ŌłåmexB | mexB-deleted plasmid from pCR-mexAB-oprM, Kmr | This study |
| pK18mobsacB | Small mobilizable vector, Kmr, sucrose-sensitive (sacB) | Sch├żfer et al. (1994) |
| pK18ŌłåmexB | pK18mobsacB with mexA-oprM, Kmr | This study |
Table┬Ā2
Oligonucleotide sequences used in this study
- TOOLS
-
METRICS

- Related articles
-
Role of
Metcalfa pruinosa as a Vector forPseudomonas syringae pv.actinidiae 2017 December;33(6)



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



