 |
 |
| Plant Pathol J > Volume 30(2); 2014 > Article |
Abstract
A comprehensive study was conducted using PPSMV resistant (BSMR 736) and susceptible (ICP 8863) genotypes to develop a segregating population and understand the inheritance of PPSMV resistance. The observed segregation was comparable to 13 (susceptible): 3 (resistant). Hence, the inheritance was controlled by two genes, SV1 and SV2, with inhibitory gene interaction.
Pigeonpea (Cajanus cajan (L.) Millsp.) is an important pulse crop for the tropical and subtropical regions of South Asia (mainly on the Indian-subcontinent), Africa, the Caribbean and Latin America, where it is serving as significant protein source to the human diet. Pigeonpea has self-compatible cleistogamous flowers, but it is often cross-pollinated by bees to an extent of 10-15 per cent. It plays an important role in food and nutritional security of human being, as it is a rich source of seed proteins (20-25 per cent), minerals and vitamins (Mallikarjuna and Saxena, 2005). Pigeonpea is relatively drought tolerant and thrives well in both deep and shallow soils. The world acreage of pigeonpea is 4.90 mha with an annual production of about 4.22 million metric tonne worth about 1.5 billion US dollars: India is the largest producer and consumer of pigeonpea with an annual production of 2.46 mt followed by Myanmar 0.54 mt and Malawi 0.16 mt (FAOSTAT 2010; http://faostat.fao.org).
The biotic stresses are the major economic concerns for pigeonpea yield, which are Fusarium wilt (FW), Pigeonpea Sterility Mosaic Disease (PSMD) and Phytopthora Blight (PB). These are serious challenges for sustainable pigeonpea production to meet the demands of resource constrained people of several African and Asian countries. PSMD is the most important foliar disease of pigeonpea in India and Nepal (Reddy and Vishwa Dhar, 2000). Kumar et al. (2000) reported a tenui-like virus of asymmetric morphology as the cause of PSMD and retained the name of virus as Pigeonpea sterility mosaic virus (PPSMV). PPSMV is transmitted by an eriophyid mite (Aceria cajani Channabasavanna). The disease is characterized by the symptoms like bushy and pale green appearance of plants followed by reduction in leaf size, excess vegetative growth, increasing number of secondary branches and mosaic mottling of leaves and finally partial or complete cessation of reproductive structures. Sometimes, some parts of the plant may show disease symptoms and other parts may remain unaffected (Kumar et al., 2003).
Development of resistant varieties is complicated in view of the genetic plasticity of the pathogens in pigeonpea (Gnanesh et al., 2011). In India, comprehensive studies on variability in the sterility mosaic pathogen have revealed the occurrence of five different isolates of the PPSMV (Reddy et al., 1993). Amongst them three distinct isolates like Bangalore, Patancheru and Coimbatore have been characterised. Wherein, the Patancheru and Coimbatore isolates have been identified as mild strains, while the Bangalore isolate as the most virulent in causing PSMD (Kulkarni et al., 2003). Knowledge of the genetic basis of yield, resistance to diseases, insect pests and abiotic stress tolerance are important factors for deciding appropriate breeding strategies for genetic improvement in pigeonpea. There are conflicting reports about the genetics of resistance to sterility mosaic disease depending on the source of resistance/cross being investigated. However, many studies have been reported susceptibility as dominant and resistance to be under the control of recessive genes (Gnanesh et al., 2011; Ganapathi et al., 2012; Nagaraj et al., 2004; Singh et al., 2003).
Development of PSMD resistant genotypes or breeding lines not only serves the direct use in breeding programmes but also in exploiting the heterosis using existing cytoplasmic genic male sterility system (Gnanesh et al., 2012; Saxena et al., 2006). Recently, Gnanesh et al. (2011) investigated the nature of resistance to PPSMV for virulent strain, Bangalore and mild strain, Patancheru isolates. In the present study, we report the nature of inheritance of PSMD resistance against Bidar isolate of PPSMV in ICP 8863 x BSMR 736 cross.
Two genotypes, BSMR 736 and ICP 8863 were used as parents to generate the appropriate segregating population to study the inheritance of resistance character against PPSMV. The BSMR 736 is known to show resistance to Gulbarga isolate of PPSMV over years and ICP 8863 is indeterminate, mid-late and susceptible genotype to PSMD (Gnanesh et al., 2011b; Saxena et al., 2010). The leaf stapling technique (Nene and Reddy, 1977) was employed to screen the parental lines for their reaction to PPSMV at ARS Gulbarga and ARS Bidar.
Selected two parents were grown separately, and individual flowers of female parent ICP 8863 were hand emasculated and pollinated with the pollen dust from resistant male parent BSMR 736 in cool hours of the day. The pollinated flowers were bagged to avoid further out crossing through honeybees and other insect pollinators. The F1 plants were raised and mature seeds were collected and sown to get advanced F2 generation. The F1 hybrids of this cross were susceptible indicating susceptibility to be dominant over resistance. A set of 225 individual plants from ICP 8863×BSMR 736 cross were grown in the field under pollination control (nylon net) cages to prevent insect entry and possible cross pollination. The mature seeds from individual F2 plants were collected to constitute F2:3 families and the same were subjected for the field evaluation.
Field evaluation of F2:3 families was done at Agriculture Research Station (ARS), Bidar; the hot-spot for PSMD of pigeonpea. The plot chosen for this experiment was positioned to have sugarcane field on one side and rice field on the other side, to favour the manifestation of PSMD. A set of 225 F2:3 families were field evaluated to assess their reaction to PPSMV infection and PSMD development. The individual families were sown in two rows each with 10-15 plants per rows (in a row of 2 m length). Individual F2:3 families were sown contiguously with ICP 8863 as check in regular intervals to serve as spreader of PPSMV. Recommended package of practices were followed to raise the crop in such a way that the manifestation of PSMD is not affected.
Mild type PPSMV Bidar isolate was used first time for artificial inoculation in the present study. The PPSMV artificial inoculation in the field was done according to “leaf stapling” and “infector-hedge” techniques (Nene and Reddy, 1976). Before actual sowing of F2:3 families of this cross, ‘infector-hedge rows’ of susceptible cultivar (ICP 8863) were established. An infector-hedge consisting of two widely spaced rows of the susceptible cultivar sown one month prior to actual planting experiment and also sown in regular intervals of test sample that serve as spreader of disease in the field. Ten days old seedlings of the hedge were inoculated with leaf stapling. The PPSMV infected pigeonpea leaves along with mites were collected and one diseased leaflet per primary leaf was generally used. The diseased leaflet was folded on the primary leaf in such a way that its lower surface came into contact with the primary leaf of the test seedlings. It was then stapled with a small paper stapler. Alternatively, two diseased leaflets were used if they were too small. The leaflets were placed in such a way that the lower surface of one of the leaves came in contact with the lower surface of the primary leaf while the lower surface of the other was in contact with the upper surface of the primary leaf. The primary leaf and the two diseased leaflets were then stapled together. As the stapled leaflets from the infected plants get dried mites from the infected leaves migrate to healthy leaf and inoculates the virus. Ratooning is known to favour the manifestation of PSMD. Hence, the experimental F2:3 families of this cross were cut from first branch position from ground and irrigated to encourage the sprouting and further vegetative development. Observations were again recorded on ratoon crop.
The infected plants were scored for PSMD incidence at 15 days interval up to 75 days before and after ratooning. The total number of plants, number of plant with and without disease symptoms in each F2:3 families were counted and observations were recorded. The data was entered in a spread sheet and mean disease incidence (in percent) was calculated. Accordingly, the F2:3 families of this cross, were classified as resistant, moderately resistant and susceptible based on the percentage of disease incidence and further grouped based on the following standard scale as 0-10 per cent of plants infected-resistant; 10.1-30 per cent of plants infected-Moderately resistant; 30.1-100 per cent of plants infected-susceptible (Singh et al., 2003).
The ratio of F2:3 families to PSMD were corroborated with respect to F2 individual plant. The Chi-square (χ2) test was performed to test the goodness of fit of the segregating F2:3 families with the expected phenotypic ratios.
Resistance and susceptibility to PSMD of both parents were conformed during 2005, 2006 and 2007. The rows of resistant parent BSMR 736 showed 100 percent resistance with no visible symptoms while, the susceptible parent rows of ICP 8863 exhibited 100 percent infection with severe PSMD symptoms (Table 1). The hedge rows coupled with artificial inoculation of diseased leaves together with the mites successfully manifested the PSMD. Over 10-15 days old hedge border rows and infector rows were infected with PPSMV using stapling of diseased leaves. These approaches could successfully manifest the disease symptoms development in more than 80 percent of the plants (Fig. 1). Large number of mites could be observed in any diseased leaf of both hedge rows and boarders in the experimental field.
The F2:3 families planted in the field were essentially derivative of respective F2 individual plants. The phenotypic observations for PSMD incidence were recorded according to the disease scale given by Singh et al. (2003). Reaction of F2:3 families of ICP 8863×BSMR 736 cross are presented in Fig. 2. A total of 225 F2:3 families were field evaluated; 37 showed resistant phenotype, 160 were moderately resistant and 28 families were observed to be susceptible for PPSMV (Table 2). The progenies recorded array of disease symptoms including bushy and pale green appearance of leaves, reduction in chlorophyll contain, reduced plant size, increased number of secondary branches, mosaic mottling of leaves and complete or partial cessation of reproductive structures etc. and plants with these symptoms were categorised as susceptible ones. Some F2:3 families were found to be segregating, among which 40-60 per cent of the progenies were susceptible and rest were resistant. Similarly, a few F2:3 families having 80-100 per cent resistant progenies within them were also recorded (Fig. 3). Finally, for convenience the F2:3 families showing moderate resistant or susceptible phenotype to PPSMV were combined together into susceptible category. The F2:3 families with resistance phenotype was categorised as resistant. The statistical analysis resulted in an observed segregation ratio of 188:37 (susceptible: resistant) (P<0.05) for studied cross, indicating more families with susceptible reaction. The segregation pattern in F2:3 families of ICP 8863×BSMR 736 cross were comparable with 13:3 (susceptible: resistant). Based on the observed segregation ratio it was suggestive that the PPSMV resistance is under two gene control with non-allelic interaction of the type ‘inhibitory gene interaction’. The proposed genotypes for resistance and susceptibility in the parents, F1 and F2 generations of this cross, are mentioned in Table 3. The goodness of fit for expected and observed values as tested by χ2 test with = 0.7 P = 3.8 are presented in Table 4.
Conventional plant breeding methods have been effective in bringing about improvement in crops but efforts are still being made to develop more efficient breeding methods to overcome specific problems. PSMD is the most destructive disease of pigeonpea (Kannaiyan et al., 1984) causing yield losses up to 95 percent (Ganapathy et al., 2011; Reddy and Nene, 1981). The early stage (<45-days old plants) of infection results into 95 to 100 percent yield losses as reported by Reddy et al. (1990); Kulkarni et al. (2003). The disease results in 100 percent yield loss when symptoms appear at the pre-flowering and podding stage. Whereas, at maturity stage loss could be of 67 percent and at pre-harvest stage up to 30 percent. Seeds from partially infected plants appear discolored, shrivelled and results into 20 percent reduction in dry weight while annual yield loss exceeds US $ 100 million in India alone (Kumar et al., 2000). Manifestation of PSMD chiefly depends on the availability of mite populations (Singh et al., 1999). The mite populations are usually positively correlated with rainfall, relative humidity and lower temperature. In the present study, the experimental plot chosen was positioned to have one side sugarcane field and another side the paddy crop. Both crops alongside to the pigeonpea experimental field are known to create favourable climatic condition for the built-up of mite populations and manifestation of PSMD. Together with ‘infector hedge row’ and ‘leaf stapling’ techniques (Nene and Reddy, 1976), the PSMD manifestation in the present experiment was excellent. In many other studies, similar efforts have resulted in manifestation of PSMD both in field and maintained conditions in pigeonpea (Gnanesh et al., 2011; Srinivas et al., 1997).
In leaf stapling technique diseased leaflet per primary leaf was stapled. As a result, the stapled leaflets from the infected plants get dried and mites leave the infected leave to migrate to healthy leaf and itransmit the virus. The PPSMV susceptible genotype, ICP 8863, was used to developed infector hedge rows on the border of the experimental field for the pathogen and mites multiplied to sufficient threshold on the hedge plants and successfully served as source of PPSMV inoculums. Generally the mites are carried through wind onto the test rows in the field. These mites served as a natural source of PPSMV inoculants in experimental field throughout the PSMD screening period. The ratooned and perennial pigeonpea is known to have highest PSMD incidence. In case of late infection of PPSMV, symptoms may not occur even if the genotype is susceptible to PPSMV. However, when plants are ratooned symptoms appear predominantly on the new growth (Jones et al., 2004; Kumar et al., 2002). The phenotyping against PSMD is destructive and was avoided in F2 generation so that seeds can be harvested from these plants to raise the advanced generation or F2:3 families. Recently, the similar experimental approaches have been employed by Gnanesh et al. (2011). All F3 progeny derived from the same F2 plant belongs to the same F2:3 family and denoted by F2:3. Observations in parents and F2:3 families indicated dominance of susceptibility over resistance.
Insufficient durability of resistance to PSMD is a major concern in pigeonpea breeding. Durable resistance that remains effective when a cultivar possessing it is widely cultivated. The knowledge of inheritance and number of genes governing the traits is essential for understanding the nature of inheritance of the characters of interest for an efficient breeding programme and development of resistant cultivars. There are conflicting reports about the genetics of resistance to sterility mosaic disease claiming both susceptibility and resistance to be dominant. However, in most cases susceptibility was shown to be dominant and resistance to be under the control of recessive genes (Singh et al., 2003). The resistance to PSMD has been reported to be controlled by single recessive gene (Ganpathy et al., 2009; Murugesan et al., 1997; Srinivas et al., 1997) and oligogenic (Gnanesh et al., 2011; Nagaraj et al., 2004; Sharma et al., 1984). Recently, Gnanesh et al. (2011) identified four QTLs for Patancheru PSMDV isolate and two QTLs for Bangalore PSMDV isolate. Present study, revealed that the susceptible F2:3 families were more compared to resistant ones. Many studies have reported the similar kind of resistance: susceptibility ratio for PSMD in pigeonpea (Amala balu, 1992; Ganapathy et al., 2009; Nagaraj et al., 2004; Singh et al., 1983; Sharma et al., 1984).
Based on the field reaction of F2:3 families, the results were suggestive that the PPSMV resistance (for Bidar isolate) is governed by two gene designated as SV1 and SV2. The dominant allele of one gene (SV1) has inhibitory action on the trait (resistance) govern by other (SV2) gene. Based on these assumption the presence of dominant allele of SV1 gene in one locus suppresses the action of dominant allele of SV2 (resistance) gene present on another locus resulting in susceptible phenotype. Only those plants with recessive allele of SV1 gene and dominant allele of SV2 gene might have shown resistant phenotype. On the basis of this hypothesis we proposed genotypes that might be possible for PPSMV resistance and PPSMV susceptible phenotypes in pigeonpea plants (Table 3).
Genetics of PSMD has been depend on the resistance source, PSMV isolates and scoring methods and hence the resistance to PSMD in pigeonpea appears to be complex (Saxena, 2008). The results of present experiment revealed that resistance to be governed by two independent nonallelic genes in studied cross. There may be additional genes for PSMD resistance in pigeonpea other than the two proposed in the present study. Understading the inheritance of PSMD is expected to aid the pigeonpea breeding and identification DNA markers linked to the PSMD resistance/susceptiblity.
Acknowledgments
Authors acknowledge the Department of Biotechnology (DBT) of the Government of India and Indian Council of Agricultural Research (ICAR) for the financial support to undertake this study. The encouragement and suggestions of Dr. N.K Singh, Dr. P.M. Salimath and Dr Rajeev Varshney are acknowledged.
Fig. 2.
Frequency distribution of per cent disease incidence for Bidar PPSMV isolate in 225 F2:3 families derived from a cross ICP 8863×BSMR 736.
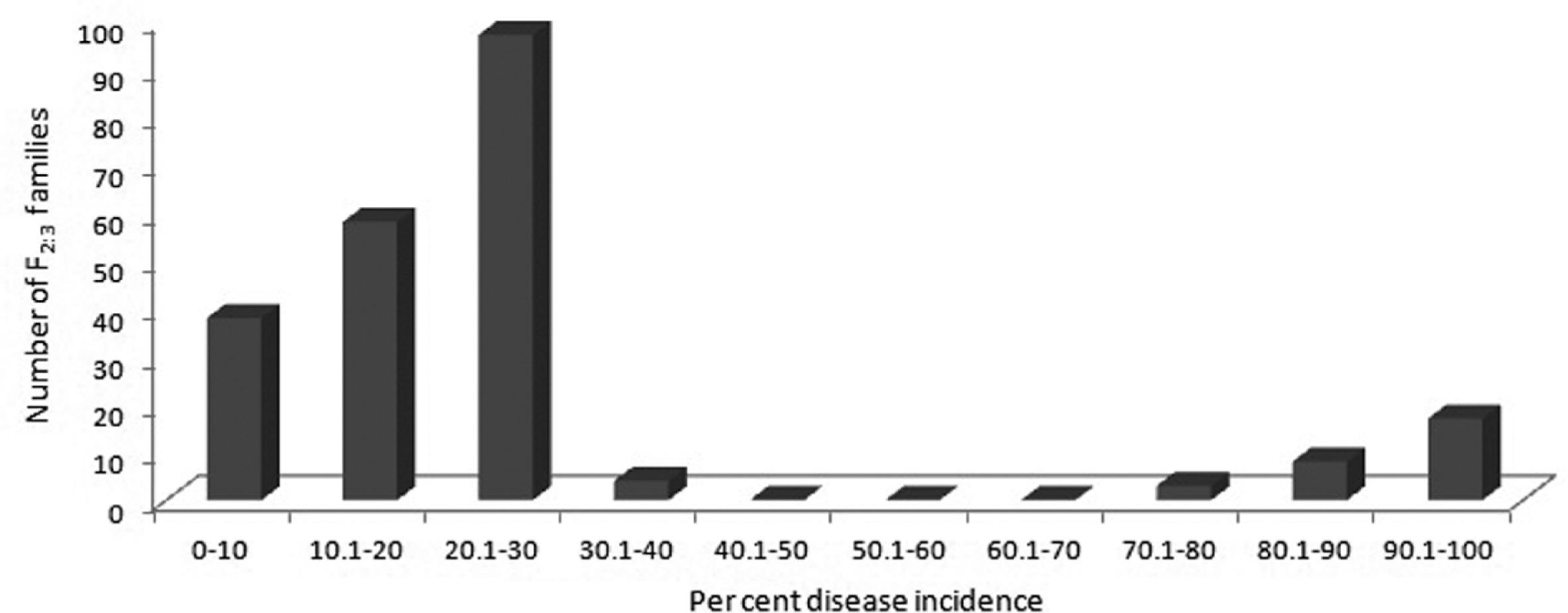
Fig. 3.
Resistance and susceptibility reactions of F2:3 families to PSMD incidence during kharif season of 2009.
(a) Resistant F2:3 families (b) Segregant F2:3 families (c) Susceptible F2:3 families (d) Typical PSMD symptoms include bushy and pale green leaves, excess vegetative growth, leaf size reduction, and mosaic and mottling of leaves.

Table 1.
Resistant and susceptible reaction of parental genotypes to PSMD during 2005, 2006 and 2007 screening experiment
Table 2.
Number of F2:3 families of the ICP 8863×BSMR 736 cross in the PSMD score range of resistant, moderately resistant and susceptible based on disease incidence during 2009
| Per cent disease incidence range | Phenotypic scored | No. of F2:3 families |
|---|---|---|
| 1 to10% of plant infected | R | 37 |
| 10.1 to 30% of plants infected | MR | 160 |
| 31.1 to 100% of plants infected | S | 28 |
|
|
||
| Total | 225 | |
Table 3.
Proposed genotypes of parents and F2 for PSMD resistance
References
Amala, BP and Rathnasamy, R 2003. Inheritance to sterility mosaic disease in pigeonpea [Cajanus cajan(L.) Millsp]. Crop Res. 25:301-304.
Ganapathy, KN, Gowda, BM, Ajay, BC, Venkatesha, SC, Gnanesh, BN, Gomashe, SS, Prasanth, BG, Girish, PS, Prasad, GN, Veerakumar, and Pati, J V 2012. Inheritance studies of sterility mosaic disease (SMD) resistance in vegetable type pigeonpea (Cajanus cajan(L.) Millsp.). Aust J Crop Sci. 6:1154-1158.
Ganapathy, KN 2009 Genetic analysis and identification of DNA markers linked to sterility mosaic disease resistance in pigeonpea [Cajanus cajan(L.) Millsp]. Ph.D (Agri.) Thesis. UAS, Bangalore.
Ganapathy, KN, Gnanesh, BN, Byre, GM, Venkatesha, SC, Gomashe, SS and Channamallikarjuna, V 2011. AFLP analysis in pigeonpea (Cajanus cajan(L.) Millsp.) revealed close relationship of cultivated genotypes with someof its wild relatives. Genet Resour Crop Evol. 58:837-847.


Gnanesh, BN, Ganapathy, KN, Ajay, BC and Byre Gowda, M 2011. Inheritance of sterility mosaic disease resistance to Bangalore and Patancheru isolates in pigeonpea (Cajanus cajan(L.) Millsp.). Electron J Plant Breed. 2:218-223.
Gnanesh, BN, Bohra, A, Sharma, M, Byregowda, M, Pande, S, Wesley, V and et al 2011b. Genetic mapping and quantitative trait locus analysis of resistance to sterility mosaic disease in pigeonpea [Cajanus cajan(L.) Millsp.]. Field Crops Res. 123:56-61.

Jones, T, Kumar, I K, Saxena, B, Kulkarni, N K and Muniyappa, V 2004. Sterility mosaic disease - the “Green Plague” of pigeonpea. ICRISAT Plant Dis. 88:436-445.

Kannaiyan, J, Nene, YL, Reddy, MV, Ryan, G and Raju, T N 1984. Prevalence of pigeonpea disease and associated crop losses in Asia Africa and America. Trop Pest Manag. 30:62-71.
Kulkarni, NK, Reddy, AS, Kumar, PL, Vijayanarasimha, J, Rangaswamy, KT, Muniyappa, V, Reddy, LJ, Saxena, KB, Jones, AT and Reddy, DV 2003. Broad based resistance to pigeonpea sterility mosaic disease in accessions of Cajanus scarbaeoides(L.) Benth. Ind J Plant Pro. 31:6-11.
Kumar, PL, Duncan, GC, Roberts, IM, Jones, AT and Reddy, DVR 2002. Cytopathology of Pigeonpea sterility mosaic virus in pigeonpea and Nicotiana benthamiana: similarities with those of eriophyid mite borne agents of undefined aetiology. Ann Appl Biol. 140:87-96.
Kumar, PL, Jones, AT and Reddy, DVR 2003. A novel mite transmitted virus with a divided RNA genome closely associated with pigeonpea sterility mosaic disease. Phytopathology. 93:71-81.


Kumar, PL, Jones, AT, Sreenivasulu, P and Reddy, DVR 2000. Breakthrough in the identification of the causal agent of pigeonpea sterility mosaic disease. J Mycol Plant Pathol. 30:249.
Mallikarjuna, N and Saxena, KB 2005. A new cytoplasmic nuclear male-sterility system derived from cultivated pigeonpea cytoplasm. Euphytica. 142:143-148.


Murugesan, S, Murugan, E and Nadarajan, N 1997. Inheritance of duration leaf colour sterility mosaic disease resistance and growth habit in Pigeonpea. Madras Agric J. 84:10-12.

Nagaraj, KM, Chikkadevaiah, and Kulkarni, RS 2004. Inheritance of resistance to sterility mosaic virus in Pigeonpea (Cajanus cajan(L.) Millsp.). Indian J Genet. 64:118-120.
Nene, YL and Reddy, MV 1976. A new technique to screen pigeonpea for resistance to sterility mosaic. Trop Grain Legume Bul. 5:23.
Nene, YL and Reddy, MV 1977. Leaf stapling technique to screen pigeonpea for resistance to sterility mosaic. Indian Phytopath. 30:153.
Reddy, MV, Raju, TN, Nene, YL, Ghanekar, AM, Amin, KS, Arjunan, G, Astaputre, JV, Sinha, BK, Reddy, SV, Gupta, RP and Gangadharan, K 1993. Variability in sterility mosaic pathogen in pigeonpea in India. Indian Phytopathol. 46:206-212.
Reddy, MV, Sharma, SB and Nene, YL 1990. Pigeonpea: Disease management. 303-347. In: The Pigeonpea, eds. by YL Nene, SD Hall and VK Sheila, CAB International, Wallingford, UK.
Reddy, MV and Nene, YL 1981. Estimation of yield loss in pigeonpea due to sterility mosaic. Proc. International workshop on Pigeonpeas. 2:15 19 December 1980. ICRISAT Center, Patancheru, AP, India. 305-312.
Reddy, MV and Vishwa Dhar, 2000. Current approaches and new dimensions in the management of pigeonpea diseases. In: Advances in management of biotic and abiotic stresses in pulse crops, eds. by M Ali, AS Asthana, YS Rathore, SN Gurha, SK Chaturvedi and S Gupta, Indian Institute of Pulses Research, Kanpur, Indian Society of Pulses Research and Development (ISPRD), Kanpur, India.
Saxena, KB, Kumar, RV, Madhavi, LK and Dalvi, VA 2006. Commercial pigeonpea hybrids are just a few steps away. Indian J Pulses Res. 19:7-16.
Saxena, RK, Saxena, K, Kumar, RV, Hoisington, DA and Varshney, RK 2010. Simple sequence repeat-based diversity in elite pigeonpea genotypes for developing mapping populations to map resistance to Fusarium wilt and sterility mosaic disease. Plant Breed. 129:135-141.

Sharma, D, Gupta, SC, Ral, GS and Reddy, MV 1984. Inherltance of resistance to sterility mosaic disease in pigeon-pea. Indian J Genet Plant Breed. 44:84-90.
Singh, AK, Rathi, YPS and Agrawal, KC 1999. Sterility mosaic of pigeonpea: a challenge of the 20th century. Indian J Virol. 15:85-92.
Singh, BV, Pandya, BP, Gautam, PL, Beniwal, SPS and Pandey, MP 1983. Inheritance of resistance to sterility mosaic virus in pigeonpea. Ind J Gene. 43:487-493.
Singh, IP and Vishwadhar, DRP 2003. Inheritance of resistance to sterility mosaic in pigeonpea (Cajanus cajan). Indian J Agric Sci. 73:414-417.
Singh, N, Gupta, D, Jayaswal, P, Mahato, A, Dutta, S, Singh, S, Bhutani, S, Dogra, V, Singh, B, Kumawat, G, Pal, J, Pandit, A, Singh, A, Rawal, H, Kumar, A, Prashat, R, Khare, A, Yadav, R, Raje, R, Datta, S, Singh, M, Wanjari, K, Fakrudin, B, Kansal, R, Dash, P, Jain, P, Bhattacharya, R, Gaikwad, K, Mohapatra, T, Srinivasan, R and Sharma, T 2011. The first draft of the pigeonpea genome sequence. J Plant Biochem Biotech. 21:98-112.


Srinivas, T, Reddy, MV, Jain, KC and Reddy, MSS 1997. Studies on inheritance of resistance and allelic relationships for strain of Pigeonpea sterility mosaic pathogen. Ann Appl Biol. 130:105-110.
- TOOLS
-
METRICS

- Related articles
-
Association Analysis of Charcoal Rot Disease Resistance in Soybean2019 June;35(3)




 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print



