Genetic Diversity of a Natural Population of Apple stem pitting virus Isolated from Apple in Korea
Article information
Abstract
Apple stem pitting virus (ASPV), of the Foveavirus genus in the family Betaflexiviridae, is one of the most common viruses of apple and pear trees. To examine variability of the coat protein (CP) gene from ASPV, eight isolates originating from 251 apple trees, which were collected from 22 apple orchards located in intensive apple growing areas of the North Gyeongsang and North Jeolla Provinces in Korea, were sequenced and compared. The nucleotide sequence identity of the CP gene of eight ASPV isolates ranged from 77.0 to 97.0%, while the amino acid sequence identity ranged from 87.7 to 98.5%. The N-terminal region of the viral CP gene was highly variable, whereas the C-terminal region was conserved. Genetic algorithm recombination detection (GARD) and single breakpoint recombination (SBP) analyses identified base substitutions between eight ASPV isolates at positions 54 and 57 and position 771, respectively. GABranch analysis was used to determine whether the eight isolates evolved due to positive selection. All values in the GABranch analysis showed a ratio of substitution rates at non-synonymous and synonymous sites (dNS/dS) below 1, suggestive of strong negative selection forces during ASPV CP history. Although negative selection dominated CP evolution in the eight ASPV isolates, SLAC and FEL tests identified four possible positive selection sites at codons 10, 22, 102, and 158. This is the first study of the ASPV genome in Korea.
Recombination is the process by which segments of genetic information switch between nucleotide strands of different genetic variants during replication (García-Arenal et al., 2001). Plant RNA viruses are known to have high mutation and recombination rates due to the absence of proofreading activity in their RNA replicase (Drake and Holland, 1999; Bruyere et al., 2000; Malpica et al., 2002). However, this mutability does not necessarily result in a high degree of genetic variability within their populations, as distribution in the virus population of genetic variants depends on two major evolutionary processes: genetic drift and selection (García-Arenal et al., 2001). Genetic drift is a random effect that occurs during the transmission of genetic traits to new generations, while selection is a directional process by which variants that are fittest in a certain environment will increase their frequency in the population (positive selection) and less fit variants will decrease their frequency (negative or purifying selection) (García-Arenal et al., 2001).
We analyzed the genetic diversity of a natural population of Apple stem pitting virus (ASPV) isolated from apple trees in Korea. ASPV is the type member of the Foveavirus genus in the family Betaflexiviridae (Martelli et al., 2007; Martelli and Jelkmann, 1998), and is one of the most common viruses of apple and pear trees worldwide (Kundu, 2003). ASPV is known to cause disease associated with stem pitting, vein yellowing, and graft incompatibility such as top-working disease in apple (Jelkmann, 1997). Previous studies have reported the genetic diversity of ASPV isolates (Komorowska et al., 2011; Schwarz and Jelkmann, 1998; Yoshikawa et al., 2000). Recently, Komorowska et al. (2011) detected recombination events and evolutionary pressures in ASPV isolates from apple and pear in Poland. In Korea, stem pitting symptoms were first reported in 1973 (Oh et al., 1973); however, genomic studies of ASPV have not been performed. To explore the evolutionary history of ASPV spread in apple trees in Korea, we surveyed ASPV infections in 251 apple trees collected from 22 apple orchards located in intensive apple growing areas through the North Gyeongsang and North Jeolla Provinces from July to September, 2012.
To detect ASPV using RT-PCR, RNA was extracted from one leaf from each tree using a CF11 cellulose (Whatman, Kent, UK) column, as described previously (Chung et al., 2009). A pair of primers was designed based on the ASPV sequence (GenBank accession no. D21829). The forward primer sequence was homologous to nucleotides 7932–7955 (5′-CAATTGCAATAGGTGCGTTCAATC-3′) of the ASPV isolate, while the reverse primer sequence was complementary to nucleotides 9201–9224 (5′-AATGCAGGAGAATTAATTAACTAA-3′) of the ASPV isolate. Complementary DNA (cDNA) synthesis was performed as follows: a mixture of 10 ng of RNA in 9 μl of nuclease-free water with 1 μl of 10 pM reverse primer was heated at 70°C for 5 min, followed by the addition of 4 μl of 5X reaction buffer, 2.5 mM MgCl2, 0.25 mM each dNTP, 1 μl of ImProm II reverse transcriptase (Promega, Madison, WI, USA), and 1 μl of RNase inhibitor (1 U/μl), with incubation at 37°C for 1 h. PCR amplification was performed in a 50-μl volume containing 20 μl of cDNA solution, 0.2 mM each dNTP, 2 mM MgCl2, 1 μl of 10 pM each primer, 2.5 U of DNA polymerase (Promega), and 1X PCR buffer. A total of 40 cycles were conducted in a PTC-0220 Perlitier Thermal Cycler (MJ Research, Waltham, MA, USA). The thermal conditions were as follows: denaturation at 94°C for 30 s (1 min for the first cycle), annealing at 45°C for 30 s, and extension at 72°C for 1 min. The final extension was performed at 72°C for 10 min.
The PCR-amplified CP gene was cloned using pGEM-T easy (Promega) according to the manufacturer’s instructions. The ligation mixture was used to transform competent Escherichia coli JM 109 cells. Recombinants were screened using the blue and white screening method (Sambrook et al., 1989). Nucleotide sequences were determined using an ABI Prism BigDye™ Terminator Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, USA). The CP gene sequences were aligned using the CLUSTAL method of DNASTAR Lasergene version 11 (Madison, WI, USA). In this report, eight ASPV isolates (SJY-HRYC1, JGH-HR-YC, NJS-HR-GW, KJH-HR-JS1, KJHHR-JS2, KJH-HR-JS3, SJY-HR-YC2, and JGS-HR-SJ) were identified and the CP gene nucleotide sequences from the isolates were registered under GenBank accession numbers KC791783–790 (Table 1). The ASPV CP gene is 1,191 nucleotides in length. The nucleotide sequence identity of the CP gene between isolates ranged from 77.0 to 97.0%, while the amino acid sequence identity ranged from 87.7 to 98.5%. The N-terminal region of the viral CP gene was highly variable, whereas the C-terminal region was conserved (Fig. 1). Among the apple trees infected with ASPV, cv. Hongro infection mixed with other apple viruses showed symptoms of chlorosis along the veins of leaves (Fig. 2), but other apple cultivars infected by only ASPV were symptomless (data not shown).
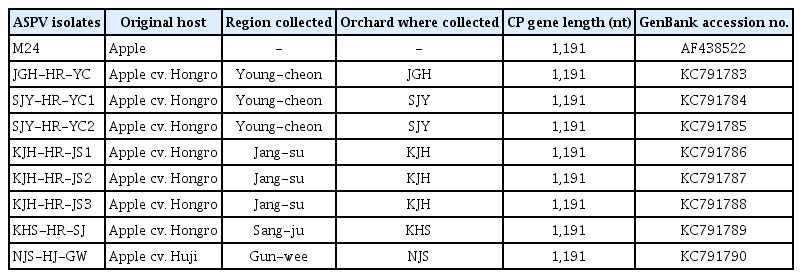
Information on the identified Apple stem pitting virus (ASPV) isolates from apple orchards in Korea. M24 was used to compare variations in the coat protein (CP) amino acid sequences

Multiple alignment of the amino acid sequences of the coat protein (CP) gene from Apple stem pitting virus. Amino acid sequence changes are indicated by a square box. The N-terminal region of the CP gene was highly variable, whereas the C-terminal region was conserved.

Apple cultivar ‘Hongro’ mixed infection with Apple stem pitting virus, Apple chlorotic leafspot virus, and Apple stem grooving virus showing chlorosis along the leaf veins.
To identify recombination events in the CP sequences, genetic algorithm recombination detection (GARD; www.datamonkey.org/GARD) and single breakpoint recombination (SBP) analyses were performed (Kosakovsky Pond et al., 2006). GARD identified possible recombination sites within the ASPV CP gene sequences at positions 54 and 57, while SBP identified the most probable recombination site at position 771 (Fig. 3). This result suggested that there were base substitutions causing the replacement of G with A at position 54, A with G at position 57, and T with C at position 771.

GARD and SBP analyses identified possible recombination sites within the ASPV CP sequences at positions 57 and 540 and position 771, respectively (replacement of A with G at position 57, replacement of G with A at position 54, and replacement of T with C at position 771).
Evolutionary pressures on proteins are often quantified based on the ratio of substitution rates at non-synonymous (amino acid altering) and synonymous (silent) sites (dNS/dS) (Kryazhimskiy and Plotkin, 2008; Nei and Gojobori, 1986). Thus, to determine whether the ASPV isolates evolved due to positive selection, the dNS/dS ratios in the CP coding regions were estimated using the GABranch method (Kosakovsky Pond et al., 2006). This method can automatically partition all branches of the phylogeny describing non-recombinant data into groups according to the dNS/dS ratio. Based on our results, all values in this GA-Branch analysis showed dNS/dS ratios below 1, indicative of negative selection during the history of ASPV CP (Fig. 4). The dNS/dS values calculated for the CP gene from the eight ASPV isolates were in the range of those calculated for CP sequences of other members of the family Flexiviridae (Komorowska et al., 2011). Despite the high variability of the CP gene sequences, all values in this GABranch analysis showed omega values below 1. Thus, high levels of variability in the CP gene sequences resulted from a high rate of synonymous mutations. Other viruses showed low dNS/dS values in the family Flexiviridae, despite the high level of variability in the CP sequences (Chare and Holmes, 2005; Teycheney et al., 2005).
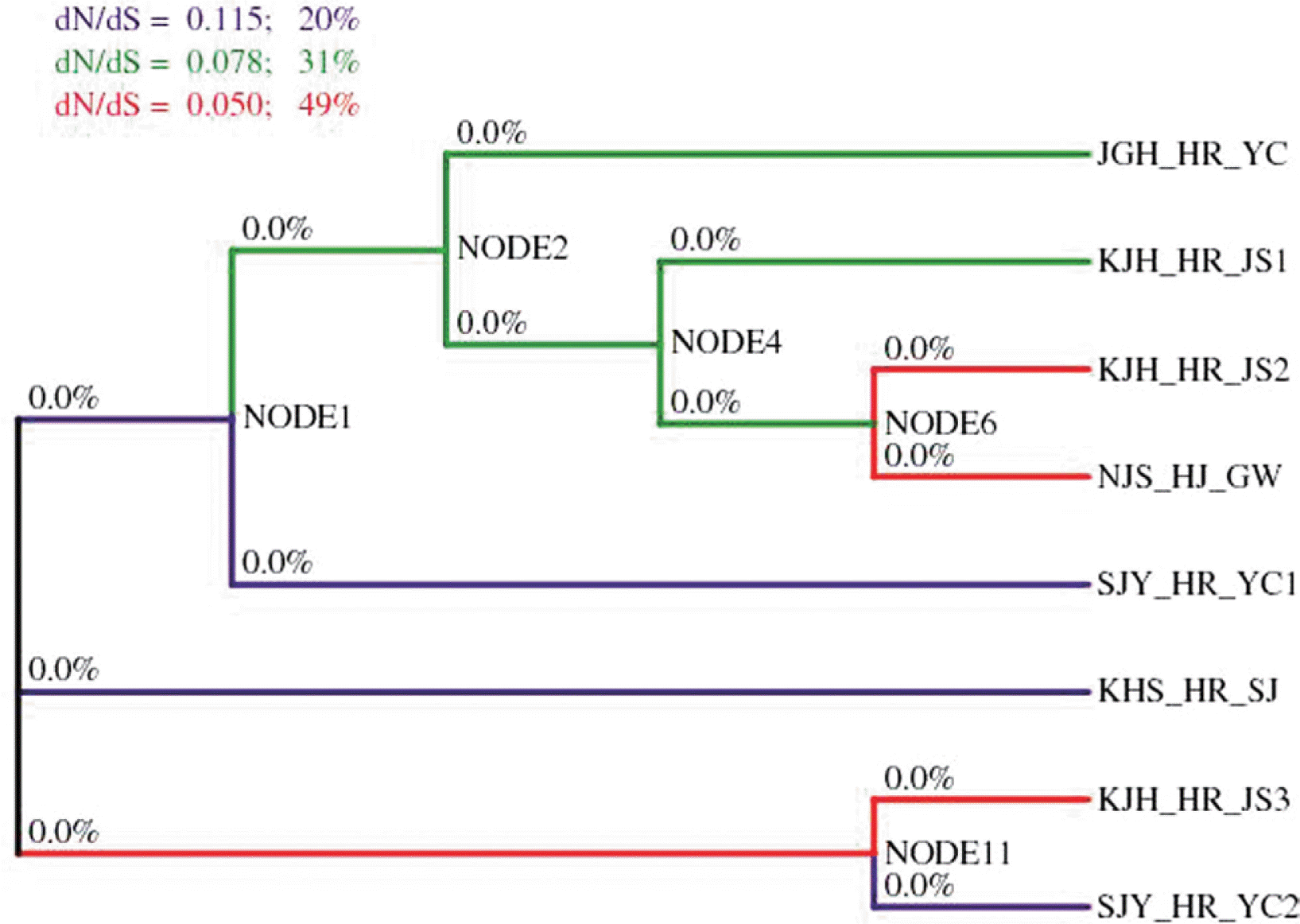
GABranch analysis showing a neighbor-joining tree with levels of omega coded by color. Purple branches represent the highest dNS/dS values, while green and red branches indicate medium and low values, respectively. All values in this GA-Branch analysis show an omega below 1, suggestive of strong purifying selection forces during the history of the ASPV CP gene.
To identify sites under positive or negative selection for each codon, SLAC, FEL, and REL were performed (Kosakovsky Pond and Frost, 2005a, 2005b). These methods can be used to estimate the ratio of non-synonymous and synonymous substitutions at each site. A consensus scoring approach was applied to determine the site most likely experiencing positive selection. Although purifying (negative) selection dominated the evolution of the CP gene in the eight ASPV isolates, all three methods showed that codons 10, 22, 102, and 158 were under positive selection (Table 2). These results suggest that various codons are under different selection forces in ASPV Korean isolates.
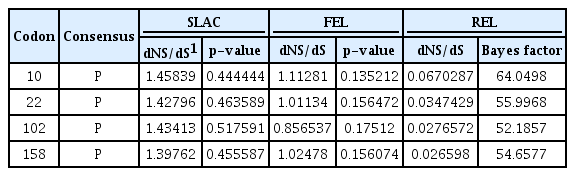
Results obtained from SLAC, FEL, and REL analyses, which were used to identify sites under positive or negative selection in the CP gene of ASPV. For each method, two values were provided (dNS/dS and p-values), with the exception of the REL method, which generates a Bayes factor instead of a p-value. The consensus column indicates a certain codon under positive selection: “P”, positive selection; “N”, negative selection
Here, we reported the genetic diversity of a natural population of ASPV originating from apple orchards located in intensive apple growing areas in Korea. Komorowska et al. (2011) reported that the majority of mutations in CP gene sequences from ASPV isolates collected from Poland were deleterious. In agreement with these results, although the Korean ASPV isolates showed strong variation in their CP gene sequences (even those originating from the same orchard), purifying (negative) selection dominated during evolution of the CP gene from eight ASPV isolates. Meanwhile, our results also show that although purifying selection dominated the evolution of the CP gene in the eight ASPV isolates, various codons were under different selection forces. This is the first study of the ASPV genome in Korea.