Rpi-blb2-Mediated Hypersensitive Cell Death Caused by Phytophthora infestans AVRblb2 Requires SGT1, but not EDS1, NDR1, Salicylic Acid-, Jasmonic Acid-, or Ethylene-Mediated Signaling
Article information
Abstract
Potato Rpi-blb2 encodes a protein with a coiled-coil-nucleotide binding site and leucine-rich repeat (CC-NBS-LRR) motif that recognizes the Phytophthora infestans AVRblb2 effector and triggers hypersensitive cell death (HCD). To better understand the components required for Rpi-blb2-mediated HCD in plants, we used virus-induced gene silencing to repress candidate genes in Rpi-blb2-transgenic Nicotiana benthamiana plants and assayed the plants for AVRblb2 effector. Rpi-blb2 triggers HCD through NbSGT1-mediated pathways, but not NbEDS1- or NbNDR1-mediated pathways. In addition, the role of salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) in Rpi-blb2-mediated HCD were analyzed by monitoring of the responses of NbICS1-, NbCOI1-, or NbEIN2-silenced or Rpi-blb2::NahG-transgenic plants. Rpi-blb2-mediated HCD in response to AVRblb2 was not associated with SA accumulation. Thus, SA affects Rpi-blb2-mediated resistance against P. infestans, but not Rpi-blb2-mediated HCD in response to AVRblb2. Additionally, JA and ET signaling were not required for Rpi-blb2-mediated HCD in N. benthamiana. Taken together, these findings suggest that NbSGT1 is a unique positive regulator of Rpi-blb2-mediated HCD in response to AVRblb2, but EDS1, NDR1, SA, JA, and ET are not required.
Many late blight resistance (R) genes were introgressed into potato cultivars from the wild species (Solanum sp.) through breeding strategies (Vleeshouwers et al., 2011). Among the potato R genes, the Rpi-blb1 and Rpi-blb2 genes were cloned from the diploid species (Solanum bulbocastanum). These genes could confer durable resistance to Phytophthora infestans (van der Vossen et al., 2003; 2005). Especially, a potato Rpi-blb2, an Mi-1 homolog conferring resistance to nematodes in tomato, was cloned from S. bulbocastanum (van der Vossen et al., 2005). Rpi-blb2 has a coiled-coil (CC)-domain in its N-terminus with nucleotide-binding and leucine-rich-repeats (NB-LRRs), and recognizes P. infestans AVRblb2, activating effector-triggered immunity (ETI; Oh et al., 2009). AVRblb2 can function in Avr activity that directly activates hypersensitive cell death (HCD) and associated disease-resistance responses mediated by Rpi-blb2 (Oh et al., 2014). HCD is a sign of induced defense responses and resembles programmed cell death in animals (Jones and Dangl, 2006). Little is known, however, about the AVRblb2 and Rpi-blb2 recognition mechanism or defense responses in plants (Bozkurt et al., 2011; Oh et al., 2009; 2014).
Mutational and knock-down analyses in plants led to the identification of proteins downstream of the R proteins (Botër et al., 2007; Kadota et al., 2010). For example, SGT1 (suppressor of the G2 allele of skp1), RAR1 (Required for Mla12 resistance), and HSP90 (Heat Shock Protein 90) interact with R proteins or are required for R functions (Kadota et al., 2010). Specifically, the ubiquitin ligase-associated protein SGT1 is required for the function of R genes (Austin et al., 2002; Azevedo et al., 2006). SGT1 homologues in Nicotiana benthamiana are indispensable for Rx-dependent HCD in response to Potato virus X (PVX) and R3a-dependent HCD in response to AVR3aKI (Azevedo et al., 2006; Bos et al., 2006; Peart et al., 2005). Rpi-blb2-mediated late blight resistance or HCD required SGT1 in N. benthamiana indicating that the knockdown of SGT1 in plants results in reduced R gene function (Oh et al., 2014).
Moreover, other resistance signaling components are enhanced disease susceptibility-1 (EDS1) and non-race-specific disease resistance (NDR1) proteins (Bhattacharjee et al., 2011; Century et al., 1997). EDS1 functions as a regulator of basal resistance and ETI mediated by Toll-interleukin-1 (TIR)-NB-LRR proteins, including RPP1 and RPP4 genes in response to oomycete pathogens or N gene in response to Tobacco mosaic virus (Aarts et al., 1998; Bhattacharjee et al., 2011; Peart et al., 2002). In contrast, NDR1 is required for disease resistance to bacterial pathogens expressing avr genes and several oomycete pathogens (Century et al., 1997; Tornero et al., 2002). In addition, NDR1 is required for R-proteins that contain the CC-NB-LRR domain, such as RPS2, RPM1, and RPP5 (Aarts et al., 1998; Century et al., 1997; Tornero et al., 2002). However, there is no information available about Rpi-blb2-mediated HCD- or resistance-signaling pathways.
Salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) play major functions in the regulation of plant defense responses to various pathogens (Glazebrook, 2005). Moreover, these molecules are involved in the regulation of different kinds of plant cell death responses (Mur et al., 2008; Rao et al., 2000). Specifically, SA acts synergistically with reactive oxygen species (ROS) accumulation to drive HCD during ETI in plants (Mur et al., 2008). Likewise, in R-gene triggered responses, SA and SA-pathway amplifications are required for HCD and defense-related gene functions, including HCD in response to many R genes (Glazebrook, 2005; Oh et al., 2014). Therefore, to elucidate the role of SA, JA, and ET signaling components in Rpi-blb2 gene-mediated HCD, we used virus-induced gene silencing (VIGS) technology with ICS1 (Isochorismate synthase 1, enzyme for SA biosynthesis), COI1 (Coronatine insensitive 1, JA-insensitive), and EIN2 (Ethylene insensitive 2) genes in Rpi-blb2-transgenic plants
To understand the signaling pathway of Rpi-blb2-mediated HCD caused by AVRblb2, we examined the molecular functions of Rpi-blb2 in N. benthamiana using Agrobacterium-mediated gene expression or VIGS methods. Through gain-of-function and loss-of-function studies, we demonstrate the necessity of SGT1, but not EDS1, NDR1, or the SA- JA-, and ethylene-mediated signaling pathways, for eliciting Rpi-blb2-mediated HCD in response to the P. infestans effector AVRblb2.
Materials and Methods
Bacteria, plants, and culture conditions
Escherichia coli DH5α and Agrobacterium tumefaciens GV3101 were grown in LB media with appropriate antibiotics at 37°C and 28°C, respectively. P. infestans 88069 was cultured on rye agar medium supplemented with 2% sucrose at 20°C. Homozygous transgenic N. benthamiana expressing Rpi-blb2 under the control of native promoters were obtained using pBINplus based plasmids described by van der Vossen et al (2005). All N. benthamiana seedlings were grown in a growth chamber and maintained at 22–25°C under a16/8 hr light-dark photoperiod.
RT-PCR analysis
Total RNA was extracted from N. benthamiana using TRI reagent, according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). RT-PCR was performed with equal amounts of total RNA using the ReverTraAce RT-PCR kit (TOYOBO Co.Osaka, Japan). Time courses of P. infestans infection on detached N. benthamiana leaves were performed using agar plugs as described by elsewhere (van der Vossen et al., 2005). Total RNA isolated from infected leaves of N. benthamiana, 6 days after inoculation (dai), from non-infected leaves (0 dai), and genomic DNA isolated from P. infestans mycelium grown in synthetic medium (My) was amplified with primer sets from the two genes. The oligonucleotides used to amplify P. infestans elongation factor 2 alpha [Table S1, (Oh et al., 2009)]. The expression of PiEF2α gene was controlled with primer pair, which is specific for the constitutively expressed NbActin gene (Table S1).
In planta HR cell death assays
For agroinfiltration assays, recombinant A. tumefaciens was grown as described elsewhere (van der Hoorn et al., 2000). A. tumefaciens carrying the respective constructs were mixed in a 2:1 ratio in inducing media (10 mM MgCl2, 10 mM MES, pH 5.6 and 150 μM acetosyringone), and then incubated at room temperature for 3 hr before infiltration. Agrobacterium solutions were infiltrated at an adjusted OD600 of 0.4 (Oh et al., 2009). Transient co-expression of Rpi-blb2 and AVRblb2 was performed by mixing the appropriate Agrobacterium in induction buffer at a ratio of 2:1 (final OD600 of 0.6:0.3). For HR assays, Agrobacterium expressing the pGR106-AVRblb2 or controls (pGR106-dGFP) were infiltrated with a final OD600 of 0.3 into transgenic N. benthamiana plants, respectively. Full length- or deleted-Rpi-blb2 clones were co-infiltrated with AVRblb2 in 20 inoculation sites per five plants.
Virus-induced gene silencing in N. benthamiana.
For the VIGS assay, cultures containing Agrobacterium with a TRV derivative vector (TRV2-dGFP, -NbSGT1, NbEDS1, NbNDR1, NbICS1, NbCOI1, and NbEIN2 constructs) and Agrobacterium GV3101 (TRV1) were transferred into LB media with antibiotics (50 mg/L kanamycin, 25 mg/L rifampicin) and grown at 28°C to an A600 of 0.8. The culture containing Agrobacterium (with TRV2-dGFP, -NbSGT1 constructs) were mixed in a 1:1 ratio (OD600=0.3) with GV3101 (TRV1) and infiltrated into the expanded leaves of 4-week-old wild type and all transgenic N. benthamiana (Liu et al., 2002). On day 21 after inoculation, transient expression of pGR106 or pGR106-AVRblb2 was performed by infiltration A. tumefaciens carrying each individual gene in an induction buffer at a ratio of 1:1 (final OD600 of 0.6). For RT-PCR analysis, total RNA was extracted from control (dGFP) and NbSGT1, NbEDS1, NbNDR1, NbICS1, NbCOI1, and NbEIN2-silenced N. benthamiana leaves using the TRIZOL solution (Invitrogen, Carlsbad, CA). Primers used to amplify NbSGT1, NbEDS1, NbNDR1, NbICS1, NbCOI1, and NbEIN2 annealed outside the VIGS target region (Table S1).
Results and Discussion
ETI strengthens basal defenses and elicits R proteins through the recognition of pathogen-derived effectors in plants (Jones and Dangl, 2006). To understand the function of Rpi-blb2-mediated HCD in response to the P. infestans effector AVRblb2, we developed gain-of-function Rpi-blb2-transgenic and loss-of-function Rpi-blb2-silenced N. benthamiana plants. The Rpi-blb2-transgenic plants were resistant to P. infestans, which is in agreement with studies in tomato and potato plants (van der Vossen et al., 2005). Therefore, we further characterized Rpi-blb2-mediated HCD using Rpi-blb2-transgenic and Rpi-blb2-silenced N. benthamiana plants.
Full-length Rpi-blb2 is required for AVRblb2-specific HCD induction and confers late blight resistance
Previously, we reported that AVRblb2 is an AVR effector that triggers Rpi-blb2-dependent HCD or resistance responses in N. benthamiana. Additionally, a 34–amino acid region in the C-terminal region of AVRblb2 is sufficient for triggering Rpi-blb2 hypersensitivity (Oh et al., 2009; 2014). However, little is known about the regions of Rpi-blb2 that govern this response. We used a N. benthamiana transient expression assay to define the regions of the Rpi-blb2 gene that are necessary for induction of cell death. We constructed full-length (CC-NBS-LRR and Rpi-blb2), N-terminal (CC domain), and C-terminal (LRR domain) deletion constructs in a 35S promoter Agrobacterium binary vector and assayed them in N. benthamiana (Fig. 1A).

The Rpi-blb2 are required to initiate HR and confer resistance to late blight disease. Deletion mutants of Rpi-blb2 were coexpressed with AVRblb2 by agro-infiltration in N. benthamiana to determine the Rpi-blb2 domains required for induction of AVRblb2–derived HR. (A) Schematic of the different mutant and deletion constructs is shown on the left. HR index with plus and minus signs indicate the presence and absence of AVRblb2-Rpi-blb2 function, respectively. Accumulation of auto fluorescent phenolic compounds associated with cell death induced by the Rpi-blb2 and AVRblb2 combination (Green) and the Rpi-blb2 and AVRblb2 combination were visualized under ultraviolet (UV) light. (B) Full-length Rpi-blb2 genomic DNA, and CC-NBS-LRR, N- (CC domain) and C-terminal (LRR domain) deletion constructs in a 35S promoter Agrobacterium binary vector. Amplification of Rpi-blb2-gDNA from pBINplus-gRpi-blb2 plasmid (Left) and RT-PCR analysis of the expression levels of pBINplus-gRpi-blb2-expressing (cDNA) N. benthamiana (Right). The NbActin gene was used as a control. (C) Typical disease symptoms after inoculation of N. benthamiana leaves with P. infestans. (D) Quantitative RT-PCR of P. infestans colonization levels in Rpi-blb2-expressing and WT plants. The expression of PiEF2a was controlled with primer NbActin, which is specific for the constitutively expressed NbActin gene. (E) Leaf area covered with disease lesion in Rpi-blb2-expressing and WT after inoculation with P. infestans. Bars indicate standard deviation (SD). All assays were repeated at least three times with similar results. CC, Coiled-coil; NBS, Nucleotide-Binding Sites; LRR, Leucine Rich Repeats; My, P. infestans mycelium.
Agrobacterium co-expression of full-length Rpi-blb2 with AVRblb2 induced HCD 3 to 4 days after infection, whereas co-expression with the control vector or the Rpi-blb2 N- and C-terminal deletions did not trigger HCD in N. benthamiana (Fig. 1A). Thus, both the CC and LRR domains, including the functional NB domain of Rpi-blb2, are necessary to induce HCD. Additionally, a full-length Rpi-blb2 genomic DNA construct was fully effective in plants and triggered Rpi-blb2-mediated HCD (Fig. 1A). RT-PCR analysis of AVRblb2 and Rpi-blb2, Rpi-blb2 N-, or Rpi-blb2 C-terminal deletion transcripts confirmed that the transcript levels of Rpi-blb2, Rpi-blb2 N-, or Rpi-blb2 C-terminal deletion were upregulated following co-expression in N. benthamiana (Fig. 1B). Although the exact mechanism of the interaction between the AVRblb2 effector and the Rpi-blb2 protein remains unclear, in planta interactions resulted in HCD in N. benthamiana (Oh et al., 2009; 2014). These results indicate that all regions of Rpi-blb2 are required for AVRblb2 recognition and induction of cell death.
Rpi-blb2-overexpressing tomato, potato, and N. benthamiana plants have heightened resistance responses against P. infestans infections (Oh et al., 2014; van der Vossen et al., 2005). To confirm the effect of Rpi-blb2 on the disease resistance response in planta, Rpi-blb2-transgenic N. benthamiana leaves were inoculated with P. infestans. Disease symptoms were visible in the control plants within 4 to 6 dai. However, the area of infection was significantly smaller in Rpi-blb2-transgenic N. benthamiana plants than in control plants, which correlates with reduced oomycete colonization (Fig. 1C). At 4–6 dai, visible disease symptoms were observed in 10–32.5% of the total leaf area in control plants, which was significantly larger than the 3.3–3.5% observed in the transgenic plants (Fig. 1D). Quantitative RT-PCR analyses revealed that PiEF2a transcript levels were significantly lower in Rpi-blb2 transgenic N. benthamiana plants than in control plants (Fig. 1E). The N. benthamiana Actin gene (NbActin) was used as a loading control and as an internal control for RNA quality. Additionally, Rpi-blb2 transcript expression was confirmed in Rpi-blb2-transgenic N. benthamiana plants (Fig. 1E). Reduced biomass may have resulted from a loss of viable tissue during disease progression. Thus, it was found that Rpi-blb2-transgenic N. benthamiana plants are resistant to P. infestans, which is in concordance with tomato and potato (van der Vossen et al., 2005). Hence, we were used for further characterization of Rpi-blb2-mediated HCD using VIGS techniques.
SGT1, but not EDS1 or NDR1, functions in Rpi-blb2-mediated HCD in response to P. infestans AVRblb2.
Knockdown analyses in N. benthamiana led to the identification of signaling components downstream of R proteins (Lu et al., 2003). EDS1, NDR1, and SGT1 are key components of the signal-transduction pathway leading to disease resistance in control plants and induce HCD in response to specific effectors (Aarts et al., 1998; Bhattacharjee et al., 2011; Kadota et al., 2010). Specifically, EDS1 functions as a regulator of basal resistance and ETI mediated by TIR-NB-LRR proteins (Bhattacharjee et al., 2011; Falk et al., 1999). CC-NB-LRR protein function requires NDR1 (Aarts et al., 1998; Shapiro et al., 2001; Tornero et al., 2002) or SGT1 (Kadota et al., 2010; Oh et al., 2014).
To examine whether HCD is induced by co-expression of AVRblb2 and Rpi-blb2, we silenced EDS1, NDR1, and SGT1 using VIGS by agro-infiltration in Rpi-blb2-transgenic N. benthamiana plants (Fig. 2A). Rpi-blb2-transgenic plants were infiltrated at the four- to five-leaf stage with mixtures of Agrobacterium carrying combinations of TRV1, TRV2-dGFP (control vector), TRV2-NbSGT1, -NbEDS1, or -NbNDR1. Twenty-one dai, plants with silenced TRV2-NbSGT1 or -NbEDS1 had abnormal phenotypes indicative of successful silencing of SGT1 and EDS1, respectively. We also confirmed EDS1, NDR1, and SGT1 silencing by RT-PCR. Expression of all specific transcripts was reduced in the silenced plants compared with control vector-containing plants (TRV2-dGFP; Fig. 2B). The silenced N. benthamiana plants were infiltrated with Agrobacterium containing AVRblb2. HCD induced by AVRblb2 was completely suppressed in SGT1-silenced Rpi-blb2-transgenic plants, but was not suppressed in plants infected with TRV2-dGFP or in EDS1- and NDR1-silenced plants. Cell death was also quantified by ion leakage from leaves after AVRblb2 infiltration (Fig. 2C).
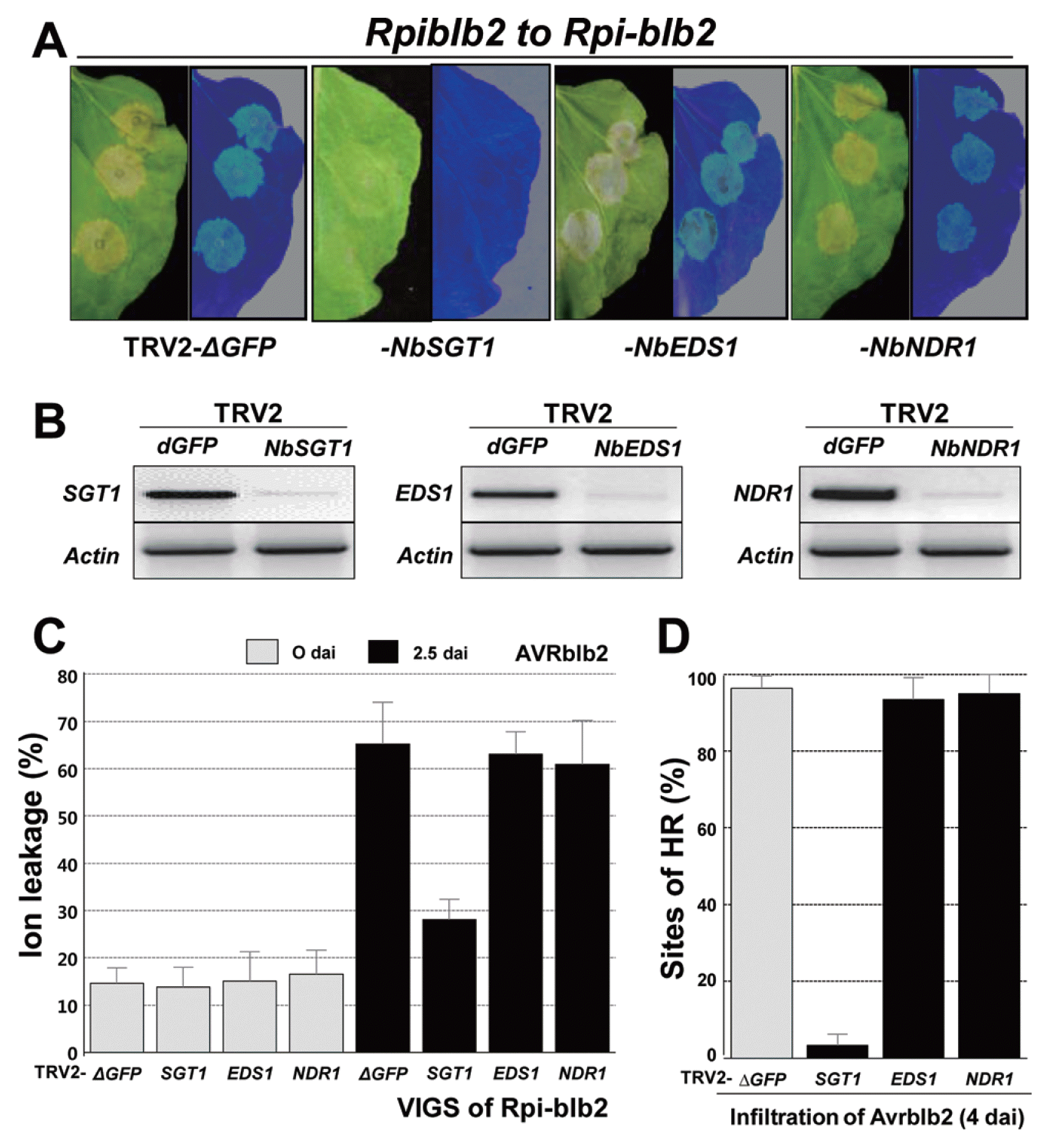
NbSGT1 functions in Rpi-blb2-mediated HR in response to AVRblb2, but not NbEDS1 or NbNDR1. (A) Rpi-blb2-transgenic N. benthamiana was inoculated with TRV2-dGFP, -NbSGT1, -NbEDS1, and -NbNDR1, respectively. After 21 days, leaves of these silenced N. benthamiana were expressed with AVRblb2-containing Agrobacterium. (B) Silencing of target genes was confirmed by RT-PCR with gene-specific primers using at least three separate leaves. The NbActin gene was used as a control. (C) After 21 days, leaves of silenced-Rpi-blb2-transgenic N. benthamiana, ion leakage from leaf discs was measured at 0 and 2.5 dai for expressed with AVRblb2-containing Agrobacterium. Values are means±SD (n=3). (D) Quantification of HR. The mean percentages of sites showing HR and the average±SD were scored from twenty infiltration sites based on five replications using N. benthamiana leaves.
In EDS1- and NDR1-silenced Rpi-blb2-transgenic plants, AVRblb2 caused visible HCD at 3–4 dai in approximately 95% of the infiltration sites in three independent experiments (Fig. 2D). However, in SGT1-silenced plants, HCD was observed in only approximately 5% of the infiltration sites (Fig. 2D). In TRV2-dGFP (vector control)-treated plants, the Rpi-blb2 and AVRblb2 combination resulted in visible HCD at 4 dai in approximately 95% of the infiltration sites (Fig. 2D). Our findings indicate that Rpi-blb2 elicits HCD through SGT1-mediated signaling in N. benthamiana. Likewise, silencing RAR1 or HSP90 in N. benthamiana plants did not abolish Rpi-blb2-mediated HCD in response to P. infestans AVRblb2 (Oh et al., 2014). Moreover, EDS1 and NDR1 are not required for HCD in response to P. infestans AVRblb2 in Rpi-blb2-transgenic N. benthamiana. Although we did not address the question of how SGT1 regulates Rpi-blb2 function here, we hypothesize that NbSGT1 affects other processes, including the translation of Rpi-blb2 and/or the maintenance of pre-activated Rpi-blb2 protein stability. We also assumed that SGT1-dependent HCD could be due to the Rpi-blb2 signaling complex in N. benthamiana in response to specific AVRblb2 effector proteins.
SA, JA, and ET are not required for HCD elicited by the Rpi-blb2-AVRblb2 interaction
Plant hormones such as SA, JA, and ET activate defense responses to pathogen attacks and cross talk with other signaling pathways (Glazebrook, 2005; Robert-Seilaniantz et al., 2011). In addition, the occurrence of HCD leads to activation of SA, JA, and ET signaling throughout the plant (Glazebrook, 2005; Overmyer et al., 2003). To functionally validate the involvement of SA, JA, and/or ET in Rpiblb2 to Rpi-blb2 -mediated cell death, we performed a TRV2-based VIGS assay. ICS1 (an enzyme for SA biosynthesis; Wildermuth et al., 2001), COI1 (a JA signaling component; Xu et al., 2002), or EIN2 (ethylene insensitive; Alonso et al., 1999) genes were independently silenced in Rpi-blb2-transgenic N. benthamiana and AVRblb2 was transiently infiltrated in the silenced plants (Fig. 3A). RT-PCR analysis of NbICS1, NbCOI1, and NbEIN2 transcripts confirmed that their levels were downregulated by VIGS (Fig. 3B). HCD was induced in NbICS1, NbCOI1, and NbEIN2-silenced plants, but was not induced in TRV2-dGPF control plants (Fig. 3C). Therefore, these results indicate that SA, JA, and ET are not essential for Rpi-blb2-mediated HCD in response to P. infestans AVRblb2.
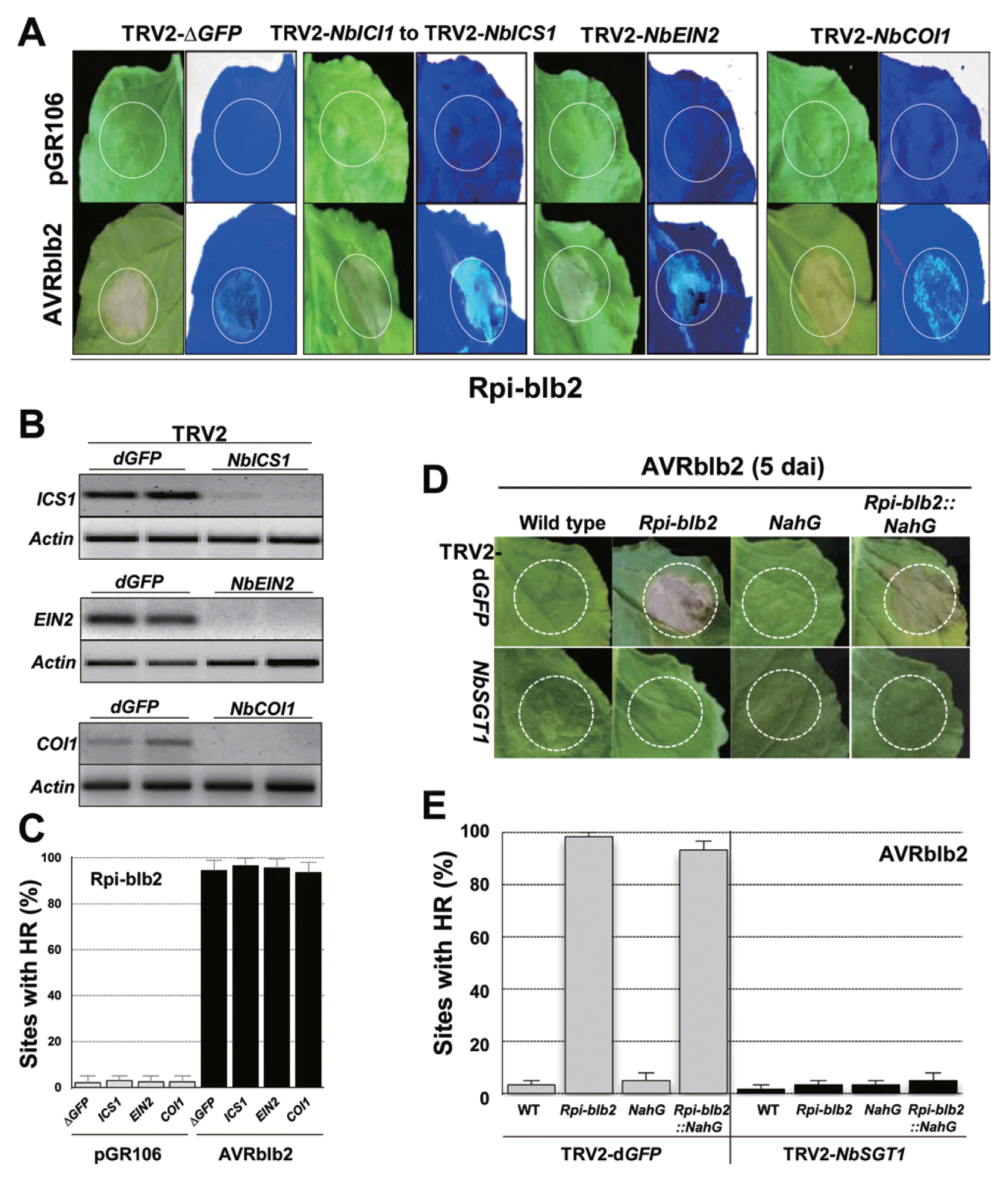
Rpi-blb2-mediated HCD in response to AVRblb2 did not require SA, JA, or ethylene signaling. (A) Evidence that each signaling component was silenced by VIGS. (B) RT-PCR was performed with 30 cycles for NbICS1 and 35 cycles for NbEIN2 and NbCOI1. Signaling components that did not affect Rpi-blb2-mediated HR in response to AVRblb2. ICS1, EIN2 or COI1 was silenced and AVRblb2 was transiently overexpressed by agro-infiltration in Rpi-blb2-transgenic N. benthamiana of leaves. (C) Percentages of infiltrated leaves with Rpi-blb2–mediated HR in response to AVRblb2. Bars indicate SD. Similar results were obtained at least twice from independent replications. Leaves of wild type, Rpi-blb2, NahG, and Rpi-blb2::NahG -transgenic N. benthamiana plants were agro-infiltrated with the pGR106-AVRblb2 vector. Rpi-blb2-transgenic plants showed HR symptoms activated by AVRblb2 at 3–4 dai. (D) Percentages of infiltrated leaves with Rpi-blb2–mediated HR. All values represent means and SD from at least three independent experiments with three replications.
Studies have demonstrated that SA is required for the full function of a number of R genes. SA signaling-associated mutants in Arabidopsis have been used to characterize the role of R gene–mediated HCD and pathogen resistance (Glazebrook, 2005; Hammond-Kosack and Parker, 2003). We also tested the effects of NahG, which encodes a salicylate hydroxylase (Delaney et al., 1994), on the HCD response to AVRblb2. We generated Rpi-blb2::NahG-transgenic plants from crosses between Rpi-blb2-transgenic- and NahG-transgenic plants (Oh et al., 2014). After infiltration by P. infestans AVRblb2, we observed that HCD lesions in wild-type (WT), Rpi-blb2-, NahG-, and Rpi-blb2::NahG-transgenic plants were similar (Fig. 3D). Silencing SGT1 on Rpi-blb2- or NahG-transgenic backgrounds completely suppressed Rpi-blb2-mediated HR in response to P. infestans AVRblb2 (Fig. 3E). These results confirmed that NbSGT1 is associated with Rpi-blb2-mediated HCD in response to AVRblb2, but NbSGT1 is not associated with the coordinated SA accumulation. Moreover, SA itself does not activate HCD, but elicits a HCD response following H2O2 accumulation (Oh et al., 2014).
Here, we demonstrate that SGT1 is required for HCD response in Rpi-blb2-transgenic N. benthamiana in response to P. infestans AVRblb2, but not EDS1 or NDR1-mediated signaling. In addition, even though SA affects Rpi-blb2-mediated resistance against P. infestans but Rpi-blb2-mediated HCD in response to AVRblb2 did not affects. We also suggest that JA and ET signaling were not required for Rpi-blb2-mediated HCD in N. benthamiana against AVRblb2. Hence, further analysis of other processes and their corresponding products will help to unravel the complexity of Rpi-blb2-mediated HCD responses to and resistance pathways against P. infestans AVRblb2 in plants.
Acknowledgements
We thank Dr. S.P. Dinesh-Kumar for providing the pTRV2 vectors. This work was supported by grants from the Agricultural Research Center program to DC (Project No. 700001-03) and The Agricultural Genome Center of the Next Generation BioGreen 21 program to S.-Y. Kwon (Project No. PJ00810304).