A Rapid and Efficient Method for Construction of an Infectious Clone of Tomato yellow leaf curl virus
Article information
Abstract
Tomato yellow leaf curl virus (TYLCV), a member of the genus Begomovirus, is responsible for one of the most devastating viral diseases in tomato-growing countries and is becoming a serious problem in many subtropical and tropical countries. The climate in Korea is getting warmer and developing subtropical features in response to global warming. These changes are being accompanied by TYLCV, which is now becoming a large problem in the Korean tomato industry. The most effective way to reduce damage caused by TYLCV is to breed resistant varieties of tomatoes. To accomplish this, it is necessary to establish a simple inoculation technique for the efficient evaluation of resistance to TYLCV. Here, we present the rolling circle amplification (RCA) method, which employs a bacteriophage using phi-29 DNA polymerase for construction of infectious TYLCV clones. The RCA method is simple, does not require sequence information for cloning, and is less expensive and time consuming than conventional PCR based-methods. Furthermore, RCA-based construction of an infectious clone can be very useful to other emerging and unknown geminiviruses in Korea.
Tomato yellow leaf curl virus (TYLCV) belonging to the Genus Begomovirus in the Family Geminiviridae (single-stranded DNA virus with twinned icosahedral virions) (Czosnek et al., 1988) is responsible for one of the most devastating viral diseases impacting tomato crops on several continents, mainly in tropical and subtropical regions. Indeed, TYLCV can cause 100% losses in tomato (Yongping et al., 2008). The first epidemic of TYLCV was reported in Israel in the early 1960s (Urbino et al., 2008), after which it was reported in other countries in Europe (Kheyr-Pour et al., 1991), Asia (Rochester et al., 1990), the south America (Brown and Bird, 1992; Harrison et al., 1991; Nakhla et al., 1994), Africa (Harrison et al., 1991) and Australia (Dry et al., 1993). In Korea, there are two TYLCV groups based on sequence and genome size, one with 2774-nucleotides and another with 2781-nucleotides. The Korea 1 group was isolated from Gyeongsang-Do and Jeollanam-Do, while the Korea 2 group was isolated from Jeju-Do and Jeollabuk-Do (Lee et al., 2010).
TYLCV is transmitted by the whitefly Bemisia tabaci to dicotyledonous plants. The management of TYLCV is mainly focused on whitefly control through application of insecticides and the use of physical barriers (Lapidot et al., 2002). However, whitefly resistance to insecticides is gradually increasing owing to their continuous use, and physical management inhibits the growth of tomato plants.
The most effective method of reducing TYLCV damage is breeding resistant varieties of tomatoes. To develop resistant varieties of tomatoes, it is necessary to establish a simple inoculation technique for the efficient evaluation of resistance to TYLCV. Specifically, rapid and easy generation of infectious clones is important to evaluation of the resistance of a variety of tomato cultivars against different TYLCV isolates. Rolling circle amplification (RCA) is a very useful technique for detection and genomics of geminiviruses (Haible et al., 2006) that has also been applied to construction of infectious clones of begomovirus (Inoue-Nagata et al., 2007; Wu et al., 2008). RCA cloning is more convenient than the PCR based- method. For example, the RCA method is simple, does not require sequence information for cloning, and is less expensive and time consuming than conventional PCR based-methods. In addition to construction of infectious clones of geminiviruses, this technique can be applied for amplification of the genome of animal pathogenic circovirus (Mankertz et al., 2004), as well as other pathogenic viruses such as nanoviruses (Gronenborn, 2004).
Geminiviruses can replicate via a rolling-circle mechanism with the virion strand replication origin (V-ori) located in an invariant motif (TAATATTAC) (Ferreira et al., 2008). Most infectious clones of geminivirus require more than one mer (unit) of the genome to produce a tandem repeat that contains two replication origins (V-ori), which increase their infectivity.
In this study, we employed RCA with a bacteriophage using phi-29 DNA polymerase to construct infectious TYLCV clones and constructed 1.9mer and 1.7mer genome-length copy of TYLCV.
Collection of leaves exhibiting TYLCV symptoms and the extraction of genomic DNA from infected tissues
Leaves from tomato plants showing typical TYLCV symptoms were collected in several tomato-growing regions in southeastern Korea, including Gimhae and Busan. Total DNA was then extracted from the collected leaves using a modified CTAB method. Briefly, approximately 100 mg of tender leaf tissues were homogenized using disposable micro pestles with 1.5 mL of extraction buffer. The homogenized samples were then incubated at 60°C for 45 min, after which the tubes were centrifuged at 13,000 rpm for 1 min. Subsequently, 400 μl of supernatant were transferred into a new tube. After adding RNase A (10 μg/ml), the tube was incubated for 45 min at 37°C. The suspension was then treated with 600 μl of chloroform: isoamylalcohol (24:1) followed by inversion and centrifugation at 13,000 rpm for 20 min. Nucleic acids in the aqueous layer were then transferred into a new tube and 300 μl of ice-cold isopropanol was added and mixed by inverting the tube gently. The tube was then incubated overnight at −20°C or for 8 hr at −80°C, after which the DNA was pelleted by centrifuging at 12,000 rpm for 15 min. The pellet was then washed with 500 μl of 70% (v/v) ethanol by inverting, which was followed by centrifugation at 12,000 rpm for 5 min. Finally, the DNA pellet was dried until all alcohol was evaporated, dissolved in 50 μl of deionized water and stored at −20°C until further use.
Amplification of 1.0mer TYLCV via rolling circle amplification (RCA) or polymerase chain reaction (PCR)
Total purified DNA from TYLCV-infected leaf samples was used as the template for RCA, which was conducted using phi-29 DNA polymerase (Ilustra™ TempliPhi Amplification Kit, GE Healthcare). Briefly, 1 pg to 10 ng of total nucleic acids template were dissolved in 5 μl sample buffer and then denatured by heating at 95°C for 3 min. The sample was subsequently cooled down to room temperature, combined with 5 μl of reaction buffer and 0.2 μl of enzyme mix and incubated at 30°C for 12–18 hours. During this process, the random hexamers (short arrows) anneal to single-stranded circular DNA templates at multiple sites (Fig. 1). Phi-29 DNA polymerase then extends each of these primers until the DNA polymerase reaches a downstream-extended primer, at which time strand displacement synthesis occurs. As the process continues, exponential isothermal amplification occurs. After this incubation, the reaction was stopped for 10 min at 65°C and the RCA products were directly cloned into the pGEM-T Easy vector. The RCA products were subsequently converted to the linear form by NcoI digestion and used directly for ligation into pGEM-T-Easy (Promega, USA) that had been digested with the same restriction enzyme, resulting in pGEM-TY-R1.0mer (Fig. 1). The pGEM-TY-1.0mer was confirmed by restriction enzyme analysis and PCR with primer pair AV494/AC1048 (Table 1), which are capable of amplifying the core CP region of many Begomoviruses. Positive recombinant plasmids were fully sequenced using a 3730XL DNA analyzer (Applied Biosystems, USA) by Macrogen (Seoul, Korea). Positive clones were denoted pGEM-TY-KH, and one of each positive clone was used in subsequent work.

Schematic diagrams of the workflow and construction of TYLCV-KH infectious clones. There were two ways to create an infectious clone, an RCA-based method and a PCR-based method.
Primers for detection of TYLCV, cloning of full-length genome of TYLCV, and sequencing of TYLCV clones
For amplification of 1.0mer TYLCV via PCR, the overlapping primer method was used to amplify a full-length TYLCV DNA genome by PCR (Patel et al., 1993) using two designed primers (Table 1), PTYC1v2182 fwd (5′-CCCAGACCATGGCCGCGCAGCGGAATAC-3′) and PTYC1c2140 rev (5′-AATAACCCATGGAGACCCATAAGTATTG-3′), which overlap a conserved NcoI site in the C1 gene of TYLCV, from genomic DNA extracted from infected leaves. The reactions were conducted in a reaction volume (50 μl) containing 5 μl of 10X PCR buffer, 1 μl of dNTPs (10 mM), 2 μl of each PTYC1v2182/PTYC1c2140 (10 pM), 2.5 μl of 5X Band Doctor, 1 μl of template DNA (diluted 1:10 in water) and 0.5 μl of 2.5 U/μl Pfu-X DNA polymerase (Solgent). The PCR conditions were 94°C for 2 min, followed by 30 cycles of 94°C for 20 s, 50°C for 40 s, and 72°C for 1 min, and then final extension at 72°C for 5 min. The DNA fragment amplified with PCR was digested with the NcoI enzyme, after which the NcoI-NcoI fragment was ligated into pGEM-T Easy (Promega, USA) that had been digested with the same restriction enzymes, resulting in the pGEM-TY-P1.0mer (Fig. 1). TYLCV-KH has a genome size of 2774-nucleotides. TYLCV-KH is belonging to The Korea 1 group. Sequencing analysis showed that TYLCV-KH shared 99.7% to 99.9% sequence identity with other TYLCV isolates, indicating that it is a typical isolate from TYLCV species.
Construction of TYLCV infectious clone and infiltration of infectious clone into tomato tissues
A 1.9mer genome-length of TYLCV clone was constructed from 1.0mer TYLCV obtained from pGEM-TY-KH. To accomplish this, a 2.5 kb EcoRI/NcoI-digested fragment obtained from pGEM-TY-KH was ligated into the EcoRI/NcoI-digested pCAMBIA0390 binary vector to create pCAMBIA0390-TY-0.9mer. Next, a 2.8 kb NcoI fragment of pGEM-TY-KH, the full-length TYLCV clone, was ligated into NcoI-linearized pCAMBIA0390-TY-0.9mer to generate pCMABIA0390-TY-1.9mer, a binary vector containing 1.9mer of TYLCV-KH (Fig. 1). For construction of pCAMBIA0390-TY-1.7mer, a 1.9 kb AseI/NcoI-digested fragment was ligated into the AseI/NcoI-digested pCAMBIA0390 binary vector to create pCAMBIA0390-TY-0.7mer. The remaining steps were the same as those used to construct the 1.9mer genome-length of TYLCV clone (Fig. 1).
To test the infectivity, pCAMBIA0390-TY-1.9mer was introduced into competent cells of Agrobacterium tumefaciens (GV3101) by heat shock transformation. The presence of the binary vector in A. tumefaciens was confirmed by colony PCR using specific primers. Transformed A. tumefaciens colonies were plated and cultivated at 28°C on Luria-Bertani (LB) medium with kanamycin and gentamycin. Bacteria was incubated at 28°C for 24 hr with shaking after inoculation into 5 ml of LB broth and then subcultured at 28°C for 10 hr in 5 ml LB broth with kanamycin and gentamycin. Bacterial cells were subsequently harvested by centrifugation at 4,000 rpm for 10 min and resuspended in 3–4 ml liquid infiltration medium supplemented with 10 mM MgCl2, 200 μM acetosyringone, and 10 mM MES to a final concentration of 0.6 OD at 600 nm, incubated for 4 hr at room temperature and used for the agroinfiltration. A. tumefaciens were inoculated into the leaf using a 1 ml syringe without the needle (de Felippes and Weigel, 2010). Plants were maintained in an insect-free containment growth chamber at 27°C with a 16 hr photoperiod.
TYLCV detection in leaves exhibiting symptoms in tomato
Typical TYLCV symptomatic leaves were sampled from tomato plants that had been inoculated by Agrobacterium-mediated infiltration. Four weeks later, TYLCV symptoms such as leaf curling, yellowing, and stunting were observed in the infiltrated leaf tissues (Fig. 2). Genomic DNA was then extracted and analyzed for the presence of the viral genome by PCR using the TYLCV primer pair PTYc787/PTYc1121 (Salati et al., 2002) and the degenerate primer pair AV494/AC1048 (Wyatt and Brown, 1996). The results clearly showed the presence of TYLCV in infected tomato, demonstrating curling and yellowing with stunting. However, TYLCV from the healthy tomato sample was not detected with either primer set (Fig. 3).

Symptoms on tomato plants inoculated with the infectious TYLCV clones. Four weeks after agroinoculation, plants become systemically infected and showed upward curling and yellowing of leaves, which are symptoms of stunting. (A, E) Tomato plants inoculated with A. tumefaciens containing empty pCAMBIA0390. (B, C, D) Tomato plants inoculated with A. tumefaciens containing pCAMBIA0390-TY-1.7mer. (F, G, H) Tomato plants inoculated with A. tumefaciens containing pCAMBIA0390-TY-1.9mer.
Detection of TYLCV DNA in tomato plants after inoculation with the infectious TYLCV clones. Agarose gel electrophoresis of amplification products from polymerase chain reactions conducted using primers (A) AV494 and AC1048 and (B) PTYv787 and PTYc1121, with nucleic acids extracted from young non-inoculated tissues of tomato 4 weeks after inoculation with A. tumefaciens having infectious clones of Tomato yellow leaf curl virus-[KH]. (A) Detection of Begomovirus in infected tomato using degenerate primers (AV494 and AC1048). (B) Detection of TYLCV using degenerate primers (PTYv787 and PTYc1121). M, 100 bp ladder molecular size marker; C, a tomato plant inoculated with A. tumefaciens containing empty pCAMBIA0390; 1–3, tomato plants inoculated with A. tumefaciens containing pCAMBIA0390-TY-1.7mer; 4–6, tomato plants inoculated with A. tumefaciens containing pCAMBIA0390-TY-1.9mer.
The infectivity of the TYLCV clones was assayed by agroinoculation. Tomato plants were inoculated with A. tumefaciens strain GV3101 harboring 1.9mer constructs or 1.7mer constructs of the TYLCV described above. The development of TYLC symptoms in some plants was first observed 2 weeks post inoculation. At 4 weeks post-inoculation, TYLCV susceptible supersunroad tomato plants inoculated with pCAMBIA0390-TY-1.9mer (or 1.7mer) showed typical symptoms of TYLCV. The presence of TYLCV in the agro-infected plants showing TYLCV symptoms was verified by PCR (Fig. 3) and digestion of restriction enzymes (data not shown).
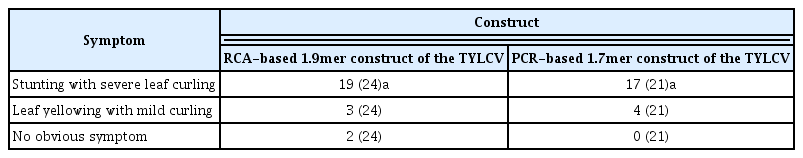
We inoculated more than 20 tomato plants with two replicates for each RCA-based 1.9mer construct and PCR-based 1.7mer construct of the TYLCV. Most of tomatoes inoculated by A. tumefaciens having each infectious construct showed typical TYLCV symptoms at 4 weeks post-inoculation. For examples, 22 out of 24 tomatoes inoculated by A. tumefaciens having RCA-based 1.9mer construct of the TYLCV exhibited symptoms. All 22 symptomatic tomatoes showed curling except for three tomatoes with mild yellowing. All 21 tomatoes inoculated by A. tumefaciens with PCR-based 1.7mer construct of the TYLCV exhibited symptoms (Table 2). Even tomatoes exhibiting mild symptoms such as yellowing changed to stunting with severe curling about 6–8 weeks post-inoculation. Tomatoes inoculated by A. tumefaciens harboring RCA-based 1.9mer construct and PCR-based 1.7mer construct of the TYLCV showed similar patterns of symptom developments.
In conclusion, the construction of the RCA-based TYLCV infectious clone is a simple and easy method because TYLCV has ~2.8 kb single circular DNA genome and one NcoI site in the genome. Thus, RAC method with general random primers can easily amplify single TYLCV genome and directly clones to the NcoI-digested vector (Fig. 1). The RCA-based construction of an infectious viral clone can be very useful to other emerging and unknown geminiviruses that have single circular DNA genome.
Acknowledgments
This study was supported by a grant (Project No. 609002-5) from the Screening Center for Disease Resistant Vegetable Crops of Agricultural Biotechnology Development Program, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea and Pusan National University Research Grant, 2011 (Y.-S. S.).