Cherry necrotic rusty mottle virus (CNRMV) and the closely related Cherry green ring mottle virus (CGRMV) are flexuous filamentous plant viruses and members of Betaflexiviridae (Adams et al., 2004). CNRMV infects stone fruit, including peach, apricot, sour cherry, and sweet cherry, and has been reported in North America, New Zealand, Europe, Chile, Japan, China, and Korea (Fiore and Zamorano, 2013; Isogai et al., 2004; Lee et al., 2012; Mandic et al., 2007; Wadley and Nyland, 1976; Zhou et al., 2013). CNRMV-infected plants typically develop brown, angular, necrotic spots, rusty chlorotic, and shot hole symptoms on leaves as well as blisters, gum pockets, and general necrosis on the bark (Rott and Jelkmann, 2001). CGRMV also infects stone fruits including sour cherry and oriental flowering cherry, with sweet cherry, peach, and apricot being symptomless hosts, and has a geographic distribution similar to CNRMV (Fiore and Zamorano, 2013; Isogai et al., 2004; Lee et al., 2012; Mandic et al., 2007; Parker et al., 1976; Zhou et al., 2011). CGRMV-infected Montmorency sour cherry shows yellow mottling and ring-like bands on its leaves, and produces misshapen, bitter, and unmarketable fruit (Parker et al., 1976).
In 2011, CNRMV and CGRMV were detected in sweet cherries in Korea (Lee et al., 2012), and abnormal cherries exhibiting ring spots on the pericarp collected from the cities of Daegu, Gyeongju and Gyeongsan were diagnosed as being co-infected with CNRMV and CGRMV (Lee et al., 2014). Asymptomatic cherry leaves from Gyeonggi province and Yeongju city harbored single infections of CNRMV and CGRMV, respectively (Cho et al., 2013; Cho et al., 2014). These reports highlight the difficulty of identifying typical symptoms of CNRMV/CGRMV by naked eye to diagnose an infection at early stages (Cho et al., 2013; Lee et al., 2012).
CNRMV and CGRMV have a similar morphology and genomic organization and share approximately 60% homology at the nucleotide level (Li and Mock, 2005). Although reverse transcriptase polymerase chain reaction (RT-PCR) methods have been developed to detect CNRMV and CGRMV using specific primer pairs (Li and Mock, 2005; Park and Kim, 2004; Rott and Jelkmann, 2001), an accurate method for detecting both viruses simultaneously would be useful for making rapid and reliable diagnoses. In the present study, a duplex RT-PCR method was developed that allows the simultaneous detection of CNRMV and CGRMV in sweet cherries, which was used to survey the incidence of these viruses in Gyeongbuk province, Korea.
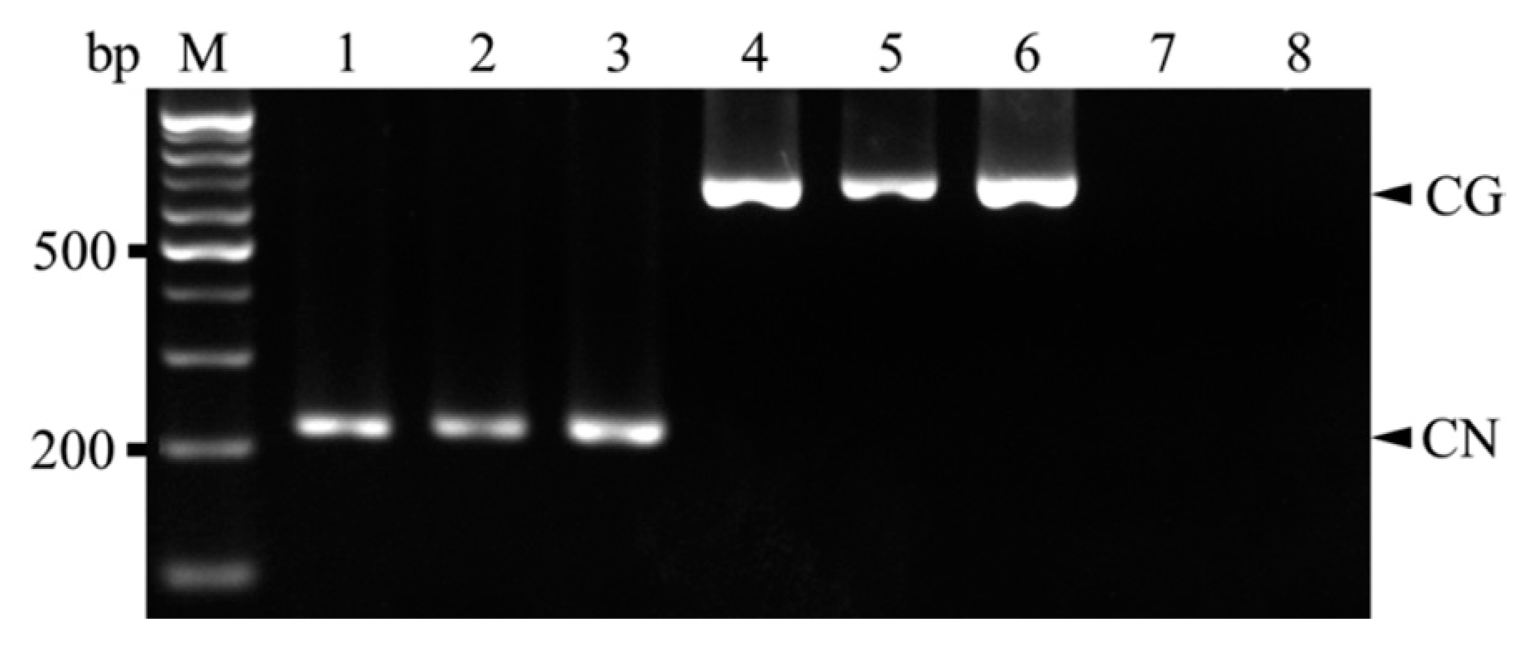
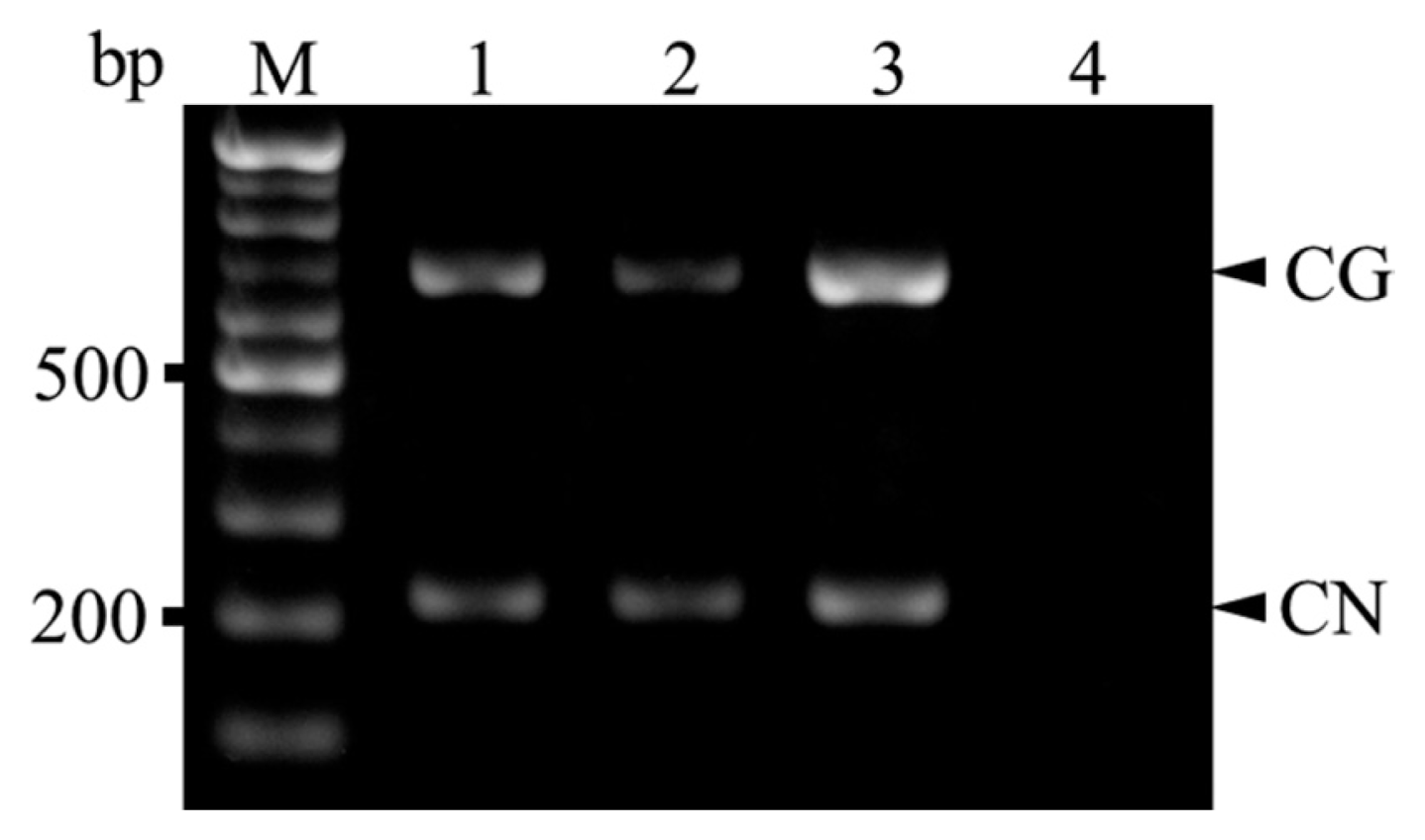
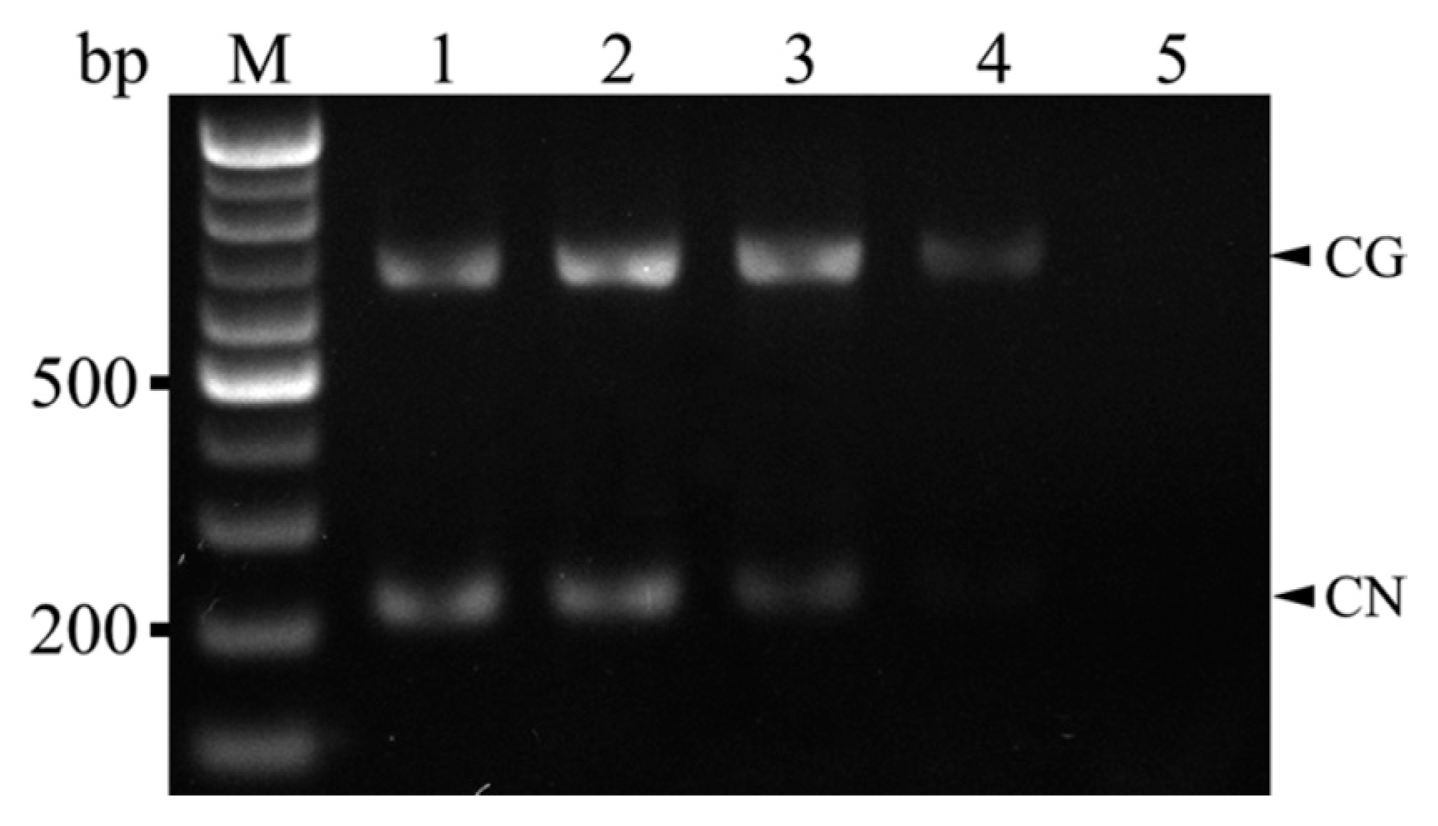
Cherry fruits with visible ring-spots or ring spot-like symptoms on the pericarp were randomly collected from sweet cherry (cv. Satonishiki) orchards located in the cities of Daegu, Gyeongju, and Gyeongsan during 2012-2013. Total RNA was extracted from samples using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions and used for RT-PCR, with CNRMV and/or CGRMV infection confirmed using the primer pair prCN4f (5′-AGC ACW RTT AGG AGC TAC TG-3′) and prCN4r (5′-TAT CAT TCA TCA CCA CCA AT-3′) that can amplify both viruses (Park and Kim, 2004). The amplification was performed using the OneStep RT-PCR kit (Solgent, Daejeon, Korea) using previously reported reaction conditions (Park and Kim, 2004) on a Veriti 96-well Thermal Cycler (Applied Biosystems, Carlsbad, CA, USA). DNA fragments of the expected size (400 bp) were amplified from 43 out of 83 samples (data not shown). The two pairs of primers for the duplex reaction were designed for specificity and compatibility based on whole coat protein sequences for CNRMV and CGRMV retrieved from GenBank of the National Center for Biotechnology Information (NCBI). Specific primers for CNRMV (CNSp-F, 5′-CTT TGA TCC CAA AAA TCC CA-3′ and CNSp-R, 5′-TGG TYT TGT CAC TTG AAC TGT T-3′) and CGRMV (CGSp-F, 5′-GCG CAA ACG GAC CCT AAG-3′ and CGSp-R, 5′-CGC CAG TCA CTT CAG TCA TT-3′) were designed to amplify fragments of ~210 and 650 bp, respectively, and each were used in simplex RT-PCR reactions to confirm the specificity of the products amplified by duplex RT-PCR. Three representative total RNA samples which confirmed as positive for CNRMV and CGRMV (Lee et al., 2014) were used in both simplex and duplex RT-PCR. Also those samples were retrieved from NCBI (Accession no. CNRMV; AB871372-AB871381 (10 strains), CGRMV; AB863720-AB863734 (15 strains)). For simplex and duplex RT-PCR, the 20-μl reaction contained 0.5 μl total RNA, 4 μl 5× RT buffer (Solgent), 1 μl of 10 mM dNTP, 1 μl forward and reverse primers (10 pM), 2 μl RT (Solgent), and 11.5 μl RNase-free water with amplification conditions as follows: reverse transcription at 50°C for 30 min, followed by 95°C for 15 min; 35 cycles of 95°C for 30s, 60°C for 45s, and 72°C for 45s; and 72°C for 5 min. RT-PCR products were visualized on a 1% agarose gel under UV light after staining with ethidium bromide. Simplex RT-PCR products of expected sizes were observed for positive samples but not for healthy leaf samples (Fig. 1). Amplicons were then purified using the HiGene Gel & PCR Purification System (Biofact, Daejeon, Korea) and sequenced yielding 176 and 623 bp sequences from each of amplified DNA fragments. A search in GenBank (NCBI) showed that the obtained 176 bp sequences for CNRMV had 99-100% identity to Daegu (AB871372), Gyeongju (AB871375), and Gyeongsan (AB871380) strains and 685 bp sequences for CGRMV had 99-100% identity to Daegu (AB863720), Gyeongju (AB863731), and Gyeongsan (AB863734) strains in nucleotide level, respectively. After duplex RT-PCR, DNA fragments of 200 and 650 bp (corresponding to CNRMV and CGRMV, respectively) were detected in each sample by gel electrophoresis (Fig. 2). Also, 10-fold serial dilutions (100-10−4) were prepared from ~10 ng total RNA to determine the detection limits of the duplex RT-PCR. According to our results it was confirmed that it can detect up to 10−2 and 10−3 dilution for CNRMV and CGRMV, respectively (Fig. 3).
A total of 43 total RNA samples (9 from Daegu, 20 from Gyeongju, and 14 from Gyeongsan) from abnormal cherries were examined using the developed duplex RT-PCR method (Fig. 4A-C). Of these, 11 were positive for CNRMV infection, 4 were infected with CGRMV, and 28 samples were co-infected with both viruses (Table 1). Among Daegu isolates, 8 were co-infected with CNRMV and CGRMV while only 1 sample had a single infection of CGMRV (Table 1). Of the 20 samples from Gyeongju, 15 were confirmed as being infected by both viruses, making this the city with the highest co-infection rate. Among Gyeongsan isolates, 7 were CNRMV and 2 were CGRMV infections, and 5 were confirmed as co-infections. Thus, the duplex RT-PCR found co-infection rates in Daegu, Gyeongju, and Gyeongsan of 29.6%, 53.6%, and 17.9%, respectively. These results demonstrate that CNRMV and CGRMV are more prevalent in these cities than in other surveyed regions in Korea (Table 1; Cho et al., 2013, 2014). In addition, cases of single infection of sweet cherry by CNRMV or CGRMV have been reported in Gyeonggi province and the Yeongju region of Korea; however, co-infections by both viruses have not been observed (Cho et al., 2013, 2014).
To date, several RT-PCR-based methods have been developed to detect CNRMV and/or CGRMV infections in cherry (Rott and Jelkmann, 2001; Isogai et al., 2004; Park et al., 2004; Fiore and Zamorano, 2013). These methods are suitable to detect CNRMV or CGRMV; however, it is impossible to differentiate between the two targeted viruses in single reaction. Multiplex RT-PCR methods are rapid, reliable, and sensitive, and have been widely used to identify more than one virus simultaneously in a single assay, for instance in viral infections of stone fruit (Sanchez-Navarro et al., 2005; Jarosová and Kundu 2010; Yardimci and Çulal-Klllç, 2011) and lily (Kwon et al., 2013). Multiplex RT-PCR has also been applied to the detection of CNRMV and/or CGRMV (Noorani et al., 2013; Zhao et al., 2013), including in sweet cherries. For CNRMV, specific primers were designed based on the replicase gene and have identified stone fruit viruses such as Cherry virus A, Little cherry virus-1, and Prune necrotic ring spot virus, in addition to CNRMV (Noorani et al., 2013). A duplex RT-qPCR method was also developed for Apple chlorotic leaf spot virus and CGRMV in peach (Zhao et al., 2013).
Co-infections by CNRMV and CGRMV have previously been confirmed in cherry orchards located in Daegu, Gyeongju, and Gyeongsan regions (Lee et al., 2014). Furthermore, it was confirmed that single infections were also occurred in Daegu, Gyeongju, and Gyeongsan city. Cho et al (2013 and 2014) also reported that single infection rate of CNRMV and CGRMV were 1.9% (3 infected samples out of 154 samples) and 3.9% (6 infected samples out of 154 samples) respectively, and there was no evidence of mixed infection. In this study, we confirmed 13.3% and 4.8% single infection rate while 33.7% mixed infection rate of CNRMV and CGRMV (Table 1). The viral infection rate especially mixed infection of CNRMV and CGRMV is more prevalent in different cities of Gyeongbuk province (Daegu, Gyeongsan, and Gyeongju city) as compared to infection rate in Gyeonggi province reported by (Cho et al., 2013; 2014). Sweet cherries were firstly introduced in Daegu and Gyeongju city where the cultivation of sweet cherry had begun in Korea. At this time, viral infected cherries could be introduced in Korea. From those orchards, many infected cherry seedlings could be spread around Daegu and Gyeongju city. Hence it is assumed that co-infection with CNRMV and CGRMV was prevalent in Daegu, Gyeongju city compare to Gyeongsan city or Gyeonggi province. Meanwhile, we also checked CNRMV and CGRMV infection rate in two cherry varieties (cvs. Skeena and Regina) collected from orchards located in Daegu city by using CNRMV/CGRMV-specific primer pair (Park et al., 2004) as well as the duplex RT-PCR method. However, there was no evidence of infection by CNRMV and CGRMV in these varieties (data not shown). Meanwhile, it was reported that not only CNRMV and CGRMV but also Cherry virus A (CVA) and Apple chlorotic leaf spot virus (ACLSV) also can infect sweet cherry in Korea (Cho et al., 2013; 2014). Therefore it is necessary to develop new detection methods which can detect and investigate the previous reported viruses in different areas of Korea such as Kimchoen, Hwaseong, Pyeongtaek and so on.
In this study, a method was developed to simultaneously detect CNRMV and CGRMV in a sample by duplex RT-PCR, which has not been previously reported in Korea. The results presented here provide useful data on infection rates among sweet cherry cv. Satonishiki in specific regions in Korea, and could be applied to a quarantine system to improve management and selection of virus-free trees.




 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print






