 |
 |
| Plant Pathol J > Volume 30(1); 2014 > Article |
Abstract
A large-scale oligonucleotide (LSON) chip was developed for the detection of the plant viruses with known genetic information. The LSON chip contains two sets of 3,978 probes for 538 species of targets including plant viruses, satellite RNAs and viroids. A hundred forty thousand probes, consisting of isolate-, species- and genus-specific probes respectively, are designed from 20,000 of independent nucleotide sequence of plant viruses. Based on the economic importance, the amount of genome information, and the number of strains and/or isolates, one to fifty-one probes for each target virus are selected and spotted on the chip. The standard and field samples for the analysis of the LSON chip have been prepared and tested by RT-PCR. The probeŌĆÖs specific and/or nonspecific reaction patterns by LSON chip allow us to diagnose the unidentified viruses. Thus, the LSON chip in this study could be highly useful for the detection of unexpected plant viruses, the monitoring of emerging viruses and the fluctuation of the population of major viruses in each plant.
Due to ecosystem changes, climate warming, the evolution and mutation of the virus, plant viruses break out with the expansion of plants and agricultural products and gradually increase the risk of emerging unknown viral diseases. Viruses have caused diseases in economically important crops and vegetables, resulting in yield loss and the reduction of commodity values. So far, viruses have been diagnosed by enzyme-linked immunosorbent assays (ELISA), polymerase chain reaction (PCR), electron microscopy (EM) and biological characteristics. However, these conventional diagnostic methods can only detect single and/or multiple within 2-6 plant viruses at one time and are time consuming for an unexpected virus. Thus, the development of oligonucleotide chip is likely to assist efficient diagnosis of known and unknown plant viruses.
In early 2000, the first reports on detecting viral pathogens by the microarray system were published (Lapa et al., 2002; Vernet, 2002; Wang et al., 2002; Wilson et al., 2002). Since then, various oligonucleotide microarrays for plant virus detection have been developed. However, these microarrays only have the ability to detect viruses from specific host or several specific viruses (Boonham et al., 2003; Bystricka et al., 2005; Lee et al., 2003; Lenz et al., 2008). Recently, a new virus microarray was developed to detect a broad spectrum of plant viruses. Tiberini et al. (2010) developed the fluorescent based microarray system by Combimatrix to detect and identify 10 tomato viruses, which are economically important. Engel et al. (2010) constructed a microarray system containing 570 unique viral oligonucleotides designed to detect 44 different grapevine viruses. Zhang et al. (2010) presented an array to detect a wide spectrum of 169 plant virus species from 13 different genera. Abdullahi et al. (2011) designed the microarray system for detection of 15 virus species infecting grapevine with 162 oligonucleotide probes. Nicolaisen (2011) constructed a microarray containing 150 probes for a subset of 52 viruses representing most of the major genera of viruses.
The microarrays are powerful tools for parallel, high-throughput detection of viral diseases (Engel et al., 2010) and have the potential to be used to detect every virus for which sequence data are available in a single test (Boonham et al., 2007). About 1,200 species of viruses have been reported to infect plants (Boonham et al., 2007). Even now, novel unrecognized viruses continue to be reported. With the tremendous advances in biotechnology, some plant viruses in many crops have also provided various applications as vectors for the production of heterologous polypeptides and as gene silencing vectors as well as plant pathogens (Sainsbury et al., 2010). Therefore, a new large scale microarray for virus detection is necessary for high-throughput and broad-spectrum strategies for detection, identification and screening of plant viruses.
In this study, a proto-type microarray system containing 115 unique viral oligonucleotides designed against 30 major plant viruses was constructed and the possibility of a large-scaled oligonucleotide chip was primarily confirmed. The Large-Scale Oligonucleotide (LSON) chip which is capable of detecting most of plant viruses with known sequence information has been constructed. This contained 3,978 probes potentially capable of detecting more than 500 plant viruses in a single experiment. The validation and application of this LSON chip was performed by testing 11 plant viruses from different genera and field sample, from which it detected a new strain of soybean virus in the soybean sample showing yellow mosaic on the leaves.
Virus samples were collected from the different geographical regions of Korea and obtained from the Korean Agricultural Culture Collection (KACC). All viral infections in this study were confirmed by RT-PCR. Samples of microarray analysis were isolated from various host plants; Kyuri green mottle mosaic virus (KGMMV) isolated from cucumber, Tobacco rattle virus (TRV) isolated from tobacco, Cowpea mild mottle virus (CPMMV) isolated from pea, Zucchini yellow mosaic virus (ZYMV) isolated from zucchini, Melon necrotic spot virus (MNSV) isolated from melon, Tomato bushy stunt virus (TBSV) isolated from tomato, Potato leafroll virus (PLRV) isolated from potato, Tobacco ringspot virus (TRSV) isolated from soybean, Alfalfa mosaic virus (AMV) isolated from goosefoot, Cucumber mosaic virus (CMV) isolated from pepper and Tomato spotted wilt virus (TSWV) isolated from pepper.
All of the probes were designed on the basis of several public databases such as NCBI and DPVWeb by using the CLC Main Workbench 6 (CLC bio, USA) according to the manufacturerŌĆÖs instructions. Each probe was forty-five to fifty nucleotides in length. The orientation of the probe sequences is sense. Thus, cDNA, which was synthesized from total RNA of a plant infected with a plant virus, can be detected by the LSON chip containing the probes. GC content of the probes was ranged from forty to fifty, and Tm (melting temperature) value was ranged from forty-five to eighty. In addition, in order to avoid secondary structures of the probes being printed on the chip, the values of the self-annealing and the secondary structure were ranged no more than forty-five, and the value of the self-end annealing was ranged no more than fifteen.
Microarray slides were printed by the GenoCheck Co. (Ansan, Korea) with a Pix5500 Arrayer (Cartesian technologies co.). Corning GAPS II slides are 25 ├Ś 75 mm glass slides coated with an aminosilane surface chemistry that enables the even immobilization of DNA. Manufactured through a proprietary process, GAPS II coated slides offer unparalleled performance in terms of uniform spot morphology, high DNA retention for maximum signal strength, and improved sensitivity through ultra-low fluorescent background. Each slide contained a total of 3,978 features spotted in duplicate to reduce positional bias of the fluorescence readout, and divided up into 16 blocks (4 ├Ś 4 blocks) of 17 rows and 15 columns each. Printed slides were exposed to UV light for cross-linking and blocked in a succinic anhydride/sodium borate solution. Excess DNA probe material was then washed off in a water bath at 95┬░C. Blocked slides were dehydrated in a bath of 100% ethanol, centrifuged, and stored in a dark cool dry location before use.
Total RNA from infected and healthy plants, used in each hybridization event, was extracted using the Tri-reagent (MRC Reagents, Inc., OH, USA) as described by the manufacturer. RNA concentration and quality was determined spectrophotometrically by NanoDrop (NanoDrop Technologies, Wilmington, DE, USA), The A260/280 ratios of all RNAs were above 1.9. The reverse transcription (RT) reactions were performed in a 30 ╬╝l volume containing 10 ╬╝g of total RNA with 2uM of N25 random primers, 500 ╬╝M dATP, 500 ╬╝M dCTP, 500 ╬╝M dGTP, 200 ╬╝M dTTP, 100 ╬╝M cy3-dUTP (targeted for virus-infected sample) or 100 ╬╝M cy5-dUTP (targeted for healthy control sample) and 300 U of RevertAid H Minus Reverse Transcriptase (Invitrogen, CA) in 1X first-strand buffer. After incubation at 42┬░C for 2 hours, the samples were incubated at 70┬░C for 15 minutes and cDNA was purified using the PCR Purification Kit (Bioneer, Deajeon, Korea) according to the manufacturerŌĆÖs protocol, quantified by NanoDrop (NanoDrop Technologies, Wilmington, DE, USA). Oligonucleotide-printed slides were outfitted with a MAUI Mixer DC hybridization lid (BioMicro, Salt Lake City, Utah) prior to hybridization. Each Cy3-labelled cDNA (target virus) probe was paired with Cy5-labelled cDNA probe (healthy plant) and hybridized to LSON chip using a Maui Hybridization System (BioMicro Systems Inc). The fluorescently labeled cDNA probes were resuspended in 2X hybridization buffer (25% formamide, 5X SSC, 0.1% SDS and Bromophenol blue), denatured at 95┬░C for 5 minutes, chilled on ice for 2 minutes and applied to the microarray. Hybridization was performed on MAUI Hybridization System (BioMicro Systems Inc.) for 16 hours at 42┬░C. After hybridization, the slides were washed twice with 2X SSC plus 0.1% sodium dodecyl sulfate for 5 minutes at 42┬░C, twice with 0.1X SSC for 1 minute at room temperature and rinsed once with distilled water. After being dried by centrifugation, the microarrays were scanned.
The hybridized slides were scanned with a GenePix 4200A scanner (Axon Instrument, Union City, CA) containing a 532 nm laser for Cy3 and a 635 nm laser for Cy5 fluorescence measurement, and for image analysis GenePixpro 6.0 software was used. The level of PMT power was selected to balance the background with the level of spots detection on a slide. The hybrid spots were then identified on the array and located using a Gal file as feature indicator, so that the measurements of fluorescence and local background fluorescence for each spot could be recorded.
As a step towards developing an efficient plant virus detection chip, a small-scaled oligonucleotide microarray was constructed as a prototype for high-throughput detection (Supplementary Fig. 1). This microarray chip was designed with probes specific for the 30 plant virus species, most of which are economically important viruses in Korea. The validation of the prototype oligonucleotide microarray was performed by hybridizing labeled cDNA from plants infected with 30 viruses from different genera. Most of the virus probes were highly specific and were able to identify virus samples to be tested (data not shown). Based on these results, a LSON chip was developed for the detection of known viruses and for the monitoring of unknown viruses.
The LSON chip which is capable of detecting most of plant viruses with known sequence information has been designed. This oligonucleotide chip contained 3,978 probes in duplicate potentially capable of detecting more than 500 plant-infecting viruses in a single experiment which includes satellite RNAs, and viroids (Table 1). Probes were spotted in 16 blocks (4 ├Ś 4 blocks) in duplicate on each slide. Individual probes are arranged in 15 ├Ś 17 grid in one block for a total of 249 or 248 spots. Each block includes three positive and negative control (buffer only) probes with virus-specific probes (Fig. 1L). Positive control probes were prepared by mixing all of the probe.
Total RNA was extracted from virus-infected plants and healthy plants, labeled with Cy3 and Cy5 and used for microarray analysis. The green spots labeled with Cy3 only indicated the detection of virus in the test plant. Probes homologous to plant genes hybridize to cDNA from both the virus-infected plants and healthy plants, thus these spots appear yellow.
The validation of the LSON chip was accomplished by hybridization of labeled cDNA from plants infected with 11 viruses from different genera. An example of 11 hybridizations on the LSON chip with 3,978 probes included is shown in Fig. 1. This LSON chip was hybridized with labeled cDNA from KGMMV in the genus Tobamovirus (A), TRV in the genus Tobravirus (B), CPMMV in the genus Carlavirus (C), ZYMV in the genus Potyvirus (D), MNSV in the genus Carmovirus (E), TBSV in the genus Tombusvirus (F), PLRV in the genus Luteovirus (G), TRSV in the genus Nepovirus (H), AMV in the genus Alfamovirus (I), CMV in the genus Cucumovirus (J) and TSWV in the genus Tospovirus (K) infected plants. Each virus species provided a powerful specific hybridization. Cucumber sample infected from KGMMV showed a hybridization pattern to the ORSV spots belonging to the same genus with KGMMV. Through the hybridization pattern to the virus belonging to the same genus, unexpected viruses can be predict the virus genus. Nine out of sixteen oligo- nucleotide probes designed for TRV did not produce a reaction signal, while seven probes exhibit a high signal strength. The probes that showed negative signal in TRV specific probes are response of different strains of TRV. This observation suggests that genotyping of several strains of TRV may be possible. In all hybridization experiments, the background signal was low which indicated a high quality of hybridization. Good quality hybridization signals show that the LSON chip has a great potential for accurate detection of various viruses.
In nationwide surveys of viral diseases of soybean (Glycine max) in Korea, various soybean samples were collected and analyzed using electron microscopy and RT-PCR with primers specific to several legume-infecting viruses. A virus with isometric particles was detected from a soybean sample showing yellow mosaic on the leaves in Sangju County of South Korea in 2005 (Nam et al., 2011a). No virus infection was detected in this samples using RT-PCR with soybean virus specific primers which was previously reported in Korea. The LSON chip developed in this study was used to identify this virus sample. Specific probes of Peanut stunt virus (PSV) are indicated with the red rectangles in block 5 (Fig. 2). The green spots in red rectangles indicate the detection of PSV in the soybean. PSV in the genus Cucumovirus of the family Bromoviridae is an economically important pathogen and occurs worldwide in legume plants. Using a LSON chip this way would enable the detection of unexpected interactions such as infection of a new host.
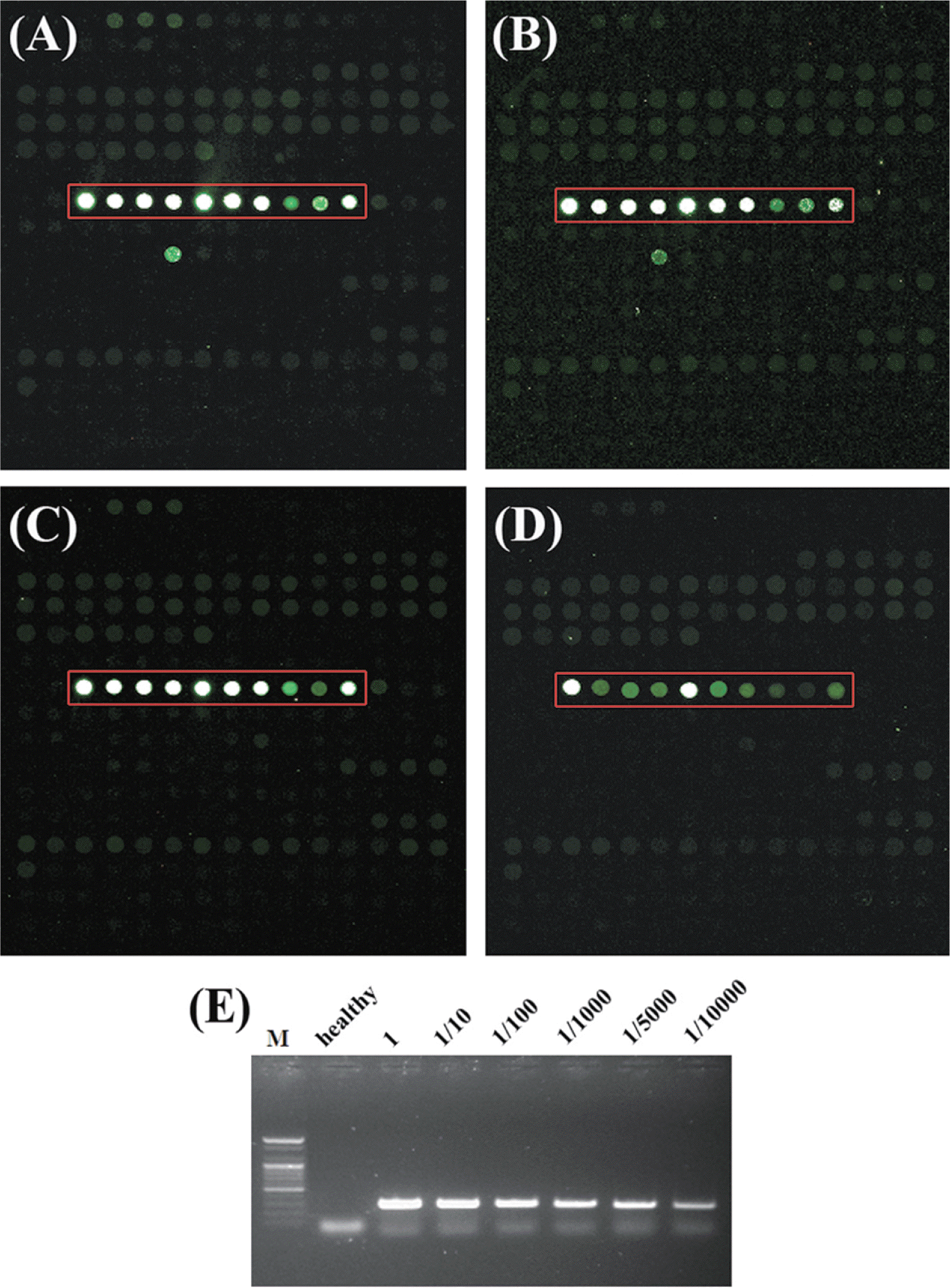
Sensitivity of a microarray is generally dependent on virus concentration in host cells and hybridization efficiency. So, it is hard to determine the exact sensitivity of a microarray. Pepper mild mottle virus (PMMoV) belongs to the plant virus genus, Tobamovirus which accumulated to high titers in plants. To test the sensitivity of the LSON chip with PMMoV, plant sap was extracted from both PMMoV-infected and healthy plants in RNA extraction buffer. Plant sap of PMMoV-infected plants was diluted with sap of healthy plants 1:10, 1:100, 1:1,000, 1:5,000 and 1:10,000 and hybridized to the microarray together with a healthy control (Fig. 3A-D). For a comparison with RT-PCR, plant material from the PMMoV infected plants was diluted 1:10, 1:100, 1:1,000, 1:5,000 and 1:10,000 and tested by RT-PCR (Fig. 3E). Comparison of the array detection of PMMoV with RTPCR showed that RT-PCR was more sensitive method for detecting virus up to the 1:10,000 dilutions, the microarray experiments revealed the presence of PMMoV in dilutions up to 1:1,000 (Fig. 3).
A new concept of diagnostic tools which can detect the unexpected virus occurring in crops and weeds and monitor virus infection in incoming and outgoing plant samples has been required unlike the conventional methods such as ELISA or PCR. A microarray system for the detection of viral infection offers a most attractive method for diagnosis of unknown viruses and monitoring of emerging viruses. However, microarrays have been criticized as expensive for routine use (Everett et al., 2010). The effective use of microarrays for viral disease requires the ability of detection with a broad-spectrum of known viruses as well as novel viruses in single experiment, while conventional methods could detect one or several viral diseases.
To construct small-scaled oligonucleotide chip (prototype chip), 115 probes were initially used for thirty different virus species selected by the importance of host crops and the occurrence for the last several years nationwide. Virus samples are obtained from the virus collections in the National Institute of Agricultural Science and Technology and fields samples nationwide. Field samples are further purified by inoculating test plants or directly applied to oligonucleotide DNA chips due to the difficulties in mechanical inoculation. All samples are carefully examined by RT-PCR and nucleotide sequencing of the products before total RNA extraction. Genome information of target viruses is obtained from several public databases, such as NCBI and DPVWeb. Approximately five thousand oligonucleotides for thirty target viruses are designed by CLC Main Workbench 6 (CLC bio, USA). Among them, 115 oligonucleotides are selected and screened. They are selected based on the transcription mechanism, titers of viruses in host plants and copy number of target genomic or subgenomic RNA. Most of the selected virus probes were highly specific and were able to identify virus samples to be tested (data not shown). Based on the performance of this proto-type chip, a large-scale oligonucleotide (LSON) chip was developed for the detection of the known viruses and monitoring of unknown viruses. The LSON chip contains a total of 3,978 probes in duplicate for 538 of targets including plant viruses, satellite RNAs and viroids. The probes consisting of isolate-, species- and genus-specific probes respectively, are designed from 20,000 of independent nucleotide sequence of plant viruses. This LSON chip could be capable of detecting a larger number of plant viruses with a single microarray than any other microarray reported so far.
Virus screening of field samples using the LSON chip could detect several viruses more rapidly and easily. There was a clinical diagnosis for abnormal symptom on paprika (Capsicum annuum var. angulosum) growing in a greenhouse in Jinju City on December 2010. The sample was analyzed by electron microscopy and RT-PCR for the known viruses in paprika in Korea, but the results were shown to be negative. In the LSON chip analysis, the agent was diagnosed with Beet western yellows virus (BWYV), a quarantine pathogen in Korea. Swift handling of BWYV could be helpful to prevent the agent from spreading. Important novel emerging diseases like, SYCMV in the genus sobemovirus, have been identified with this LSON chip (Nam et al., 2012b). The soybean sample was analyzed by electron microscopy and RT-PCR for the known soybean viruses in Korea, but it failed to detect any other viruses. In the LSON chip analysis, the microarray analysis exhibited positive hybridization with conserved probes for the genus Sobemovirus. The virus sample was diagnosed with an unknown sobemovirus. In addition, many unexpected viruses have been detected effectively using LSON chip.
In conclusion, we have developed a large-scale oligonucleotide (LSON) chip microarray for detection and identification of 538 species of plant virus. This LSON chip will be able to detect most known plant RNA viruses and offer a most attractive method for the detection of unknown viruses and the monitoring of emerging viruses and strains. The probes using this microarray will be expanded through ongoing improvement to detect emerging viruses and strains. Thus, the LSON chip in this study could be highly useful for the detection of known and unknown plant viruses, the monitoring of emerging viruses and strains and the fluctuation of the population of major viruses in each plant.
Acknowledgments
This research was supported by Kyungpook National University Research Fund, 2013 and by a grant from the Next-Generation BioGreen 21 Program (PJ009033), Rural Development Administration, Republic of Korea.
Fig.┬Ā1.
Hybridization patterns of the LSON chip analysis for detection. For each array, plant samples infected with 11 viruses from different genus were used. (A) Kyuri green mottle mosaic virus (KGMMV) in the genus Tobamovirus, (B) Tobacco rattle virus (TRV) in the genus Tobravirus, (C) Cowpea mild mottle virus (CPMMV) in the genus Carlavirus, (D) Zucchini yellow mosaic virus (ZYMV) in the genus Potyvirus, (E) Melon necrotic spot virus (MNSV) in the genus Carmovirus, (F) Tomato bushy stunt virus (TBSV) in the genus Tombusvirus, (G) Potato leafroll virus (PLRV) in the genus Luteovirus, (H) Tobacco ringspot virus (TRSV) in the genus Nepovirus, (I) Alfalfa mosaic virus (AMV) in the genus Alfamovirus, (J) Cucumber mosaic virus (CMV) in the genus Cucumovirus, (K) Tomato spotted wilt virus (TSWV) in the genus Tospovirus (L).

Fig.┬Ā2.
Detection of a new virus strain of Peanut stunt virus (PSV) in the soybean using the LSON chip. Specific probes of Peanut stunt virus (PSV) are indicated with red rectangles in block 5. Using a LSON chip would enable the detection of unexpected interactions such as infection of a new strain of PSV in the soybean, which has not been reported in Korea yet.
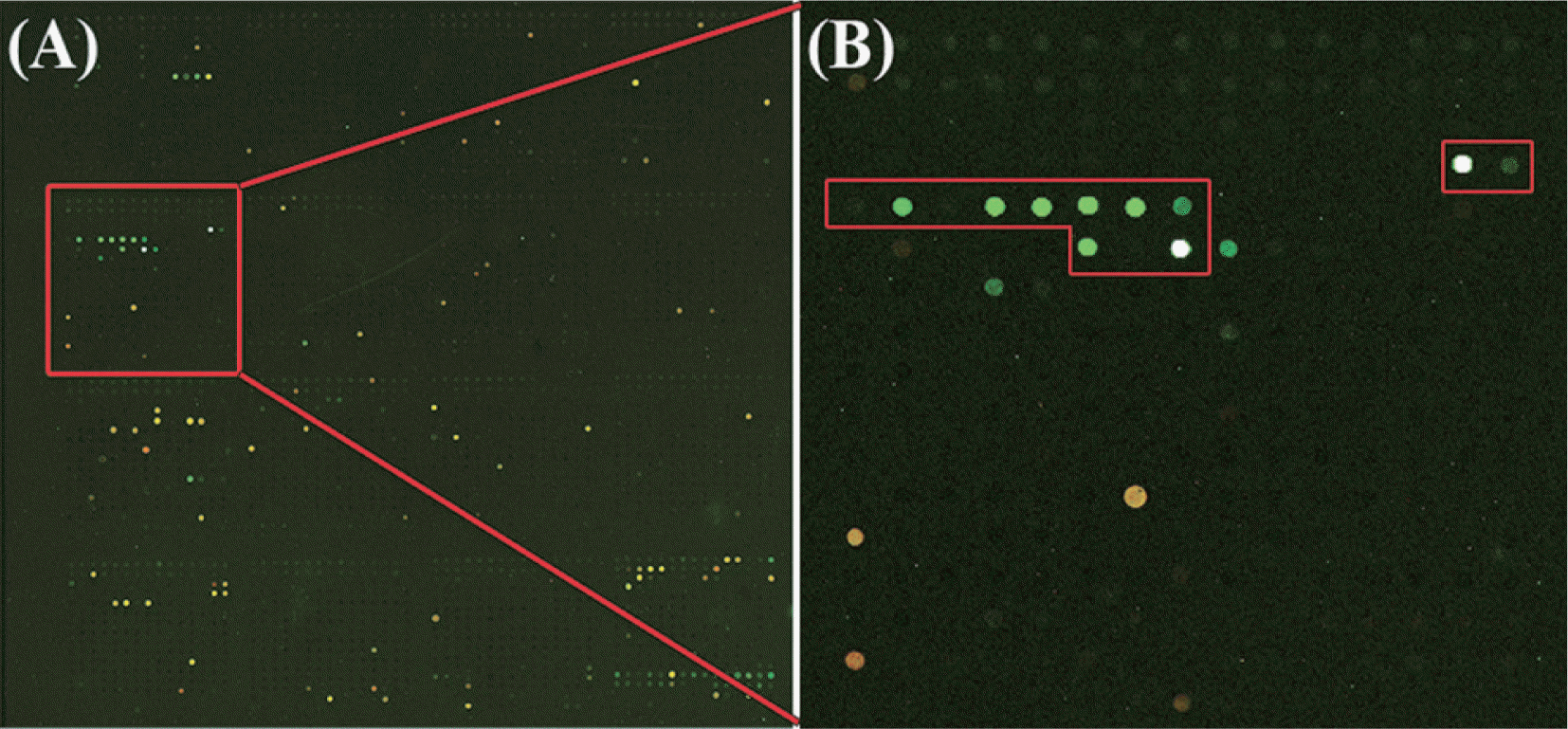
Fig.┬Ā3.
Sensitivity comparison using plant dilution sap extracted from both Pepper mild mottle virus (PMMoV)-infected and health plants between the LSON chip and PCR methods. Specific probes of PMMoV are indicated with red rectangles in block 14. PMMoV sample for sensitivity test was diluted with (A) undiluted sample, (B) 1:10, (C) 1:100, (D) 1:1000, (E) Sensitivity of RT-PCR with PMMoV specific primers.
Table┬Ā1.
Lists of target viruses in the LSON chip
References
Abdullahi, I, Gryshan, Y and Rott, M 2011. Amplification-free detection of grapevine viruses using an oligonucleotide microarray. J. Virol.Methods. 178:1-15.


Boonham, N, Tomlinson, J and Mumford, R 2007. Microarrays for rapid identification of plant viruses. Annu Rev Phytopathol. 45:307-328.


Boonham, N, Walsh, K, Smith, P, Madagan, K, Graham, I and Barker, I 2003. Detection of potato viruses using microarray technology: towards a generic method for plant viral disease diagnosis. J. Virol.Methods. 108:181-187.


Bystricka, D, Lenz, O, Mraz, I, Piherova, L, Kmoch, S and Sip, M 2005. Oligonucleotide-based microarray: a new improvement in microarray detection of plant viruses. J. Virol.Methods. 128:176-182.


Engel, EA, Escobar, PF, Rojas, LA, Rivera, PA and Fiore, N 2010. A diagnostic oligonu-cleotide microarray for simultaneous detection of grapevine viruses. J. Virol.Methods. 163:445-451.


Everett, KR, Rees-George, J, Pushparajah, IPS, Janssen, BJ and Luo, Z 2010. Advantages and disadvantages of microarrays to study microbial population dynamics-a minireview. New Zealand Plant Protect. 63:1-6.


Lapa, S, Mikheev, M, Shchelkunov, S, Mikhailovich, V, Sobolev, A, Blinov, V, Babkin, I, Guskov, A, Sokunova, E, Zasedatelev, A, Sandakhchiev, L and Mirzabekov, A 2002. Species-level identification of orthopoxviruses with an oligonucleotide microchip. J Clin Microbiol. 40:753-757.




Lee, GP, Min, BE, Kim, CS, Choi, SH, Harn, CH, Kim, SU and Ryu, KH 2003. Plant virus cDNA chip hybridization for detection and differentiation of four cucurbit infection Tobamoviruses. J. Virol.Methods. 110:19-24.

Lenz, O, Petrzik, K and Spak, J 2008. Investigating the sensitivity of a fluorescence- based microarray for the detection of fruit-tree virus. J. Virol.Methods. 148:96-105.

Nam, M, Park, SJ, Kim, YJ, Kim, JS, Park, CY, Lee, JS, Choi, HS, Kim, JS, Kim, HG and Lee, SH 2012a. First report of Peanut stunt virus on Glycine max in Korea. Plant Pathol J. 28:330.

Nam, M, Kim, JS, Park, SJ, Park, DY, Lee, JS, Choi, HS, Kim, JS, Kim, HG, Lim, SM, Moon, JS and Lee, SH 2012b. Biological and molecular characterization of soybean yellow common mosaic virus, a new species in the genus Sobemovirus. Virus Res. 163:363-367.


Nicolaisen, M 2011. An oligonucleotide-based microarray for detection of plant RNA viruses. J. Virol.Methods. 173:134-143.

Sainsbury, F, Canizares, MC and Lomonossoff, GP 2010. Cowpea mosaic virus: The plant virus-based biotechnology workhorse. Annu Rev Phytophthol. 48:437-455.

Tiberini, A, Tomassoli, L, Barba, M and Hadidi, A 2010. Oligonucleotide microarray-based and identification of ten major tomato viruses. J. Virol.Methods. 168:133-140.

Wang, D, Coscoy, L, Zylberberg, M, Avila, PC, Boushey, HA, Ganem, D and DeRisi, JL 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci.USA. 99:15687-15692.

- TOOLS



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




