Molecular Characteristics of Pseudomonas syringae pv. actinidiae Strains Isolated in Korea and a Multiplex PCR Assay for Haplotype Differentiation
Article information
Abstract
The molecular features of Pseudomonas syringae pv. actinidiae strains isolated in Korea were compared with strains isolated in Japan and Italy. Sequencing of eight P. syringae pv. actinidiae and three P. syringae pv. theae strains revealed a total of 44 single nucleotide polymorphisms across 4,818 bp of the concatenated alignment of nine genes. A multiplex PCR assay was developed for the detection of P. syringae pv. actinidiae and for the specific detection of recent haplotype strains other than strains isolated since the 1980s in Korea. The primer pair, designated as TacF and TacR, specifically amplified a 545-bp fragment with the genomic DNA of new haplotype of P. syringae pv. actinidiae strains. A multiplex PCR conducted with the TacF/TacR primer pair and the universal primer pair for all P. syringae pv. actinidiae strains can be simultaneously applied for the detection of P. syringae pv. actinidiae and for the differentiation of new haplotype strains.
Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit, was first described in Japan in 1989 (Takikawa et al., 1989) and subsequently reported in Korea (Koh et al., 1994). The bacterial canker was highly destructive, devastating kiwifruit orchards and causing economic losses in both countries. Although this pathogen was isolated in Italy in 1992, it had a limited economic impact with a low incidence on kiwifruit plants until 2008 (Scotichini, 1994). However, severe epidemics have been reported in central Italy since 2008, first in Actinidia chinensis and then in A. deliciosa (Balestra et al., 2009; Ferrante and Scortichini, 2009). The bacterial canker disease found in central Italy appeared to have been caused by a different population from that reported in 1992 (Ferrante and Scortichini, 2010). A severe form of P. syringae pv. actinidiae was then isolated in other European countries, including Portugal (Balestra et al., 2010), Spain (Balestra et al., 2011), France (Vanneste et al., 2011), and Turkey (Bastas and Karakaya, 2012). This pathogen was also found in both New Zealand and Chile in 2010 (Everett et al., 2011; Anonymous, 2011). Indeed, the worldwide kiwifruit industry is seriously threatened by this new pathogen.
P. syringae pv. actinidiae was suspected of being introduced into Korea from Japan through imported seedlings of kiwifruit, because bacterial canker was first observed in the late 1980s in Korea (Koh et al., 1994), several years after its first occurrence in Japan. However, the Korean strains did not have the coding sequences for phaseolotoxin, which is present in the Japanese strains; instead they have the coronatine biosynthesis genes (Han et al., 2003). The genes for coronatine biosynthesis were not found in P. syringae pv. actinidiae isolates from Japan, Italy, New Zealand, and Chile (Chapman et al., 2012). To date, the coronatine biosynthesis genes have only been detected from the Korean strains. Lee et al. (2005) distinguished Korean strains from Japan strains using the random amplified polymorphic DNA (RAPD) analyses and suggested that Korean and Japanese strains of P. syringae pv. actinidiae may have different phylogenetic origins.
The P. syringae pv. actinidiae population consists of different haplotypes that show different levels of virulence toward kiwifruit and their ability to rapidly spread. P. syringae pv. actinidiae strains recently isolated in Europe, New Zealand, and Chile belong to a single genetic lineage and are considered more virulent than the Italian strains isolated in 1992 (Chapman et al., 2012; Mazzaglia et al., 2012). In 2011 P. syringae pv. actinidiae strains showing the rep-PCR pattern similar to those from recent Italian outbreaks were isolated from a kiwifruit orchard in Korea (Koh et al., 2012). These strains do not possess gene clusters coding for the toxins coronatine or phaseolotoxin, which were retained by the previous Korean and Japanese strains, respectively (Han et al., 2003). The effector genes, hopA1 and hopH1, present in the strains from recent outbreaks in Italy (Ferrante and Scortichini, 2010) were detected from these isolates. These strains were therefore considered new type of P. syringae pv. actinidiae closely related to those recently isolated from Italy (we will refer to these strains as “new haplotype strains” throughout this article).
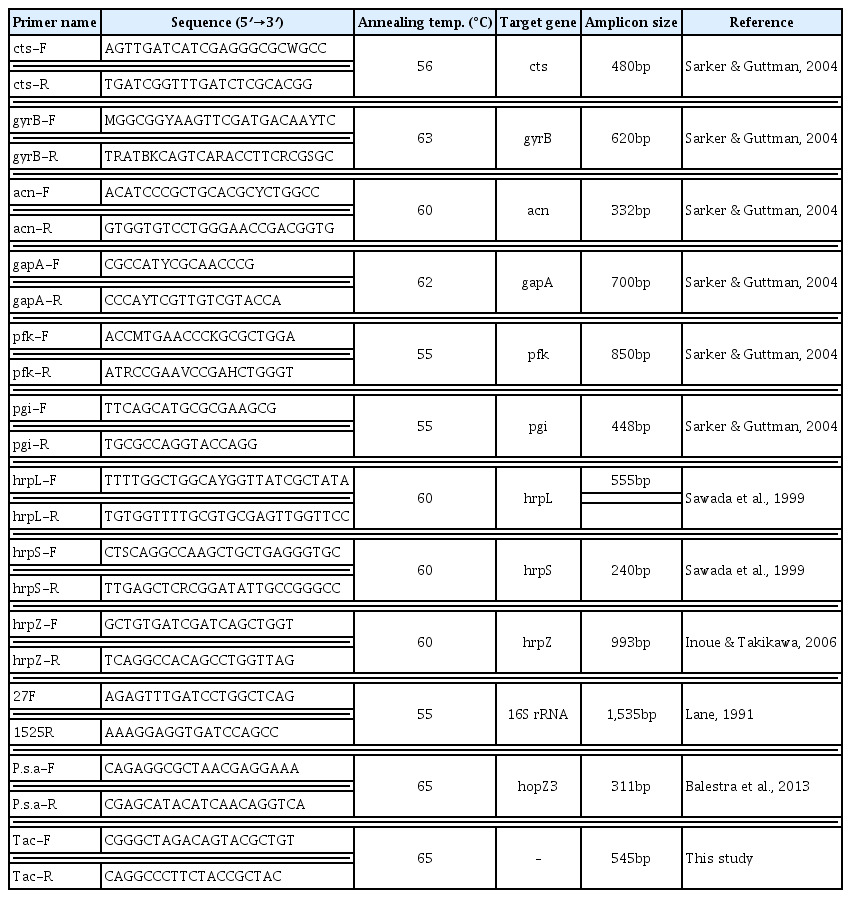
In this work, two Korean and two Japanese strains (obtained in 1999 and 1989, respectively), two Italian strains (isolated in 2011), two “new haplotype” of Korean strains (isolated in 2011), and three P. syringae pv. theae strains were used for multilocus sequence analysis (MLSA). DNA fragments of six housekeeping genes, cts, gyrB, acnB, gapA, pfk, and pgi, and three effector genes, hrpL, hrpS, and hrpZ, were amplified and sequenced (Table 1). The nucleotide sequences of PCR primers and annealing temperature used for MLSA are listed in Table 2. Forward and reverse sequences of PCR products were obtained by direct sequencing method using the primers for each locus.

Single nucleotide polymorphisms (SNPs) that distinctively recognize Pseudomonas syringae pv. actinidiae and P. syringae pv. theae strains
Sequencing of eight P. syringae pv. actinidiae and three P. syringae pv. theae strains revealed a total of 44 single nucleotide polymorphisms (SNPs) across 4,818 bp of the concatenated alignment of nine genes (Table 1). Two Italian and two “new haplotype” of Korean strains were identical in sequences at nine loci determined. Consequently, these strains may belong to the same genetic lineage.
The numbers of SNPs between the new haplotype strains (SYS1, SYS4) and Korean (CJW3, JYG6) and Japanese strains (Kw11, PaB1) previously isolated were 11 and 4, respectively. These data indicate that the new haplotype strains are more closely related to the Japanese strains than the Korean strains. This result also confirmed previous report that the strains from the recent European outbreaks belong to separate branch in phylogenetic tree with the Korean and the Japanese strains (Mazzaglia et al., 2011).
In addition, the number of SNPs between Korean (CJW3, JYG6) and Japanese strains (Kw11, PaB1) was 13, suggesting these two populations belong to separate genetic lineages. Of these 13 nucleotide variations, eleven sites were in the housekeeping genes, cts, acnB, and gapA, which are essential for survival and have a low probability of horizontal genetic exchange. Mazzaglia et al. (2012) estimated a divergence time of the Korean and Japanese strains from their common ancestor on the order of hundreds of years.
Chapman et al. (2012) also elucidated the phylogenetic relationships between P. syringae pv. actinidiae isolates worldwide using MLSA of housekeeping, type III effector, and phytotoxin genes. They showed that at least four MLSA groups, Psa1-4, are present in P. syringae pv. actinidiae populations globally. The Japanese strains were genetically identical to Italian strains isolated in 1992 and designated as Psa1. The P. syringae pv. actinidiae population present in Korea was genetically very similar to Japanese strains but formed a distinct genetic lineage named Psa2. The Psa3 group represented isolates from Italy (2008–2009 outbreaks), New Zealand, and Chile. The Psa4 strains have been isolated from kiwifruit plants in New Zealand and Australia that showed no bacterial canker symptoms. MLSA results of this work suggested that the new haplotype of Korean strains isolated in 2011 also belong to the Psa3 MLSA group.
The number of SNPs between type strains of P. syringae pv. actinidiae (Kw11) and P. syringae pv. theae (LMG 5092) was 36, while that of Kw11 and two other P. syringae pv. theae strains (MAFF 302851 and 302852) was 31. A total of nine SNPs were identified between type strain LMG 5092 and two other P. syringae pv. theae strains, illustrating genetic variability at the intra-pathovar level (Table 1). P. syringae pv. theae and P. syringae pv. actinidiae both belong to genomospecies 8 sensu Gardan et al. (1999) and are very closely related (Scortichini et al., 2002; Ferrante and Scortichini, 2011). In fact, PCR primers designed to be complementary to a portion of the 16S–23S rRNA intertranscribed spacer regions are unable to distinguish P. syringae pv. actinidiae from P. syringae pv. theae (Rees-George et al., 2010). For the further characterization of strains belonging to the two pathovars, the 16S rRNA gene sequences were compared. PCR amplification and sequencing of the 16S rRNA gene were carried out as described previously (Lee et al., 2012). The 16S rRNA gene sequence of a type strain of P. syringae pv. theae (LMG 5092) was identical to the new haplotype strains but showed three base differences with the pathotype strain of P. syringae pv. actinidiae (Kw11). However, the 16S rRNA gene sequences of three P. syringae pv. theae strains determined in this work disclosed some differences. The nucleotides “ATC” were found for pathovar type strain LMG 5092, while the nucleotides “GAT” were found for the other two strains at position 471–473 of Escherichia coli numbering (Brosius et al., 1978). Although the 16S rRNA gene sequence in a type strain of P. syringae pv. theae revealed 100% similarity with that of the new haplotype of P. syringae pv. actinidiae, the other two pv. theae strains had identical sequences with Korean and Japanese strains of P. syringae pv. actinidiae previously isolated. As the type strain of P. syringae pv. theae LMG 5092 formed distinct clusters with other strains in ribotyping analysis in a previous study (Gardan et al., 1999), Bull et al. (2010) suggested that this strain is not suitable as a pathotype strain. Although the number of strains examined in this study is not enough to make a definite conclusion, the MLSA and the 16S rDNA sequence results also revealed that strain LMG 5092 could not represent P. syringae pv. theae as a pathotype strain.
The new haplotype strains isolated in a Korean orchard in 2011 have not spread to other orchards to date. Instead, P. syringae pv. actinidiae strains recently isolated from other orchards in Korea were identical to those first reported in 1994 (Koh et al., 1994). It is interesting to note that although the new haplotype strain found in Italy had rapidly spread, that from Korea was limited to the orchard where it first occurred. To monitor the spread of the new haplotype strain in Korea, a multiplex PCR method which can detect P. syringae pv. actinidiae and differentiate the new haplotype strains, was developed in this study.
The first primer pair was previously designed to specifically detect P. syringae pv. actinidiae (Balestra et al., 2013). This primer pair was based on the sequence of the hopZ3 gene which is conserved among all P. syringae pv. actinidiae strains determined but different from other P. syringae pathovars including pv. theae. The primer pair “P. syringae pv. actinidiae F/R” amplifies a 311-bp product from P. syringae pv. actinidiae strains. However, three additional primer pairs which were designed by Balestra et al. (2013) to distinguish European, Chinese, and Japanese/Korean P. syringae pv. actinidiae strains did not produce the expected size of amplicons with DNA of new haplotype strain isolated in Korea (data not shown). The second primer set was designed based on a RAPD marker specific to the new haplotype strain. Random primer OPA-01 (Operon Biotechnologies, USA) was used as a single primer. A specific RAPD product was excised from the agarose gel and the DNA was purified from the gel using an AccuPrep gel purification kit (Bioneer, Korea). The purified DNA was ligated into a pGEM-T Easy vector (Promega, USA) following the manufacturer’s instructions. The nucleotide sequence of the inserted fragment was determined by SolGent Co. (Korea) and was deposited in GenBank under accession number KF772879. A RAPD marker was converted into a sequence characterized amplified region (SCAR) marker using sequence information. The SCAR primer pair designated as Tac F/R was expected to amplify a 545-bp DNA fragment with genomic DNA of the new haplotype strains. Nucleotide sequences of PCR primers used for multiplex PCR are listed in Table 2.
The multiplex PCR was performed with a DNA Thermal Cycler (Takara Shozo, Japan) under the following conditions: an initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 65°C for 30 s, extension at 72°C for 30 s, and final extension at 72°C for 7 min. Each reaction mixture contained a 1x PCR buffer (10 mM Tris-HCl, 40 mM KCl, 1.5 mM MgCl2, pH 9.0), 0.2 mM dNTPs, 10 pmol of each primers, 1.25 U of Top DNA polymerase (Bioneer, Korea), and 10 ng of template DNA in a volume of 50 μl. Results employing two primer pairs in a multiplex PCR with eight P. syringae pv. actinidiae and three P. syringae pv. theae are shown in Fig. 1.
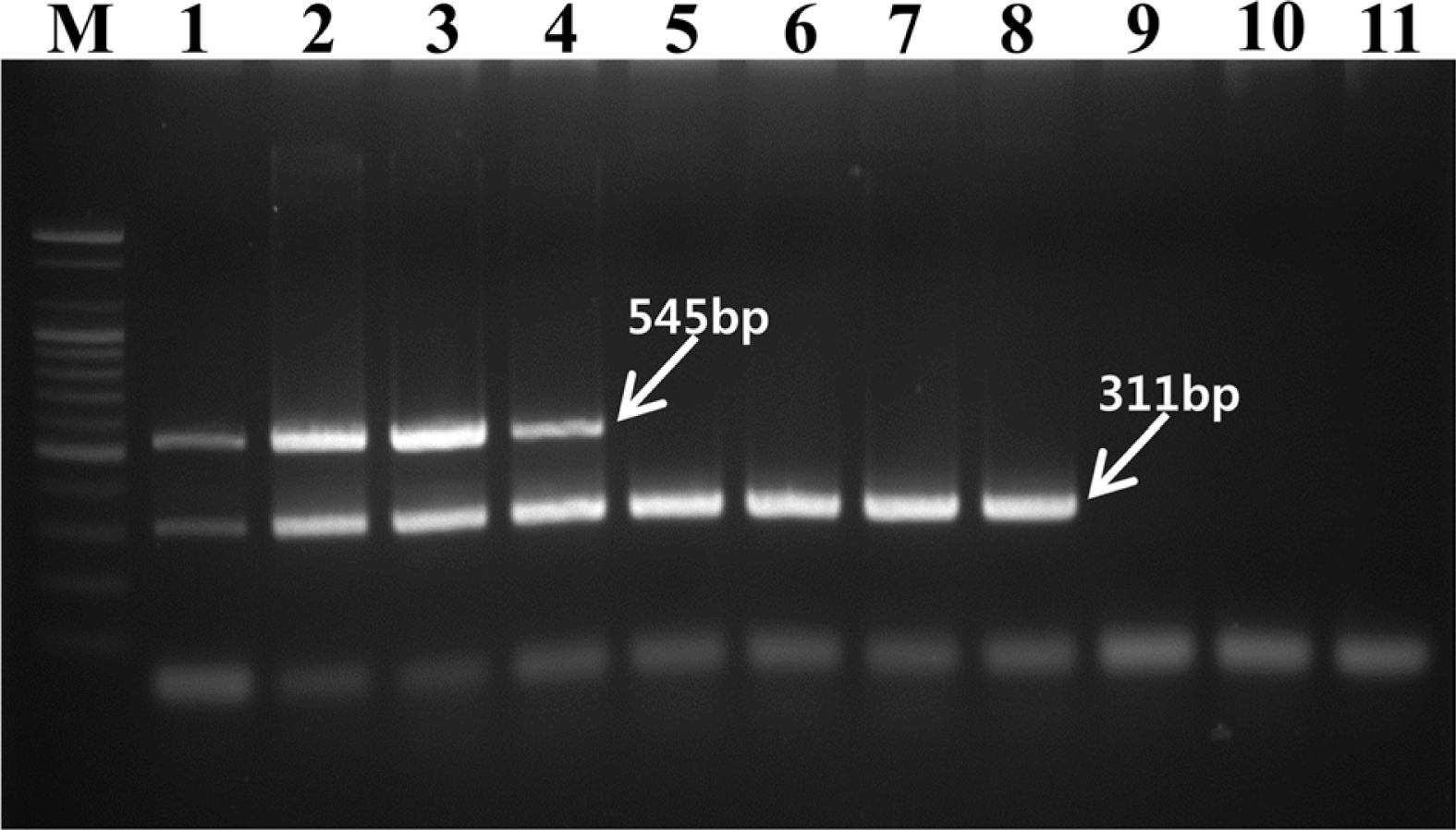
Agarose gel electrophoresis patterns of PCR products amplified by the multiplex PCR assay. Lanes 1–8 were Pseudomonas syringae pv. actinidiae strains and lanes 9–11 were P. syringae pv. theae strains. Lane M, 100 bp marker (Bioneer); lane 1, SYS1; lane 2, SYS4; lane 3, IHL1; lane 4, IKB4; lane 5, CJW3; lane 6, JYG6; lane 7, Kw11; lane 8, PaB1; lane 9, LMG 5092; lane 10, MAFF 302851; lane 11, MAFF 302852.
The expected fragments of 311 bp were amplified from the all eight P. syringae pv. actinidiae strains (lanes 1 to 8) but the three P. syringae pv. theae strains did not produce any amplicon (lanes 9 to 11). The four new haplotype strains, including two strains isolated in Korea (SYS1, SYS4) and two obtained from Italy (IHL1, IKB4), produced an expected 545-bp amplicon (lanes 1 to 4), while the two Korean (CJW3, JYG6) and two Japanese strains (Kw11, PaB1) did not amplify this DNA segment with primer pair Tac F/R (lanes 5 to 8). The multiplex PCR method with two primer pairs, P. syringae pv. actinidiae F/R and Tac F/R, can be used simultaneously to detect P. syringae pv. actinidiae and to distinguish the new haplotype strains from other haplotype strains which have been isolated in Japan and Korea since the 1980s. Therefore, the multiplex PCR assay designed in this study could be a valuable tool for monitoring the spread of the new haplotype strain in Korea.
Acknowledgements
This study was carried out with the support of the “Cooperative Research Program for Agricultural Science and Technology Development (PJ009326)”, Rural Development Administration, Republic of Korea.