Analysis of Fungicide Sensitivity and Genetic Diversity among Colletotrichum Species in Sweet Persimmon
Article information
Abstract
Anthracnose, caused by Colletotrichum gloeosporioides (C. gloeosporioides; Teleomorph: Glomerella cingulata), is the most destructive disease that affects sweet persimmon production worldwide. However, the biology, ecology, and genetic variations of C. gloeosporioides remain largely unknown. Therefore, in this study, the development of fungicide resistance and genetic diversity among an anthracnose pathogen population with different geographical origins and the exposure of this population to different cultivation strategies were investigated. A total of 150 pathogen isolates were tested in fungicide sensitivity assays. Five of the tested fungicides suppressed mycelial pathogen growth effectively. However, there were significant differences in the sensitivities exhibited by the pathogen isolates examined. Interestingly, the isolates obtained from practical management orchards versus organic cultivation orchards showed no differences in sensitivity to the same fungicide. PCR-restriction fragment length polymorphism (RFLP) analyses were performed to detect internal transcribed spacer regions and the β-tubulin and glutamine synthetase genes of the pathogens examined. Both the glutamine synthetase and β-tubulin genes contained a complex set of polymorphisms. Based on these results, the pathogens isolated from organic cultivation orchards were found to have more diversity than the isolates obtained from the practical management orchards.
Sweet persimmon (Diospyros kaki L.) is cultivated as a major economic fruit crop in Korea. The pomiculture of sweet persimmon has been steadily increasing, and currently has reached nearly 29,000 hectares. Correspondingly, sweet persimmon constitutes a large portion of the domestic market in Korea, and it is also steadily increasing as an overseas export. However, anthracnose, caused by Colletotrichum gloeosporioides (C. gloeosporioides) in sweet persimmon, is a devastating disease that affects fruit production (Kwon and Park, 2004). In general, Colletotrichum spp. infect various plants and are responsible for considerable economic losses, including those from sweet persimmon crops. Currently, at least two Colletotrichum spp. have been identified as causal anthracnose agents in sweet persimmon, C. gloeosporioides and C. acutatum (Zhang et al., 2005). Anthracnose occurs on the young twigs, tissues of stems, flowers, and fruits, with minimal effects on leaves (Kim et al., 2004). Anthracnose disease not only reduces fruit yield and quality, but can also cause severe postharvest disease.
Currently, chemical control is considered the best method for treating crop disease. For the cultivation of sweet persimmon, fungicide spray is applied 6–8 times to provide disease control. However, despite the regular application of fungicide spray, many sweet persimmon orchards are still severely damaged by anthracnose disease. This may be due to the development of tolerance by fungicide-resistant pathogen strains (Wharton and Deiguez-Uribeondo, 2004). In a previous study, anthracnose pathogens were found to be effectively controlled by prochloraz and tebuconazole, with up to 91% control achieved (Lim and Choi, 2006). In contrast, propineb only inhibited 46.4% of a hypha growth strain (Lim et al., 2009). Lim and Choi (2006) further demonstrated that the frequency of pathogen isolates that exhibit propineb resistance has increased, and pathogens that were continually exposed to the fungicide developed resistance. Moreover, the resistance isolates became more dominant within the pathogen population. For each isolated pathogen, geological separation also contributed to the differences observed in fungicide sensitivity (Kumar et al., 2007).
Morphological characteristics of anthracnose pathogens represent a complicated relationship among the genus. For taxa identification in fungi, sequencing of the internal transcribed spacer (ITS) is most commonly used. Gene variations in both the ITS regions and the β-tubulin gene sequences have been used for the identification of C. acutatum versus C. capsici and C. gloeosporioides species (Phoulivong et al., 2010). However, the genus, Colletotrichum, is a genetically variable and complex pathogen, and is considered to be a group species (Sutton, 1992). Therefore, the identification of gene variations is not useful for identifying the Colletotrichum species. Recent studies of anthracnose have focused on the complex relationship, or genetic diversity, among pathogen populations (Xie et al., 2010). Molecular markers have been used for characterizing race, phylogenetic relationships, and/or diversity among pathogen populations (Lopez and Lucas, 2010). In particular, restriction fragment length polymorphism (RFLP) patterns of ribosomal DNA have been used to differentiate Colletotrichum species (Ramdeen et al., 2013), and often, β-tubulin or glutamine synthetase (GS) gene sequences are employed to identify or determine Colletotrichum genetic variations (Guerber et al., 2003).
The present studies were performed to elucidate the relationship between fungicide sensitivity and genetic variations among sweet persimmon anthracnose pathogens. In addition, the effect of different methods of cultivation on the development of fungicide resistance strains among the Colletotrichum population was investigated.
Materials and Methods
Origin and isolation of the anthracnose pathogens
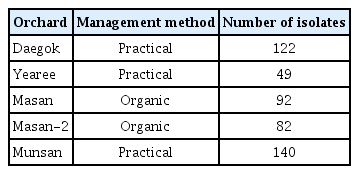
During the cultivation season between 2012 and 2013, five sweet persimmon orchards in the Gyeongnam province were visited. Three orchards were maintained with practical management methods (Daegok, Munsan, Yearee) and two orchards (Masan, Masan-2) were maintained with organic farming methods. Typical anthracnose disease symptoms were observed on twigs and fruit, and infected samples were collected and transported to the lab within three hours of collection. Disease lesions were cut from the surface of the fruit, with a depth less than 1 cm. The surface of each lesion was sterilized with 1% sodium hypochlorite followed by 70% ethanol and then the specimens were washed in sterilized distilled water three times. The specimens were dried for 15 min under a fume hood and were placed in water agar media (WA) at 28°C until spores developed. Conidia were observed using a microscope (Olympus BX15). From the five orchards, a total of 485 pathogens were isolated (Table 1).
Identification of the pathogen isolates with species-specific PCR
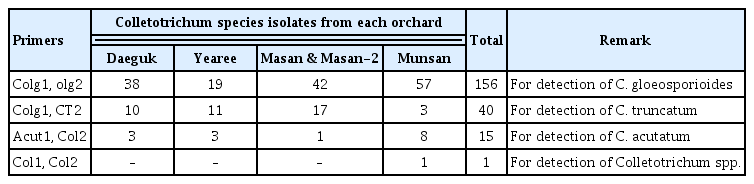
Genomic DNA was extracted from fresh mycelium by the CTAB method (Ford et al., 2000). DNA from each isolate was amplified using the following species-specific primer pairs: Colg1 (5′-AACCCTTTGTGAACATACC-3′) and Colg2 (5′-CCCTCCGGATCCCAG-3′) (443-bp) for C. gloeosporioides; and Colg1 and CT 2 (5′-CTT TAA GGG CCT ACG TCA A-3′) (375-bp) for C. truncatum (Chen et al., 2006). The PCR conditions included an initial denaturation step at 94°C for 5 min, followed by 34 cycles of 94°C for 30 sec, 63°C for 30 s, and at 72°C for 1 min. A final extension step heated samples to 72°C for 6 min. The primer set, col2 (d5′-TTA CTA CGC AAA GGA GGC T-3′) and acut1 (d5′-CCG GAG GAA ACC AAA CTC TAT TTAC-3′) (462-bp) were used to detect C. acutatum, while the primer set, col1 (d5′-AAC CCT TTG TGA ACR TAC CTA-3′) and col2 (318-bp), were employed for all other Colletotrichum spp. (Martinez et al., 2003). The same PCR conditions were applied for the identification of C. acutatum and Colletotrichum spp. samples, except that the annealing temperature was 56°C for the col2 and col2 primers. The PCR mixtures for analysis included 10 μg of template DNA, 1U Taq DNA polymerase, 0.1 mM dNTPs, 2×PCR reaction buffer, and 10 pmol of each primer. The complete ITS region of the ribosomal DNA were amplified with the primers, ITS1 (5′-GCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3) (Kumar et al., 2005). Sequencing was subsequently performed by Solgent Co. (Daejon, Korea). Sequences were compared to previously deposited Colletotrichum spp. sequences in GenBank (www.blast.ncbi.nlm.nih.gov/).
Evaluation of the fungicide sensitivity of the anthracnose pathogens
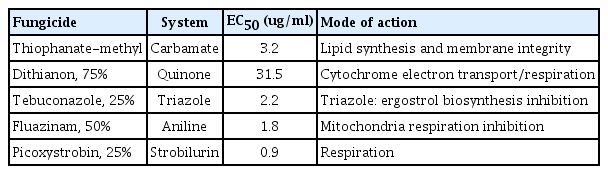
Five fungicides were selected based on their mode-of-action and standard operating procedure (SOP) (Table 2). Thiophanate-methyl (Carbamate), dithianon (Quinone), tebuconazole (Triazole), fluazinam (Aniline), and picoxystrobin (Strobilurin) were evaluated for effective concentration 50 (EC50) using a standard pathogen strain (C. gloeosporioides JH-1). Briefly, six-well plates were filled with PDA media containing amended with half-log scale concentrations of each fungicide (0 μg/ml, 0.1 μg/ml, 0.213 μg/ml, 0.457 μg/ml, 1 μg/ml, 2.13 μg/ml for fluazinam and thiophanate-methyl; 0 μg/ml, 0.213 μg/ml, 0.457 μg/ml, 1 μg/ml, 2.13 μg/ml, and 4.57 μg/ml for picoxystrobin, dithianon, and tebuconzaole) and a 4-mm plug of actively growing mycelium was placed at the center of each agar plate. Mycelium growth was measured every 24 h up to 5 days. EC50 values were calculated using a Probit model with curve slope value.
The sensitivity of each isolate (n=150, with 30 isolates from each orchard) to the fungicides examined was tested by determining the EC50 concentrations for each fungicide. Mycelia growth was measured 3, 5, and 7 days after the inoculation of fungi on the fungicide amended with 1/5×PDA at 28°C. Sensitivity or tolerance of all of the tested isolates was evaluated based on the mean growth value±standard deviation (SD).
PCR-RFLP and phylogenetic analysis
The ITS region β-tubulin, and GS (intron 2) genes were amplified for PCR-RFLP analysis. The following primer sets were used: ITS1 (5′-GCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Martin et al., 2004) for ITS, TubG1 (5′-TCTCGATGTTATCCGCCG-3′) and TubGR (5′-TGAGCTCAGGAATGACG-3′) (MacKenzie et al., 2009) for β-tubulin, and GSF1 (5′-ATGGCCGAGTACATCTGG-3′) and GSR1 (5′-AACCGT CGAAGTTCCAC-3′) (Liu et al., 2007) for GS. PCR assays were performed using PCR PreMix reagent (Bioneer, Korea) with 10 pmol of each primer and 80 ng of fungal genomic DNA. The PCR conditions included an initial denaturation step at 94°C for 7 min, followed by 30 cycles of 94°C for 1 min, 45°C for 2 min, and 72°C for 3 min. A final extension step at 72°C for 7 min was also performed. The resulting PCR amplicons were subjected to enzymatic digestion with the restriction enzymes, AluI, HeaIII, TeqI, and MspI (NEB, UK) in a 20 μl total reaction volume according to the manufacturer’s protocol. After 2 h at 37°C, the enzymes digests were inactivated at 70°C for 15 min. The fragmented DNAs were separated on 3% agarose gels at 90 V or 200 mA for 40 min in 1× TAE buffer. The gels were stained with ethidium bromide to visualize the bands.
The fragmented products were scored according to the presence (score of 1) and absence (score of 0) of particular bands. These distances were used to cluster the isolates according to the unweighted pair group method with averages (UPGMA) to determine genetic distances among the isolates (Nei et al., 1973). A phylogenetic tree was generated using the XLSTAT program (www.xlstat.com).
Results
Species-specific identification of pathogens by PCR
From the Daegok, Yearee, Masan, Masan-2, and Munsan orchards, a total of 122, 49, 92, 82, and 140 anthracnose pathogens were isolated, respectively (Table 1). Four sets of species-specific primers were employed to identify the following pathogens: Colg1 and Colg2 for C. gloeosporioides, col1 and acut1 for C. acutatum, Colg1 and CT2 for C. tuncatum, and col1 and col2 for Colletotrichum spp. (Supplementary Fig. 1). Based on the PCR results obtained for the anthracnose pathogens isolated from each orchard, 156 were identified as C. gloeosporioides, 40 were identified as C. tuncatum, 15 were identified as C. acutatum, and 1 was identified as Colletotrichum spp. (Table 3). The conidia morphology for all of the isolates were typical of C. gloeosporioides, and included a cylindrical shape with a rounded end. The average spore diameter was 4.5 μm (Supplementary Fig. 2).
Evaluation of fungicide sensitivity for the isolated pathogens
Effective concentration 50 (EC50) values were calculated for five fungicides which are commonly used to control anthracnose disease in sweet persimmon orchards. As a standard, the strain, C. gloeosporioides JH-1, was used. The EC50 values were 31.5 μg/ml for dithianon, 0.9 μg/ml for picoxystrobin, 3.2 μg/ml for thiophanate-methyl, 1.8 μg/ml for fluazinam, and 2.2 μg/ml for tebuconazole in a Probit model of mycelium growth (Table 3). The five tested fungicides also exhibited a reliable inhibition of mycelium growth against the standard strain (C. gloeosporioides JH-1) (Fig. 1).
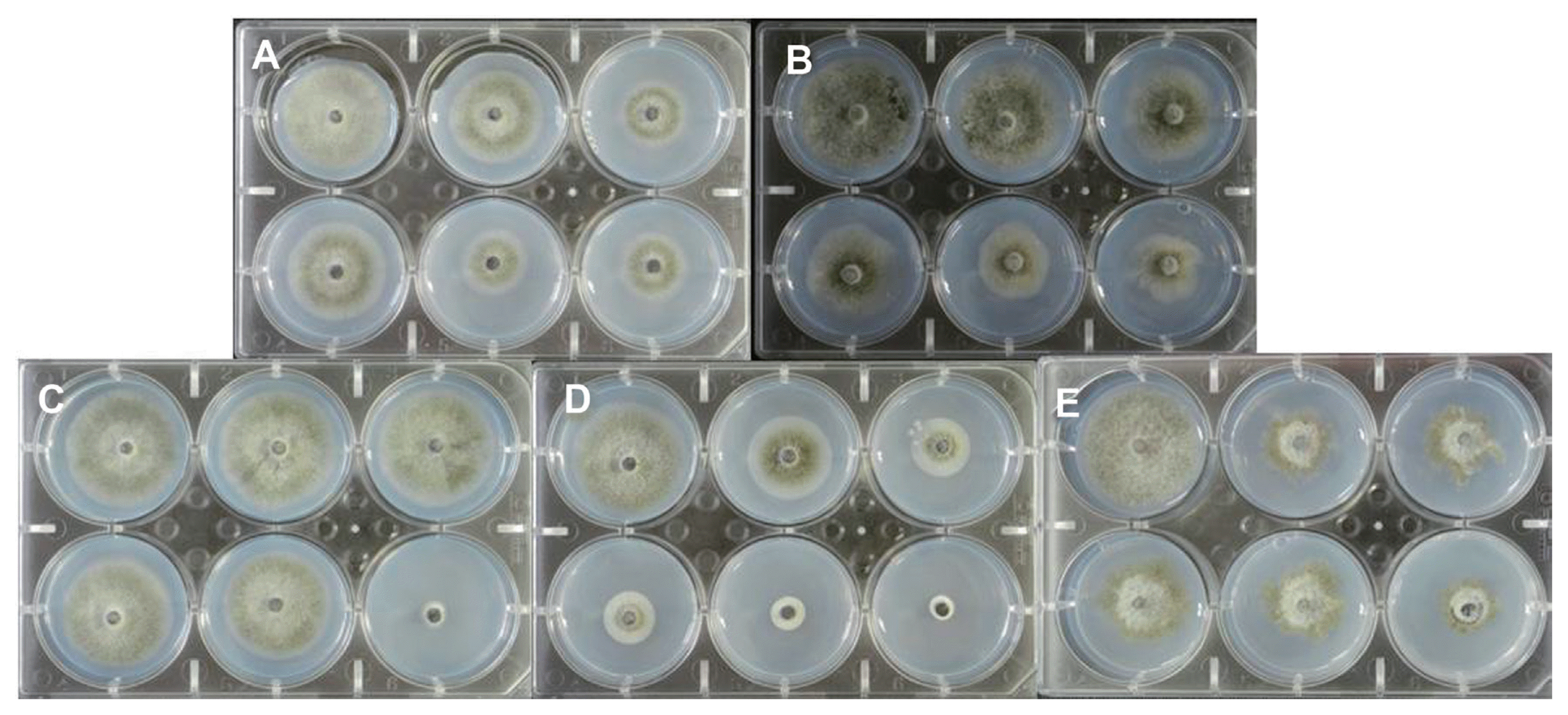
The standard pathogen strain, C. gloeosporioides JH-1, was used to calculate predicted EC50 values for the following fungicides: (A) Picoxystrobin; (B) Dithianon; (C) Thiophanate-methyl; (D) Fluazinam; and (E) Tebuconazole. Fungicides were amended in 1/5 × PDA at 28°C and the strains were incubated for 5 d.
Thirty isolates from each orchard (a total of 150 isolates) were randomly selected to be tested in the fungicide sensitivity assay. All tested concentrations of the fungicides were based on the EC50 values determined for each fungicide. The mean mycelium growth diameters were 10.752 mm for picoxystrobin, 10.265 mm for dithianon, 2.929 mm for thiophanate-methyl, 1.269 mm for fluazinam, and 4.821 mm for tebuconazole. Fluazinam provided the best inhibition of pathogen growth. However, the sensitivities of the other fungicides were found to vary significantly against the pathogen isolates. Based on the EC50 concentrations that were determined, the isolates showed tolerance against picoxystrobin and dithianon, yet were sensitive to thiophanate-methyl, fluazinam, and tebuconazole, regardless of their orchard of origin. It is of note that the Musan orchard has been severely damaged by anthracnose disease for the past couple years (data not shown). No resistance isolate against fluazinam was detected. It was also observed that the fungicide sensitivities of the organic orchard isolates differed from those of the practical orchard isolates (Fig. 2). Most of the Colletotrichum isolates exhibited minimal fungicide resistance, while thiophanate-methyl and fluazinam effectively inhibited pathogen growth in vitro regardless of each pathogen origin.

Mycelium growth assays to detect fungicide sensitive and resistant isolates from each orchard. Four-direction radial mycelium growth was measured. (A) Picoxystrobin - 10.752±2.973 mm; (B) Dithianon - 10.265±2.332 mm; (C) Thiophanate-methyl - 2.929±5.395 mm; (D) Fluazinam - 1.269±2.373 mm; (E) Tebuconazole - 4.821±5.171 mm. Each line separates the Daegok, Yearee, Masan, Masan-2, and Munsan orchards, respectively.
PCR-RFLP and phylogenetic analysis
A total of 127 isolates were analyzed by PCR-RFLP to analyze genetic variations. Digests of the resulting ITS gene products with AluI, HeaIII, and MspI produced multiple bands in the isolates obtained from the practical and organic orchards (Supplementary Fig. 3). The AluI-ITS and MspI-ITS digests produced two bands (400 bp, 200 bp) in most of the samples analyzed. In contrast, the HeaIII-ITS digests generated three bands (300 bp, 200 bp, 100 bp), while digests of the practical orchard isolates with TaqI did not produce any fragmentation of the original product band (400 bp). For the organic orchard isolates, two band fragments (400 bp, 200 bp) were generated for the ITS regions digested with TaqI.
The β-tubulin PCR-RFLP patterns were similar to those of the ITS regions. The anthracnose isolates obtained from the practical orchards (Daegok, Munsan, Yearee) showed a simple pattern, while the isolates from the organic orchards (Masan and Masan-2) showed a diverse pattern (Supplementary Fig. 4). Greater diversity was observed in the analysis of the GS gene in the organic orchard and practical orchard isolates (Supplementary Fig. 5). For example, when the GS gene in the practical orchard isolates was digested with AluI, two bands (200 bp, 190 bp) were produced. When the GS gene product was digested with MspI, five bands (300 bp, 210 bp, 150 bp, 120 bp, 100 bp) were generated. The digests with HeaIII also generated five bands (300 bp, 210 bp, 150 bp, 130 bp, 100 bp). The digests containing TaqI only showed the original PCR product (1.5 kb) was present. Most of the GS regions that were amplified from the organic orchard isolates produced four bands following a digest reaction with TaqI (500 bp, 400 bp, 300 bp, 200 bp). Among the four restriction enzyme assays, MspI and HaeIII provided clearer resolution patterns than those observed following the digests with AluI and TaqI, regardless of the genes involved. The PCR-RFLP results showed that the isolates from the organic orchards exhibit more diverse patterns at the genetic level than the isolates from the practical orchards.
A phylogenetic analysis of the ITS-RFLP (Fig. 3a) and β-tubulin-RFLP (Fig. 3b) results showed that they are not distinguishable from the origin of the pathogen. In contrast, the phylogenetic tree that was generated from the GS-RFLP data shows reliable grouping between the pathogen isolates from the practical orchards and the organic orchards (Fig. 3c). However, the PCR-RFLP data did not distinguish the fungicide sensitive versus fungicide-resistant isolates (data not shown).
Discussion
The primary objective of this study was to elucidate the relationship between anthracnose pathogen genetic diversity and farming management strategy of sweet persimmon. Based on the species-specific PCR results, 156 isolates were identified as C. gloeosporioides, 40 were identified as C. tuncatum, 15 were identified as C. acutatum, and 1 was identified as Colletotrichum spp. However, the morphological characteristics of all of the observed conidia were matched with type-species of C. gloeosporioides. Moreover, while C. tuncatum is a pathogen which has been shown to cause anthracnose disease in soybean, it has not previously been reported as a pathogen in sweet persimmon. An analysis of the ITS region sequences obtained in the present study further suggest that the isolates are C. gloeosporioides (data not shown). Consistent with the previous results of Phoulivong (2011), the morphological characteristics and molecular identification of the isolated anthracnose pathogens in the present study did not match between two criteria.
A total of five fungicides (thiophanate-methyl, dithianon, tebuconazole, fluazinam, and picoxystrobin) were tested for their effectiveness against the isolated sweet persimmon anthracnose pathogens. Of the 150 isolates tested (30 from each of five orchards), thiophanate-methyl and fluazinam were found to effectively suppress pathogen growth under in vitro conditions regardless of each pathogen’s origin. Furthermore, based on the EC50 values obtained for the fungicides, thiophanate-methyl and fluazinam exhibited the greatest inhibition of pathogen growth with EC50 values of 3.2 μg/ml and 1.8 μg/ml, respectively. However, there was no difference between the isolates from the practical orchards and the isolates from the organic orchards in terms of fungicide sensitivity. These results suggest two important points. First, the development of fungicide resistance appears to be largely due to pathogen genetics rather than management strategy. Second, an appropriate fungicide spray may increase the effective control of anthracnose disease. Hidenrand et al. (1988) demonstrated that the use of a effect of cross-talk fungicide could prevent the emergence of fungicide-resistant strains. Later studies reported similar results (Kumar et al., 2007; Maymon et al., 2006; Sanders et al., 2000; Shin et al., 2014).
PCR-RFLP analysis has been used to characterize population diversity and relationships among many plant pathogenic fungi, and RFLP of DNA has been used to differentiate among Colletotrichum species (Ramdeen and Rampersad, 2013). In a study of a Colletotrichum population, the ITS region and the β-tubulin and GS genes were found to distinguish the distance of a population (Weir et al., 2012). ITS sequences have been proposed to serve as an official barcoding marker for fungi (Schoch et al., 2012). However, PCR-RFLP analysis of ITS regions has not consistently distinguished Colletotrichum species (Martin et al., 1999). Furthermore, Weir et al. (2012) showed that an analysis of the ITS sequences of C. gloeosporioides did not provide a reliable identification of pathogen species or population diversity. In the present study, ITS PCR-RFLP phylogenetic trees showed no significant difference between the practical orchard and organic orchard isolates. Taken together, these results suggest that the ITS region may not provide sufficient information to classify the Colletotrichum population, or even the 22 species in the genus, Colletotrichum.
The β-tubulin gene has often been used to study a pathogen population or to identify a particular pathogen (Ramdeen and Rampersad, 2013). For example, a sequence analysis of ITS and a partial analysis of the β-tubulin gene were used to identify C. gloeosporioides among anthracnose stains that were isolated from strawberry plants (Kim et al., 2006). Chung et al. (2010) later showed that use of the β-tubulin gene in PCR-RFLP can differentiate benzimidazole-resistant isolates among a C. gloeosporioides population. However, additional studies of the Colletotrichum genus and other fungi were needed to confirm the precision and stability of the TubGF1 and TubGR primers. In the present study, β-tubulin PCR-RFLP and a phylogenetic analysis found no significant differences among the sweet persimmon anthracnose pathogens examined. Alternatively, the coffee berry disease pathogen, C. kahawae, has been distinguished and identified using GS sequences (Weir et al., 2012). In the present study, GS PCR-RFLP patterns and phylogenetic tree analysis showed that the sweet persimmon anthracnose pathogen population could be separated according to the management strategy of the orchards from which the pathogens were isolated (e.g., practical vs. organic). These results demonstrate the potential for pathogens to be detected and differentiated based on the degree of variability detected in the polymorphic markers of these pathogens. If this approach is confirmed, the diagnosis and management of pathogen disease could be improved.
In conclusion, there were no significant differences in the fungicide sensitivity of the isolates that were obtained from orchards maintained with a practical management strategy versus an organic management strategy. These results suggest that disease control is mainly determined by whether a compatible fungicide has been used or not. However, it was observed that isolates from the practical orchards exhibited less genetic diversity than the isolates from the organic orchards, and this finding support the hypothesis that organic orchards provide greater microbial diversity. The phylogenetic analyses performed did not distinguish fungicide sensitivity or tolerance for the isolates examined. However, PCR-RFLP of GS may represent a reliable tool for diversity studies and investigations of population genetics for sweet persimmon anthracnose pathogens.
Acknowledgments
This research was performed with the support of the “Cooperative Research Program for Agriculture Science & Technology Development (PJ010904)” from the Rural Development Administration of Korea.