 |
 |
| Plant Pathol J > Volume 31(4); 2015 > Article |
Abstract
Surveys of yellowing viruses in plastic tunnels and in open field crops of melon (Cucumis melo cultivar catalupo), oriental melon (C. melo cultivar oriental melon), and cucumber (C. sativus) were carried out in two melon-growing areas in 2014, Korea. Severe yellowing symptoms on older leaves of melon and chlorotic spots on younger leaves of melon were observed in the plastic tunnels. The symptoms were widespread and included initial chlorotic lesions followed by yellowing of whole leaves and thickening of older leaves. RT-PCR analysis using total RNA extracted from diseased leaves did not show any synthesized products for four cucurbit-infecting viruses; Beet pseudo-yellows virus, Cucumber mosaic virus, Cucurbit yellows stunting disorder virus, and Melon necrotic spot virus. Virus identification using RT-PCR showed Cucurbit aphid-borne yellows Virus (CABYV) was largely distributed in melon, oriental melon and cucumber. This result was verified by aphid (Aphis gossypii) transmission of CABYV. The complete coat protein (CP) gene amplified from melon was cloned and sequenced. The CP gene nucleotide and the deduced amino acid sequence comparisons as well as phylogenetic tree analysis of CABYV CPs showed that the CABYV isolates were undivided into subgroups. Although the low incidence of CABYV in infections to cucurbit crops in this survey, CABYV may become an important treat for cucurbit crops in many different regions in Korea, suggesting that CABYV should be taken into account in disease control of cucurbit crops in Korea.
Cucurbits are economically important vegetable crops in the world. In Korea, an array of squashes (Cucurbita pepo), cucumbers (Cucumis sativus), pumpkins (Cucurbita moschata), bitter gourds (Momordica charantia), and melons (Cucumis melo) are commercially grown in plastic tunnels as cash crops and in home gardens. Yellowing symptoms caused by virus infections are often attributed to nutritional disorders mainly to the deficiency of Mg2+ or Mn2+ ion. In addition, the toxicities of pesticides and insecticides frequently cause yellowing symptoms in cucurbit plants including melon. However, agronomical approaches such as addition of nutrients in melon plants showing yellowing symptoms aggravate the early appearance of some melon plants with chlorotic spots to rapid spread of yellowing symptoms in the whole leaves of melon plants. The presence of high aphid populations illustrates that these yellowing symptoms are strongly associated with infections of plant viruses. In recent decades, at least 39 different virus species are known to infect naturally cucurbit plants worldwide (Lecoq, 2013; Provvidenti, 1996).
Of these virus species, the three most frequent and economically important viruses in the world are Papaya ringspot virus (PRSV, formerly known as WMV-1), Watermelon mosaic virus (WMV, formerly known as WMV-2), and Zucchini yellow mosaic virus (ZYMV) (Lecoq, 2013). The three viruses fail to produce the yellowing symptoms in melon, but the viruses cause severe mosaic, stunting, wilt, and malformation of leaves and fruits. Of candidate viruses, yellowing symptoms in melon plants are more similar to those caused by an aphid-transmissible Cucurbit aphid-borne yellows virus (CABYV). The yellowing symptoms are even similar to those caused by Beet pseudo-yellows virus (BPYV) or Cucurbit yellows stunting disorder virus (CYSDV) (Celix et al., 2006; Desbiezm et al. 2003; Wisler et al., 1998; Wintermantel et al., 2009), but these two viruses are transmitted by the whitefly species (Bemisia tabaci and Traleurodes vaporariorum).
With the recent ratification by the International Committee of Taxonomy of Viruses (ICTV) of Carrot red leaf virus (Huang et al., 2005), Chickpea chlorotic stunt virus (Abraham et al., 2006), Melon aphid-borne yellows virus (Xiang et al., 2008) and Tobacco vein-distorting virus (Mo et al., 2003), it is currently accepted virus species of the genus Polerovirus (DŌĆÖArcy and Domier, 2005; King et al., 2011). In addition, three tentative species, Cotton leaf roll dwarf virus in cotton (Corr├¬a et al., 2005), Suakwa aphid-borne yellows virus in cucurbits (Shang et al., 2009) and Wheat yellow dwarf virus-GPV (Zhang et al., 2009) in cereals were tentatively classified into the genus Polerovirus by the ICTV.
CABYV that belongs to the genus Polerovirus (the family Luteoviridae) causes a complex of yellowing symptoms in melon. The virus was first reported in France where it has caused regular epidemics since 1982 (Lecoq et al., 1992). Since then, CABYV has been identified worldwide in temperature, subtropical areas and Mediterranean basin (Lecoq, 2013). CABYV infects most cucurbit crops including melon, cucumber and watermelon (Lecoq et al., 1992) and CABYV was found in three weed species (Capsella bursa-pastroris, Lamium amplexicaule, and Senecio vulgaris) and in Ecballium elaerium (Knierim et al., 2010; Lecoq et al., 1992; Mnari-Hattab et al., 2009). CABYV is transmitted in a circulative, non-propagative manner by specific aphid vectors, but are not mechanically transmissible (DŌĆÖArch and Domier, 2005; Lecoq et al., 1992). The virus often found in mixed infections with other cucurbit-infecting viruses (Abou-Jawdah et al., 2000; Boubourakas et al., 2006; Kassem et al., 2007; Knierim et al., 2010; Mnari-Hattab et al., 2009).
Polerovirus particles have an icosahedral symmetry of about 25 nm and a core of a single-stranded (ss) positive sense RNA molecule of 5.7 kb (DŌĆÖArcy and Domier, 2005). The genome contains 7 open reading frames (ORFs). A non-coding intergenic region of approximately 200 nucleotides separates RNA-dependent RNA polymerase (ORF2) from coat protein (ORF3) (DŌĆÖArcy and Domier, 2005). The members of the genus Polerovirus are distinguished from those of the two other genera Luteovirus and Enamovirus in the Luteoviridae by terms of different genome organizations. The difference includes the presence or absence of ORF0 (possible membrane-linked replication factor and RNA silencing suppressor), ORF4 (movement protein) and ORF6 (assigned as a RNA silencing suppressor) (DŌĆÖArcy and Domier, 2005; Kozlowska-Makulska et al., 2010; Liu et al., 2012; Mangwende et al., 2009; Pfeffer et al., 2002). The ORF1 and ORF2 of poleroviruses and enamoviruses show more close relationship to those of sobemoviruses, whereas those of luteoviruses are more associated with the family Tombusviridae (DŌĆÖArcy and Domier, 2005). Though the ORF7 product has the activity of nucleic acid binding, the protein function in host plants is unclear (Ashoub et al., 1998). Species in the Luteoviridae are distinguished when at least one gene product shows more than 10% amino acid sequence difference and when luteoviruses have different biological and serological properties (DŌĆÖArcy and Domier, 2005; King et al., 2011).
In this study, disease surveys in 2014 were performed in plastic tunnels to identify the causal agent of the observed yellowing disease of melon plants (cultivar cantalupo) as well as viral disease-like oriental melon and cucumber. To achieve this purpose, reverse transcription-polymerase chain reaction (RT-PCR) was applied for detection of CABYV. Aphid transmission assays were carried out to characterize biological properties of Korean isolate of CABYV during CABYV isolation.
Leaf samples from melon plants showing interveinal chlorosis, yellowing symptoms, and thickening of older leaves were collected from two areas in Korea. Samples with some asymptomatic leaves were also collected from plastic tunnels in 2014. Samples from oriental melon and cucumber plants were also collected from farms near the plastic tunnels cultivated in melon. For subsequent analysis or virus transmission experiments, the cucurbit leaves including melon were maintained in a plastic bag at 4┬║C for approximately 10 days or stored by desiccation of small amounts of infected leaf materials over calcium chloride at 4┬║C.
Total RNA was extracted from 1 g leaves of the symptomatic or asymptomatic melon, oriental melon and cucumber using Plant RNeasy Mini kit (Qiagen, USA), according to the manufacturerŌĆÖs instructions. One-step RT-PCR analysis was carried out to synthesize the full-length cDNA of CABYV coat protein (CP) using a pair of primers (CABYV-CP-For : 5ŌĆ▓-atgaatacggccgcggctagaaatc-3ŌĆ▓, CABYV-CP-Rev : 5ŌĆ▓-ctatttcgggttctggacctggca-3ŌĆ▓) synthesized newly in the study, based on CP gene sequences of some reference CABYV isolates available in GenBank databases.
The thermo-cycling conditions were as follows: 60 min at 45┬║C for RT, 5 min at 95┬║C (1 cycle), 94┬║C, 30 s, 55┬║C, 30 s and 72┬║C, 60 s (40 cycles), and a final extension at 72┬║C for 7 min. RT-PCR product was analyzed in 1.2% agarose gel and visualized after soaking in ethidium bromide solution.
Apterous adults of nonviruliferous Aphis gossypii maintained in our laboratory were reared in cucumber. From 10 to 20 nonviruliferous young Apterous adults of A. gossypii were allowed to feed for 30 to 36 h on symptomatic melon leaves. At least two successive transmission periods of 30 to 36 h were performed to avoid all non-persistently transmitted viruses (for example, WMV). Subsequently, the aphids were transferred to each healthy seedling of melon, oriental melon and cucumber with 2 to 3 leaves grown under individual cages. After this period, aphids were killed with insecticide (Dongbu Farm Corp., Korea) and plants were transferred to an insect-proof greenhouse which was maintained at 20 to 28┬║C. Production of typical CABYV-like symptoms was monitored for 4 to 5 weeks following inoculation by aphids. Successful transmission of CABYV was confirmed using RT-PCR analysis with a pair of primers (CABYV-CP-For and CABYV-CP-Rev) described above.
RT-PCR analysis for CABYV isolates was carried out as described above and the resulting RT-PCR fragment (the full-length CP gene) for CABYV was cloned into pCR4TOPO vector (Invitrogen, USA). The candidate cDNA clones were selected using digestions of restriction endonucleases, according to standard protocols. The entire cDNA insert was sequenced using a BigDye termination cycle sequencing kit (Applied Biosystems, USA) with M13 forward/reverse primers. The comparisons of determined nucleotide sequence and deduced amino acid sequence were analyzed using the Jukes-Cantor index and the neighbor-joining method with 1000 bootstrap replications in the MEGA 6.0 and DNASTAR package software (Choi et al., 2011; Tamura et al., 2013). Phylogenetic tree of CABYV CP sequences was constructed using DNASTART package software.
To determine a causal agent that produces yellowing symptoms in melon, we investigated two different farms where melon plants were grown in plastic tunnels in Korea, 2014. Various symptoms (for instance, yellowing, chlorotic spot of lower leaves, yellowing mosaic, necrotic, or malformation of melon fruits) were observed in the two farms in Korea (Table 1 and Fig. 1). A total of 1,200 symptomatic and asymptomatic melon leaves were collected from two farms. Based on the observed melon symptoms, all symptomatic melon plants suspected to be infected with CABYV were analyzed using RT-PCR analysis with specific primers (CABYV-CP-For and CABYV-CP-Rev). The RT-PCR result showed an expected product of 600 bp that was amplified using the primers (CABYV-CP-For and CABYV-CP-Rev) (Fig. 2A). All symptomatic samples showed positive to CABYV. Further RT-PCR analysis with asymptomatic melon leaves indicated that most of asymptomatic melon samples showed no amplification of RT-PCR products similar to healthy melon plants (Fig. 2A). However, CABYV was detected from a few asymptomatic melon leaves (approx. 1%) in a farm (Fig. 2A). These results suggest that CABYV can be distributed throughout the Korea in terms of space and timing of CABYV. Since BPYV and CYSDV induce similar yellowing symptoms in cucurbits, all symptomatic and asymptomatic samples were checked using RT-PCR analyses specific to primers of BPYV (BPYV-For: 5ŌĆ▓-tcgaaagtccaacaagacgt-3ŌĆ▓, BPYV-Rev: 5ŌĆ▓-ctgatggtgcgcgagtg-3ŌĆ▓) or primers of CYSDV (CYSDV-For: 5ŌĆ▓-atggacatgcctaactgttactt-3ŌĆ▓, CYSDV-Rev: 5ŌĆ▓-atagctgctgcagatggttc-3ŌĆ▓). None of samples showed the synthesis of RT-PCR products (data not shown), indicating that all melon samples collected were not infected with BPYV and CYSDV.
We further analyzed all symptomatic and asymptomatic samples using primers specific to Cucumber mosaic virus (CMV) (Choi et al., 1999) and Melon necrotic spot virus (MNSV) (MNSV-For: 5ŌĆ▓-aggttagcaatggatactgg-3ŌĆ▓, MNSV-Rev: 5ŌĆ▓-tctcgtttgatgtggaagac-3ŌĆ▓). Less 1% melon plants were infected with CMV or MNSV in symptomatic samples, but not in asymptomatic samples (data not shown). These results suggest that RT-PCR analysis is one of reliable methods for the diagnosis of yellowing-inducing viruses and chronically problematic viruses in case that antisera specific to cucurbit-infecting viruses are unavailable. Taken together, these results clearly showed that CABYV was the causal agent for yellowing symptoms in melon plants.
To verify whether the Korean isolate of CABYV can be transmitted by aphid species, Apterous adults of nonviruliferous A. gossypii adults were reared on the melon leaves showing yellowing symptoms. The aphids were allowed to acquire CABYV isolate for 30-36 h. Most of inoculated plants (melon, cucumber, squash and watermelon) were infected with CABYV (Table 2), showing various symptoms. RT-PCR analysis confirmed CABYV infections in the inoculated plants, suggesting that the CABYV Korean isolate can be transmitted by aphid vectors. Viral disease survey was extended to oriental melon and cucumber plants near the plastic tunnels for melon cultivation. Oriental melons showing chlorotic spots and interveinal yellowing and cucumber showing mild faint chlorotic spots or symptomless were collected (Fig. 2A). To verify the observation from systemic symptoms, RT-PCR analysis using the above pairs of the primers specific to five cucurbit-yellowing viruses including CABYV was carried out. An expected 600 bp RT-PCR product specific to CABYV CP gene was amplified from the symptomatic oriental melon and cucumber plants (Fig. 2C). CABYV was not detected from healthy oriental melon and cucumber plants used as negative controls, suggesting that CABYV only infected oriental melons and cucumber plants. These results suggest that CABYV can be found in a great diversity of areas and environments and the virus is a potential epidemic virus in cucurbit crops in Korea.
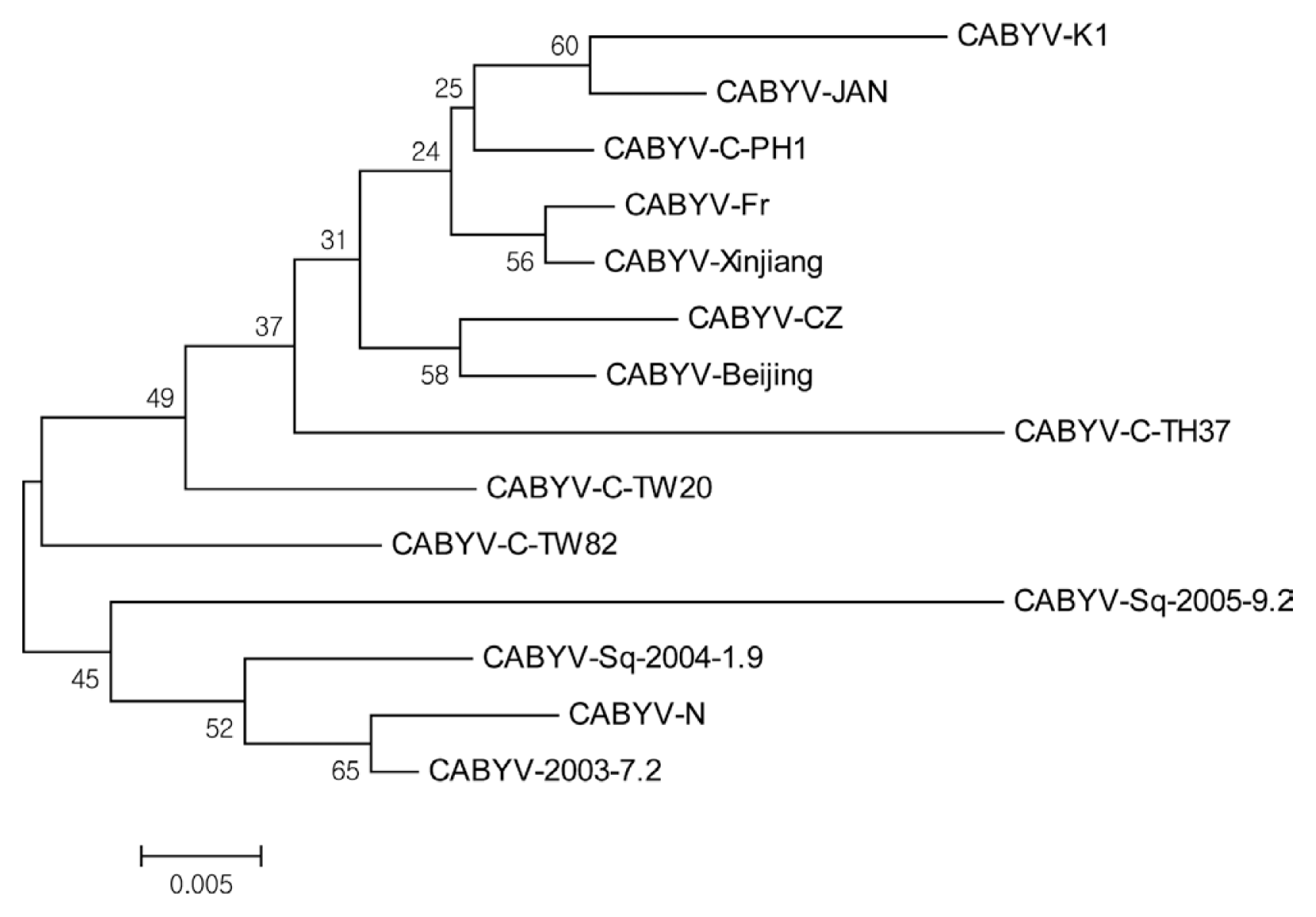
To determine the sequences of CABYV CP gene from melon, all RT-PCR products (600 bp) were directly introduced into the pCR4TOPO vector (Promega, USA), resulting in pGEM-CABYV-CP. Sequencing analysis showed the CP sequences of CABYV was truly 600 bp in length and CP sequences of CABYV was 100% nucleotide identity between the cDNA clones. It is likely that one isolate of CABYV (named CABYV isolate K1, CABYV-K1) infected melon plants in the plastic tunnels. CABYV-K1 shared amino acid sequence identity of 92-98% with other CABYV isolates. These results suggest that CABYV-K1 is a typical isolate from cucurbits. The complete nucleotide sequence of CABYV-K1 CP gene has been deposited in NCBI Genbank under accession number LC060147. Sequence alignment of CP amino acids between CABYV-K1 and 13 CABYV isolates (from Japan, China, Taiwan, France etc.) showed that N-terminal region of the CP was more divergent that C-terminal region. The central region of CABYV CPs was highly conserved (Fig. 3). Phylogenetic tree analysis using the neighbor-joining method showed CABYV-K1 was more closely related to CABYV Japan isolate and showed no clear relationships with geographical origins of the isolates and isolation host species (Fig. 4). These observations were supported by the phylogenetic tree regardless of the analyzed methods (for example, Maximum Likelihood).
In this study, we first identified CABYV infections in melon, cucumber, and oriental melons in Korea. Using aphid transmission assay, we confirmed that CABYV was the authentic virus that caused yellowing symptoms in melon. In addition, we found that cucurbit plant species infected with CABYV showed various symptoms or symptomless. CABYV infection was found in about 5% of the melon samples with yellowing symptoms, compared with about 30-50% CABYV infections (Bananej et al., 2009; Kassem et al., 2007; Shang et al., 2009; Xiang et al., 2008). This difference could be caused by the presence of other species causing this type of yellowing symptoms. For example, criniviruses producing yellowing symptoms of cucurbits, such as BPYV are not detected in Korea yet. In spite of the low incidence of CABYV and none of cucurbit-infecting criniviruses, the rate of infections in samples with symptoms can be increased. CABYV-associated yellowing symptoms on cucurbits have been wrongly attributed to nutritional deficiency, natural senescence and other abiotic stresses, leading to underestimation of the CABYV incidence. In addition, it canŌĆÖt be completely excluded the possibility that other yellowing viruses which were not tested in this study could be responsible for such yellowing symptoms. Undetected yellowing viruses could have been detrimental to stable productions of cucurbits in Korea. Control of CABYV and other poleroviruses through use of insecticides to control aphid vectors is effective. However, this control method is unreliable due to high level appearance of aphids resistant to insecticides and is potentially hazardous to farmers and the environment. Cultivation of resistant cultivars is safer and more effectively sustainable alternative. Procedures for selection of breeding lines and resistance evaluation against CABYV are needed to be established.
Many studies for field survey of CABYV incidence showed enzyme linked immunosorbent assay (ELISA) using antibody specific to CABYV was effective for the CABYV detection method (Abou-Jawdah et al., 2000; Deng et al., 1997; Kassem et al., 2007; Mnari-Hattab et al., 2005). In recent reports, ELISA is not sensitive enough to detect the presence of CABYV and other luteoviruses in plants (Figueira et al., 1997; Oritz et al., 2005; Van den Heuvel et al., 1989). We set up the molecular detection method, RT-PCR, for diagnosis of CABYV in this study and showed the RT-PCR analysis is highly reliable to detect CABYV in suspicious samples. These findings were supported by some studies that demonstrated to detection level of CABYV (Celix et al., 1996; Knierim et al., 2010; Mnari-Hattab et al., 2005; 2009).
The comparison of CP amino acid sequences between CABYV-K1 and other CABYV strains revealed no distinct subgroups due to high identity (over 92%). This result is different from reports published previously that CABYV isolates can be divided into 2 or 3 subgroups. These differences are caused by viral proteins that used for phylogenetic tree analyses. Phylogenetic trees of the complete CABYV CPs published previously showed no lineage of CABYV isolates (Knierim et al., 2010; Mnari-Hattab et al., 2009). With movement protein (MP) or partial RNA-dependent RNA polymerase (RdRp) of CABYV, a few claudes were made from the sequences of MP or RdRp in the phylogenetic trees. However, CP is the most important protein for polerovirus taxonomy. Based on the sequence alignment and the phylogenetic tree analysis, it is not plausible that CABYV isolates are not divided into subroups. This conclusion remains to be determined using a large number of CABYV samples collected from various geographical nations for a long time.
Acknowledgements
This work was supported by a grant from the Basic Research Program (Project no. PJ008472022015) of National Institute of Horticultural and Herbal Science, Rural Development Administration, Republic of Korea.
Fig.┬Ā1
Symptoms of CABYV in Cucumis melo (cultivar cantalupo). (a) An uninfected melon fruit. (b) A melon fruit infected with CABYV. (C) to (f) Foliar symptoms of melon plants grown in plastic tunnels.
Fig.┬Ā2
RT-PCR analysis for detection of CABYV. (A) A single 600 bp RT-PCR product was solely synthesized from symptomatic (indicated as Yellow) melon leaves using CABYV-specific viruses, but not from asymptomatic (indicated as Symptomless). The synthesized CABYV CP gene is indicated by an arrow and DNA size marker is shown in the left side. (B) Various symptoms in cucurbit crops naturally infected with CABYV. Acronym is presented in the parenthesis. (C) RT-PCR analysis for the validation of CABYV infections in cucurbits naturally infected. yM, melon (cultivar cantalupo) infected with CABYV (as a positive control); HyOM, healthy oriental melon (as a negative control); yOM, oriental melon infected; HC, healthy cucumber (as a negative control); csCu, cucumber infected.

Fig.┬Ā3
Multiple alignment of CP amino acids of CABYV Korean isolate and other isolates from different countries. Consensus sequence obtained with the CLUSTAL algorithm is shown above the alignment as a majority. The aa identical to the consensus are indicated by dashes within the alignment. The aa positions are indicated on the both sides. Sequence variability was indicated by red letters. Accession numbers analyzed for this study are as follows: CABYV-K1 (LC060147), CABYV-C-PH1 (GQ700823), CABYV-C-TH37 (KF815678), CABYV-C-TW80(JQ700305), CABYV-R-TW82 (JQ700306), CABYV-CZ (HQ439023), CABYV-Fr (GQ221223), CABYV-Xinjiang (EU636992), CABYV-Beijing (EU000535), CABYV-N (NC_003688), CABYV-Sq-2003-7.2 (JF939812), CABYV-Sq-2005-9.2 (JF939814), and CABYV-Sq-2004-1.9 (JF939814). The isolate sequences were named using the initials of the isolation host and country of origin or were adopted from GenBank database.

Fig.┬Ā4
Phylogenetic tree analysis of a Korean CABYV isolate (CABYV-K1) and other isolates based on CP amino acid sequences. The neighbor-joining tree was constructed from the sequence alignment of 14 isolates. Multiple sequence alignments were generated by using the DNASTAR program, and the phylogenetic tree was constructed by the neighbor-joining algorithm, based on calculations from pairwise amino acid sequence distances for amino acid analysis derived from the multiple-alignment format. The scale bar represents 0.005 divergence of the Jukes and Cantor dissimilarity index. Boostrap analysis was done with 1000 replicates of the starting tree. Bootstrap values are shown in each branch.
Table┬Ā1
Infection of Cucurbit aphid-borne yellows virus (CABYV) on melon plants in two different areas in 2014, Korea
| Virus | Survey Area | |
|---|---|---|
| Cheonyang | Jinju | |
| CABYV | 159/600a (6.5%)b | 24/600 (4.0%) |
Table┬Ā2
Transmission analysis of CABYV by A. gossypii. Melon leaves showing yellowing symptoms were used as a source for aphid transmission. Aphids fed on the diseased melon leaves were transferred to each assay plant species. Infections of CABYV from the inoculated plants were confirmed using RT-PCR analysis
| Plant | No. of plants inoculated | No. of plants infected |
|---|---|---|
| Melon | 6 | 6 |
| Cucumber | 6 | 4 |
| Squash | 6 | 4 |
| Watermelon | 6 | 5 |
References
Abou-Jawdah, Y, Sobh, H, El-Zammar, S, Fayyad, A and Lecoq, H 2000. Incidence and management of virus diseases of cucurbits in Lebanon. Crop Protection. 19:217-224.

Ashoub, A, Rohde, W and Dirk Pr├╝fer, D 1998. In planta transcription of a second subgenomic RNA increases the complexity of the subgroup 2 luteovirus genome. Nucl Acids Res. 26:420-426.



Boubourakas, IN, Avgelis, AD, Kyriakopoulou, PE and Katis, NI 2006. Occurrence of yellowing viruses (Beet pseudo-yellows virus, Cucurbit yellow stunting disorder virus and Cucurbit aphid-borne yellows virus) affecting cucurbits in Greece. Plant Pathol. 55:276-283.

Bananej, K, Desbiez, C, Wipf-Scheibel, C, Vahdat, I, Kheyr-Pour, A, Ahoonmanesh, A and Lecoq, H 2006. First report of Cucurbit aphid-borne yellows virus in Iran causing yellows on four cucurbit crops. Plant Dis. 90:526

Celix, A, Lopez-Sese, A, Almarza, N, Gomez-Guillamon, ML and Rodriguez-Cerezo, E 1996. Characterization of Cucurbit yellow stunting disorder virus, a Bemisia tabaci transmitted Closterovirus. Phytopathology. 86:1370-1376.
Choi, SK, Choi, JK, Park, WM and Ryu, KH 1999. RT-PCR detection and identification of three species of cucumoviviruses with a genus-specific single pair of primers. J Viol Mehods. 83:67-73.
Corr├¬a, RL, Silva, TF, Sim├Ąes-Ara├║jo, JL, Barroso, PA, Vidal, MS and Vaslin, MF 2005. Molecular characterization of a virus from the family Luteoviridae associated with cotton blue disease. Arch Virol. 150:1357-1367.



DŌĆÖArcy, CJ and Domier, LL 2005. Luteoviridae. eds. by CM Fauquet, MA Mayo, J Maniloff, U Desselberger and LA Ball, In: Virus Taxonomy, the 8th Report of the ICTV, London, UK. Elsevier/Academic Press, 891-900.
Deng, TC, Tsai, CH, Chen, YF, Chang, CA, Hsiao, CH and Lecoq, H 1997. First report of cucurbit aphid-borne yellows luteovirus in Taiwan. Plant Prot Bull Taiwan. 39:395-396.
Desbiez, C, Lecoq, H, Girard, M, Cotillon, AC and Schoen, L 2003. First report of Cucurbit yellow stunting disorder virus in commercial cucumber greenhouses in France. Plant Dis. 87:600

Figueira, AR, Domier, LL and DŌĆÖArcy, CJ 1997. Comparison of techniques for detection of Barley yellow dwarf virus-PAV-IL. Plant Dis. 81:1236-1240.


Kassem, MA, Sempere, RN, Ju├Īrez, M, Aranda, MA and Truniger, V 2007. Cucurbit aphid-borne yellows virus is prevalent in field-grown cucurbit crops of southeastern Spain. Plant Dis. 91:232-238.


King, AMQ, Adams, MJ, Carstens, EB and Lefkowitz, EJ 2011. Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses. Amsterdam. Elsevier Academic Press.
Knierim, D, Deng, TC, Tsai, WS, Green, SK and Kenyon, L 2010. Molecular identification of three distinct Polerovirus species and a recombinant Cucurbit aphid-borne yellows virus strain infecting cucurbit crops in Taiwan. Plant Pathol. 59:991-1002.

Kozlowska-Makulska, A, Guilley, H, Szyndel, MS, Beuve, M, Lemaire, O, Herrbach, E and Bouzoubaa, S 2010. P0 proteins of European beet-infecting poleroviruses display variable RNA silencing suppression activity. J Gen Virol. 91:1082-1091.


Lecoq, H, Bourdin, D, Wipf-Scheibel, C, Bon, M, Lot, H, Lemaire, O and Herrbach, E 1992. A new yellowing disease of cucurbits caused by a luteovirus, Cucurbit aphid-borne yellows virus. Plant Pathol. 41:749-761.

Lecoq, H 2003. Cucurbits. eds. by G Loebenstein and G Thottapilly, In: Virus and Virus-like Diseases of Major Crops in Developing Countries, Dordrecht, the Netherlands. Kluwer Academic Publishers, 665-687.

Liu, Y, Zhai, H, Zhao, K, Wu, B and Wang, X 2012. Two suppressors of RNA silencing encoded by cereal-infecting members of the family Luteoviridae. J Gen Virol. 93:1825-1830.


Mangwende, T, Wang, M-L, Borth, W, Hu, J, Moore, PH, Mirkov, TE and Albert, HH 2009. The P0 gene of Sugarcane yellow leaf virus encodes an RNA silencing suppressor with unique activities. Virology. 384:38-50.


Mnari-Hattab, M, Kummert, J, Roussel, S, Ezzaier, K, Zouba, A and Jijakli, MH 2005. First report of Cucurbit aphid-borne yellows virus in Tunisia causing yellows on five cucurbitacious species. Plant Dis. 89:776

Mnari-Hattab, M, Gauthier, N and Zouba, A 2009. Biological and molecular characterization of the Cucurbit aphid-borne yellows virus affecting cucurbits in Tunisia. Plant Dis. 93:1065-1072.


Ortiz, V, Castro, S and Romero, J 2005. Optimization of RT-PCR for the detection of Bean leaf roll virus in plant hosts and insect vector. J Phytopathol. 153:68-72.

Pfeffer, S, Dunoyer, P, Heim, F, Richards, KE, Jonard, G and Ziegler-Graff, V 2002. P0 of beet Western yellows virus is a suppressor of posttranscriptional gene silencing. J Virol. 76:6815-6824.




Shang, QV, Xiang, HY, Han, CG, Li, DW and Yu, JL 2009. Distribution and molecular diversity of three cucurbit-infecting poleroviruses in China. Virus Res. 145:341-346.


Tamura, K, Stecher, G, Peterson, D, Filipski, A and Kumar, S 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725-2729.



Van den Heuvel, JFJM and Peters, D 1989. Improved detection of Potato leafroll virus in plant material and in aphids. Phytopathology. 79:963-967.

Wintermantel, WM, Hladky, LL, Cortez, AA and Natwick, ET 2009. A new expanded host range of Cucurbit yellow stunting disorder virus includes three agricultural crops. Plant Dis. 93:685-690.





 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




