Establishment of an Agrobacterium-mediated Inoculation System for Cucumber Green Mottle Mosaic Virus
Article information
Abstract
The infectious full-length cDNA clones of Cucumber green mottle mosaic virus (CGMMV) isolates KW and KOM, which were isolated from watermelon and oriental melon, respectively, were constructed under the control of the cauliflower mosaic virus 35S promoter. We successfully inoculated Nicotiana benthamiana with the cloned CGMMV isolates KW and KOM by Agrobacterium-mediated infiltration. Virulence and symptomatic characteristics of the cloned CGMMV isolates KW and KOM were tested on several indicator plants. No obvious differences between two cloned isolates in disease development were observed on the tested indicator plants. We also determined full genome sequences of the cloned CGMMV isolates KW and KOM. Sequence comparison revealed that only four amino acids (at positions 228, 699, 1212, and 1238 of the replicase protein region) differ between the cloned isolates KW and KOM. A previous study reported that the isolate KOM could not infect Chenopodium amaranticolor, but the cloned KOM induced chlorotic spots on the inoculated leaves. When compared with the previously reported sequence of the original KOM isolate, the cloned KOM contained one amino acid mutation (Ala to Thr) at position 228 of the replicase protein, suggesting that this mutation might be responsible for induction of chlorotic spots on the inoculated leaves of C. amaranticolor.
Cucumber green mottle mosaic virus (CGMMV), a member of the genus Tobamovirus, is a rod-shaped virus with approximately 300 nm in length and contains a plus-sense single strand RNA (ssRNA) genome of 6.4 kb (Ugaki et al., 1991). The CGMMV genome encodes at least four proteins, including the 5′ terminal 129 kDa protein, its translational read-through product of 186 kDa, a 29 kDa cell-to-cell movement protein, and a 17.4 kDa coat protein (Ugaki et al., 1991). The 129 kDa and 186 kDa proteins are replication-associated proteins. The 129 kDa protein harbors a methyltransferase-like domain in its N-terminal region and a helicase-like domain in its C-terminal region, while the read-through part of the 186 kDa protein contains a polymerase-like domain (Ugaki et al., 1991). In Korea, CGMMV was isolated firstly in 1989 (Lee et al., 1990) and caused widespread ‘blood flesh’ disease in watermelons and considerable economic damage in 1995 in Korea (Lee, 1996). According to the virus isolates and hosts, CGMMV causes various symptoms, including mottling and systemic mosaic symptoms on leaves and deterioration on fruits of watermelon, oriental melon, cucumber, and zucchini (Lee et al., 1990). In our previous study, we investigated symptomatic characteristics of two CGMMV isolates, KW and KOM, which were isolated from watermelon and oriental melon, respectively, and determined their full-genome sequences (Kim et al., 2003).
Plant RNA viruses accumulate mutations easily because of their error-prone replication activity (Domingo et al., 1985). Constructing infectious cDNA clones is, therefore, needed to maintain molecular and biological characteristics of RNA viruses and to study their genetic aspects. Infectious cDNA clones of many kinds of plant RNA viruses, including Tobacco mosaic virus (TMV), Potato virus X (PVX), and Soybean mosaic virus (SMV), have been constructed under the control of the cauliflower mosaic virus (CaMV) 35S promoter or T7 RNA polymerase promoter (Dawson et al., 1989; Ruiz et al., 1998; Seo et al., 2009b). Previously, a full-length infectious cDNA clone of the chb isolate of CGMMV has been constructed for in vitro transcription and shown to have high infectivity in Chenopodium amaranticolor and cucumber (Zhong et al., 2015). However, preparation of viral in vitro transcripts is cost-ineffective and laborious. On the other hand, Agrobacterium-mediated inoculation of viral infectious cDNA clones only requires bacterial cultivation and easily infiltrated onto plants. In this stuty, we established an Agrobacterium-mediated inoculation method for CGMMV. The construction of infectious cDNA clones of CGMMV-KW and -KOM is described and virulence and symptomatic characteristics of the cloned CGMMV isolates were investigated.
Two CGMMV isolates, KW and KOM (Kim et al., 2003) were propagated in Cucumis melo L. (oriental melon) in a greenhouse. Total RNA extraction was carried out from the Cu. melo L. leaves infected with either CGMMV isolate KW or KOM using the TRI Reagent (MRC, USA) according to the protocols provided by the manufacturer. The extracted total RNA was used for cDNA synthesis of CGMMV-KW or -KOM. Specific primers (CGMMV-SacI-F, 5′-CGAGCTCGTTTTAATTTTTATAATTAAACAAAC AACAACAACAAC-3′ and CGMMV-R, 5′-TGGGCC CCTACCCGGGGAAA-3′) were designed based on previously reported sequences of CGMMV-KW and -KOM (GenBank accession numbers AF417243 and AF417242, respectively). cDNAs of two CGMMV isolates were synthesized using the SuperScript III reverse transcriptase (Invitrogen, USA) with CGMMV-R. The resulting cDNAs were subjected to amplify full-length genomes of CGMMV-KW and KOM using the Pfu Ultra II DNA polymerase (Agilent Technologies, USA) with CGMMV-SacI-F and CGMMV-R. The amplified full-length products of CGMMV-KW and -KOM were then digested with SacI and inserted into between the SacI and SmaI sites in pSNU1 vector (Park and Kim, 2006), which is a modified binary vector containing the CaMV 35S promoter. The resulting constructs were named pCGMMV-KW and pCGMMV-KOM, respectively (Fig. 1). The full-length nucleotide sequences of the cloned CGMMV-KW and KOM were determined by the dideoxy nucleotide termination method and an ABI PRISM 3700 XL DNA Analyzer (Applied Biosystem, USA) located at the National Instrumentation Center for Environmental Management (NICEM, Seoul National University).

Schematic representation of the construction of infectious cDNA clones of CGMMV. The modified pSNUI vector contains, in sequential order, a left border of T-DNA (LB), a double CaMV 35S promoter (35S), a cis-cleaving ribozyme sequence (RZ), a NOS terminator (NOSt), and a right border of T-DNA (RB). The restriction enzyme cleavage sites used to make the constructs are shown in gray boxes. Full-length in vivo transcripts of CGMMV were produced under the control of 35S promoter.
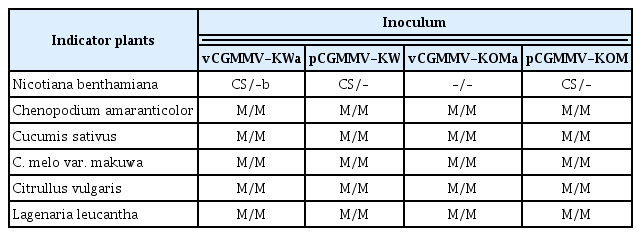
The plasmid DNAs of pCGMMV-KW and -KOM were transformed into Agrobacterium tumefaciens strain GV 2260. The Agrobacterium transformants were selected on YEP medium plates containing 100 mg/l of kanamycin and 50 mg/l of rifampicin. After screening by colony PCR, the Agrobacterium transformants carrying pCGMMV-KW or -KOM were incubated for overnight at 28ºC with shaking in YEP liquid medium containing 100 mg/l of kanamycin and 50 mg/l of rifampicin. The Agrobacterium cells were centrifuged at 4,000×g for 10 minutes and resuspended in the infiltration buffer (10 mM MES, 200 μM acetosyringone, and 10 mM MgCl2) to a final OD600 of ~0.5. The resuspended cells were incubated with shaking for 4 hrs at 28ºC to activate the Agrobacterium Vir genes. To verify whether CGMMV full-length in vivo transcripts generated from pCGMMV-KW or KOM are infectious and the virulences are the same as previously reported (Kim et al., 2003), the Agrobacterium cells were inoculated into Nicotiana benthamiana leaves by agroinfiltration. At 12 days post inoculation (dpi), both pCGMMV-KW and -KOM induced typical systemic mosaic symptoms in N. benthamiana (Fig. 2A). This result demonstrates that pCGMMV-KW and -KOM are fully infectious. To examine symptomatic characteristics of pCGMMV-KW and -KOM, the saps of N. benthamiana infected with pCGMMV-KW or -KOM were used as inoculums to mechanically inoculate indicator plants, including C. amaranticolor, Cu. sativus, Cu. melo var. makuwa, Citrullus vulgaris, and Lagenaria leucantha. In the previous study, it has been shown that the CGMMV isolate KOM could not infect C. amaranticolor, while the isolate KW induced chlorotic spots on the inoculated leaves of C. amaranticolor (Kim et al., 2003). However, both of the cloned CGMMV-KW and KOM induced chlorotic spots on the inoculated leaves of C. amaranticolor (Fig. 2B and Table 1). In addition, as same as previously described (Kim et al., 2003), both of the cloned CGMMV-KW and KOM systemically infected Cu. sativus, Cu. melo var. makuwa, Ci. vulgaris, and L. leucantha plants and induced mosaic symptoms in these indicator plants (Table 1). Virus replication in the inoculated plants was confirmed by subjecting total RNAs extracted from the inoculated and upper un-inoculated leaves to RT-PCR detection using the CGMMV-specific primers (5′-AGTTACAAGTATAATAGCGGATGT-3′ and 5′-TCAAATACTTGAAAACCGG-3′) (data not shown).
Symptom appearance in indicator plants inoculated with either pCGMMV-KW or pCGMMV-KOM. (A) Nicotiana benthamiana plants were infiltrated with Agrobacterium cells harboring either pCGMMV-KW or pCGMMV-KOM. (B and C) Chenopodium amaranticolor plants were mechanically inoculated with saps of N. benthamiana infected with pCGMMV-KW or -KOM. Both of pCGMMV-KW and -KOM induced chlorotic spots on the inoculated leaves of C. amaranticolor (B), while no infection was observed on the upper uninoculated leaves (C). Photographs were taken at 12 days post-inoculation.
Because the cloned CGMMV-KOM showed different symptomatic characteristic on C. amaranticolor from the original virus isolate, we determined full genome sequences of the cloned CGMMV isolates KW and KOM to examine if mutations are introduced into the cloned CGMMV genomes. The determined deduced amino acid sequences of the cloned CGMMV genomes were compared together with the previously reported original sequences of the CGMMV isolates KW and KOM (GenBank accession numbers AF417243 and AF417242, respectively). Amino acid sequence comparison result was summarized in Table 2. Two amino acids (Lys to Gln at position 7 of the 129 kDa protein and Cys to Ser at position 1572 of the 186 kDa protein) were mutated in the cloned KW and only one amino acid (Ala to Thr at position 228 of the 129 kDa protein) was substituted in the cloned KOM when compared with the sequences of their original isolates. In addition, only four amino acids (at positions 228 and 699 of the 129 kDa protein and at positions 1212 and 1238 of the 186 kDa protein) differ between the cloned isolates KW and KOM. No amino acid sequence difference was observed in the MP and CP regions among the cloned and original CGMMV genomes. Sequence comparison suggests that the mutation A228T in the 129 kDa of the cloned KOM might be responsible for induction of chlorotic spots on the inoculated leaves of C. amaranticolor, because this mutation is the unique difference between the original and cloned KOM genomes. However, when compared with the sequence of the original KOM isolate, the cloned KW contained same Ala at position 228 of the 129 kDa protein but three different amino acids at position 699 of the 129 kDa protein and at positions 1212 and 1238 of the 186 kDa protein (L699I, K1212N, and K1238R). This further suggests that the amino acid at position 228 of the 129 kDa protein is not solely responsible for symptomatic change of CGMMV but other amino acid mutations in the replicase protein region could affect on disease response of CGMMV. In all, our results suggest that the replication-associated proteins (the 129 kDa and/or 186 kDa proteins) may play a role as elicitors inducing chlorotic spots in C. amaranticolor. To confirm this possibility, we are currently examining by introducing amino acid substitutions at positions 228 and 699 of the 129 kDa protein and at positions 1212 and 1238 of the 186 kDa protein into the cloned CGMMV isolates.
Construction of infectious cDNA clones of plant RNA viruses is useful for the investigation of the molecular biology of the viruses including pathogenesis, replication, genome expression, and viral gene functions. In this study, we constructed infectious full-length cDNA clones of two isolates of CGMMV, KW and KOM. Infectious in vivo transcripts of the cloned CGMMV genomes were produced under the control of the 35S promoter of CaMV and processed by a self-cleaving ribozyme sequence and a nopaline synthase poly(A) signal to generate authentic 3′ end of a inserted cDNA sequence. These viral transcription and processing signals enable to produce infectious in vivo transcripts by bypassing the difficulties of in vitro RNA transcription.
It is generally known that RNA viruses exist as quasispecies because of the lack of proofreading activity of viral RNA-dependent RNA polymerase (RdRp) (Domingo et al., 1985). It has been demonstrated that serial passages of a cloned plant RNA virus can result in accumulation of mutations in viral progeny populations (Hajimorad et al., 2003). This genetic heterogeneity of virus population caused by error-prone replication of plant RNA viruses is advantageous to overcome selection pressures that limit virus survival. We firstly determined and reported full-genome sequences of CGMMV isolates, KW and KOM, in 2003. Since then, the virus isolates have been propagated via several passages in susceptible host plants. The full-length cDNA clones of these CGMMV isolates were finally constructed in this study and sequence comparison revealed that several mutations have been introduced into the CGMMV genomes during the passages. Two out of six nucleotide mutations resulted in synonymous substitutions in the cloned KW genome, while one out of three nucleotide mutations resulted in synonymous change in the cloned KOM genome (Table 2 and data not shown). Previous studies have shown that a single amino acid substitution introduced by error-prone replication of RNA viruses can result in emergence of resistance-breaking variants (Hebrard et al., 2006; Seo et al., 2009a; Seo et al., 2011). Indeed, one amino acid mutation introduced in the cloned KOM genome seemed to affect original symptomatic characteristic of the isolate in C. amaranticolor (Fig. 2B and Table 1). Since the cloned CGMMV-KOM infected N. benthamiana and other susceptible hosts successfully, it is likely that the replication-associated proteins (the 129 kDa and/or 186 kDa proteins) of CGMMV functions in elicitation of chlorotic spots in C. amaranticolor. Further studies are needed to characterize the mode of resistance against CGMMV in C. amaranticolor and involvement of the replication-associated proteins in activation of resistance.
Acknowledgments
This work was supported in part by grants from the Agenda Programs (PJ00922904 & PJ01130602) funded by the Rural Development Administration and the Vegetable Breeding Research Center (No. 710001-07-05) through the Agriculture Research Center program from the Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea.