 |
 |
| Plant Pathol J > Volume 36(6); 2020 > Article |
|
Abstract
Although limited progress have been made about pathogen system of Hibiscus rosa-sinensis and Hibiscus latent Fort Pierce virus (HLFPV), interaction between plant host and pathogen remain largely unknown, which led to deficiency of effective measures to control disease of hibiscus plants caused by HLFPV. In this study, infection of HLFPV in Hibiscus rosa-sinensis was firstly confirmed for the first time by traditional electron microscopy, modern reverse transcription polymerase chain reaction and RNA-seq methods in China (HLFPV-Ch). Sequence properties analyzing suggested that the full-length sequences (6,465 nt) of HLFPV-Ch had a high sequence identity and a similar genomic structure with other tobamoviruses. It includes a 5′-terminal untranslated region (UTR), followed by four open reading frames encoding for a 128.5-kDa replicase, a 186.5-kDa polymerase, a 31-kDa movement protein, 17.6-kDa coat protein, and the last a 3′-terminal UTR. Furthermore, HLFPV-Ch-derived virus-derived siRNAs (vsiRNAs) ant its putative target genes, reported also for the first time, were identified and characterized from disease Hibiscus rosa-sinensis through sRNA-seq and Patmatch server to investigate the interaction in this pathogen systems. HLFPV-Ch-derived vsiRNAs demonstrated several general and specific characteristics. Gene Ontology classification revealed predicted target genes by vsiRNAs are involved in abroad range of cellular component, molecular function and biological processes. Taken together, for first time, our results certified the HLFPV infection in China and provide an insight into interaction between HLFPV and Hibiscus rosa-sinensis.
Hibiscus plants (Hibiscus rosa-sinensis) are common ornamentals in subtropical and tropical areas, including China. Hibiscus rosa-sinensis are widely used for hedges and miniascape in private and public gardens due to their dense growth, beautiful and attractive flowers (Adkins et al., 2003, 2006; Kamenova and Adkins, 2004). However, disease caused by virus pathogens has become a huge obstacle or threaten for Hibiscus rosa-sinensis-related industry. Hibiscus latent Fort Pierce virus (HLFPV; genus Tobamovirus and family Virgaviridae) has been shown to infect Hibiscus rosa-sinensis in the worldwide, including United States (Adkins et al., 2003, 2006; Kamenova and Adkins, 2004), Thailand (Adkins et al., 2006), Indonesia (Matsui et al., 2005), Japan (Matsui et al., 2005; Yoshida et al., 2014), Brazil (Gao et al., 2016) and Italy (Nerva et al., 2018), but excluding China until reported in this study. Hibiscus rosasinensis infected with HLFPV normally showed severe abnormal symptoms, such as chlorotic spots, ringspots and chlorotic mottle on plant leaves (Adkins et al., 2003). HLFPV possess a single-stranded plus-sense RNA genome. As other tobamoviruses, the genomic structure of HLFPV includes a 5′-ternimal untranslated region (UTR), followed by four open reading frames (ORFs) encoding respectively for a 128-kDa replicases protein with methyltransferase and RNA helicase domains, a 188-kDa readthrough protein with polymerase domain, a 38-kDa movement protein and 18-kDa coat protein, and a 3′-ternimal UTR (Adams et al., 2009). Although limited progress, such as virion morphology, genome organization (Adkins et al., 2003; Gao et al., 2016; Nerva et al., 2018; Yoshida et al., 2014), host range and transmission efficiency of different inoculation (Kamenova and Adkins, 2004), have been made about this pathogen-systems, interaction between hibiscus plants host, Hibiscus rosa-sinensis and HLFPV pathogen remain largely unknown, which led to deficiency of effective measures to control disease of hibiscus plants caused by HLFPV.
Recently, next generation high-throughput parallel sequencing platforms of small RNA (sRNA-seq) have proved to be highly efficient in study of interactions between viruses and its hosts (Mandadi and Scholthof, 2015; Prabha et al., 2013; Rubio et al., 2015; Sharma et al., 2013; Vaucheret, 2006; Wang, 2015). This approach exploits a natural and fundamental antiviral defense mechanism called RNA interference (RNAi). In eukaryotes, triggered by virus infection, RNAi employs Dicer (DCL) enzymes to cleave viral RNAs into small interfering RNAs (siRNAs) with sizes about 21 nucleotides (nt), which are further amplified by RNA dependent RNA polymerases. These siRNAs are loaded into Argonaute (AGO) proteins to form the RNA induced silencing complex to specifically silence target genes (Baulcombe, 2004; Ding, 2010). Thus, happening of interaction between virus and host is characterized by the generation of siRNA derived from the viral genome (virus-derived siRNAs, vsiRNA). In this study, we firstly confirmed the infection of HLFPV in hibiscus plants, Hibiscus rosa-sinensis for the first time in China and characterized its sequence properties of genome; we furthermore identified and characterized HLFPV-derived vsiRNAs ant its putative target genes from disease hibiscus plants, Hibiscus rosa-sinensis, through sRNA-seq and Patmatch server to investigate the interaction between HLFPV-Ch and its host plant, Hibiscus rosa-sinensis.
Hibiscus rosasinensis were sampled from Taiwan Farmers Entrepreneurship Park in Zhangzhou, Fujian. Total RNA was extracted from both healthy and virus-infected leaves of Hibiscus rosa-sinensis with Trizol reagent (Invitrogen, Carlsbad, CA, USA) as instruction. Concentrations and integrity of total RNA were detected using a spectrophotometer (Nanodrop 2000, Thermo Fisher Scientific, Wilmington, DE, USA) and a BioAnalyser 2100 (Agilent, Palo Alto, CA, USA), respectively. To detect infection of HLFPV, primers (available upon requested) were designed and synthesized in SANGON BIOTECH Co., Ltd. (Shanghai, Beijing, China) basing on the sequences of coat protein (CP) gene of the HLFPV Japanese isolate (Yoshida et al., 2014). Reverse transcription polymerase chain reaction (RT-PCR) was conducted with the primers to amplify the CP gene with FastKing One Step RT-PCR Kit (TIANGEN BIOTECH, Beijing, China) as instruction. To characterize the sequences properties of HLFPV genome, 11 primers pairs (available upon requested) for amplifying 11 sequence segments with overlap regions were designed and synthesized in SANGON BIOTECH Co., Ltd. basing on the HLFPV Japanese isolate (Yoshida et al., 2014). Then, 11 products of RT-PCR were cloned into pMD18-T vector. The obtained recombined vectors were sequenced and the obtained sequences were combined into a full sequence with software DNAMAN. ORFs were predicted on the NCBI website, https://www.ncbi.nlm.nih.gov/orffinder/. The phylogenetic tree was established by maximum likelihood method with MEGA 5.1 software with 1,000 replicates. Sequence similarity comparison of HLFPV ORFs was performed with NCBI Blast. Sequence similarity comparison of HLFPV 5′-terminal and 3′-terminal regions was performed with software BioEdit. Prediction of secondary structures of the full-length viral genome and its 3′-terminal UTR was performed with RNAfold server at the website, http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi.
Leaf dips of both healthy and virusinfected leaves of hibiscus plants, Hibiscus rosa-sinensis were made on 200 mesh Formvarcoated copper grids and stained with 5% (w/v) uranyl acetate. Virus particles were observed under H-7650 Hitachi transmission electron microscope (Hitachi, Tokyo, Japan) at 80 kV as described previously (Lan et al., 2016).
Total RNA was extracted from virus-infected and healthy leaves of hibiscus plants, Hibiscus rosa-sinensis using Trizol Reagent (Invitrogen) as introduction. Concentrations and integrity of total RNA were detected using a spectrophotometer (Nanodrop 2000) and a BioAnalyser 2100 (Agilent Technologies), respectively. Small RNA libraries were constructed and sequenced with the Solexa protocol (Gen Denovo, Guangzhou, China) as described previously (Lan et al., 2015, 2016). Briefly, small RNA molecules (18-32 nt) were purified and Solexa adaptors were ligated to 5′-and 3′-terminals. Then, these small RNA molecules were amplified with the adaptor primers for 25 cycles and fragments of about 90 bp (small RNA and adaptors) were isolated from the agarose gel. The purified DNA was utilized directly for small RNA sequencing analysis with Illumina’s Solexa Sequencer. Raw data sets for the small RNA were analyzed. In brief, adaptor sequences were trimmed and small RNA reads without an identifiable linker were removed. The remaining reads were filtered by length. Reads of >32 or <18 nt were discarded. To identify vsiRNAs, using the software Bowtie v.12.7 with a parameter of 0 mismatch, we aligned all the cleaned reads to the HLFPV genome. The downstream analyses of vsiRNAs were performed using Perl scripts and Excel. Target genes were predicted with the small RNA target analysis server Patmatch_v1.2 with a parameter of 0 mismatch, minimum free energy ≥90% of free energy when vsiRNAs perfectly binds to target genes. The Gene Ontology (GO) annotation analysis of the predicted target genes of the vsiRNAs was performed using BLAST2GO software (Lan et al., 2019).
Hibiscus plants samples, Hibiscus rosa-sinensis showing severe chlorotic spots, ringspots and chlorotic mottle on plant leaves (Fig. 1A) for about 10 days were collected from Taiwan Farmers Entrepreneurship Park in Zhangzhou, Fujian. The infection of HLFPV was concurrently confirmed by RT-PCR and by H-7650 Hitachi transmission electron microscopy. Our results showed that CP genes (about 500 bp) of HLFPV were successfully amplified by RT-PCR with specific primers (available upon requested) from plant leaves showing symptom but not healthy plant leaves (Fig. 1B). The obtained positive RT-PCR products were sequenced and the results of NCBI Nucleotide Blast confirmed that it matched with CP gene of HLFPV genome (data not shown). The phylogenetic tree of the CP gene showed that HLFPV-Ch isolate clustered with analogue of other HLFPV isolates (Supplementary Fig. 1), implying high homology among them. Furthermore, electron microscopy by negative staining method revealed that approximately 300 nm rod-shaped virus particles, perhaps representing for HLFPV, existed in leaf dips prepared from viruses-infected leaves of Hibiscus rosa-sinensis (Fig. 1C) but not from healthy leaves of Hibiscus rosa-sinensis (data not shown). Thus, these results preliminary proved Hibiscus rosa-sinensis may be infected by HLFPV. This is reported for HLFPV infection in China for the first time.
By being cloned, sequenced and joined, the complete genomic sequence of HLFPV isolated from China was 6,465 nucleotides (nt) in length (Fig. 2A). It contained four ORFs encoding proteins of 128.5, 186.5, 31.1 and 17.6 kDa in ORF1 (nt 47-3,463), ORF2 (nt 47-4,987), ORF3 (nt 4,968-5,822), and ORF4 (nt 5,794-6,270), respectively (Fig. 2A) by being predicted on the NCBI website. ORF3 overlaps partially with ORF2 and ORF4 by 20 and 29 nt, respectively (Fig. 2A). These results implied the similarity in genomic organization or arrangement of tobamoviruses. The four ORFs of HLFPV isolated from China were 99.4-100%, 99.4-100%, 99.3-100%, and 99.2-100% identical at the nucleotide level and 99.4-100%, 99.4-100%, 99.7-100%, and 98.7-100% identical at the amino acid level to counterparts of other four HLFPV isolates, respectively (Table 1). These results suggested that the detected HLFPV isolated from China in Hibiscus rosa-sinensis was a tobamovirus. Subsequently, we designated this China isolate of HLFPV as HLFPV-Ch.
The length of the 5′-ternimal UTR of HLFPV-Ch was 46 nt, which is shorter than HLFPV Japanese and Brazil isolates but longer than Italy isolate (Gao et al., 2016; Nerva et al., 2018; Yoshida et al., 2014). The 5′-UTR shared 21.7-43.7% identity at the nucleotide level with other HLFPV isolates and showed the highest identity to that of HLFPV Italy isolate (Table 1). Eight copies of CAA triplets, which are conserved in the 5′-ternimal UTR regions of other tobamoviruses, were found in the 5′-ternimal UTR of HLFPV-Ch. Furthermore, the 5′-proximal 10 nucleotides of all HLFPV isolates including HLFPV-Ch was GUAUGUUUUA, implying its importance in the infection cycles of virus.
The 3′-ternimal UTR of HLFPV-Ch was 195 nt in length. It was 36.9-49.7% identical at the nucleotide level to counterparts of other three HLFPV isolates, showing the highest (49.7%) identity to that of HLFPV isolated from Brazil (Table 1). The 3′-ternimal UTR of HLFPV-Ch contained a 79 nt poly(A) region but not the pseudoknot region (Fig. 2A), both of which was conserved in most tobammoviruses (Yoshida et al., 2014), at the downstream of the stop codon in ORF4. More and more evidences have shown that poly(A) region of plant viruses plays an important role in the infection cycles of viruses by interactions with many host proteins, such as poly(A)-binding proteins (Beauchemin and Laliberté, 2007). It has been shown that downstream of the poly(A) region of the 3′-UTR of HLFPV isolated from Japan was predicted to contain a tRNA-like secondary structure (Yoshida et al., 2014). Using RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi), downstream of the poly(A) region of the 3′-UTR of HLFPV-Ch was also predicted to contain a tRNA-like secondary structure (Supplementary Fig. 2), implying their the similar functions in virus infection cycle. The 3′-proximal sequence of HLFPV-Ch was also CCCA, suggesting the sequence conservatism among tobamoviruses.
Following determination of HLFPV-Ch infection, small RNA libraries were constructed and sequenced with the Solexa protocol (Gen Denovo) with total RNA extracted from HLFPV-Ch-infected and healthy Hibiscus rosa-sinensis. A total of 10,839,244 and 8,733,689 sRNAs reads were sequenced from HLFPV-Ch-infected and healthy Hibiscus rosa-sinensis, respectively. It showed that 21-, 24-, and 22-nt sRNAs were most dominant in HLFPV-Ch-infected and healthy Hibiscus rosasinensis (Fig. 3A). Furthermore, both 21-nt, 22-nt and 23-nt sRNAs reads increased markedly, but 24-nt sRNA reads decreased significantly in HLFPV-Ch-infected Hibiscus rosa-sinensis (Fig. 3A), which is similar to previous reports for the same Tobamovirus genus (Lan et al., 2019; Li et al., 2016). These results suggested that the infection of virus, HLFPV-Ch, modulated the distribution pattern of sRNAs in Hibiscus rosa-sinensis.
After mapped to the HLFPV-Ch genome, 30,925 unique vsiRNAs (18-32 nt) were identified in HLFPV-Ch-infected Hibiscus rosa-sinensis, accounting for 0.29% of siRNAs reads. Conversely, only 571 unique vsiRNAs (18-32 nt), accounting for 0.007% of siRNAs reads, were identified in healthy Hibiscus rosa-sinensis. In HLFPV-Ch-infected Hibiscus rosa-sinensis, the majority (71%) of vsiRNAs were 19-24 nt in length, with relative average abundance for different sized species although 21-nt and 22-nt vsiRNAs being most abundant orderly (Fig. 3B); however, the majority of vsiRNAs in many virus-plant systems were 21-nt and 22-nt in length (Kreuze et al., 2009; Lan et al., 2019; Li et al., 2016; Mitter et al., 2013; Xu and Zhou, 2017; Yan et al., 2010; Yang et al., 2014). Thus, these results implicated that the homologues of DCL4 and DCL2 in Hibiscus rosa-sinensis may also be the predominant DCLs involved in the biogenesis of 21-nt and 22-nt vsiRNAs respectively which functions as the predominant antiviral silencing mediators (Blevins et al., 2006; Deleris et al., 2006; Ding, 2010; Donaire et al., 2008; Liu et al., 2018; Niu et al., 2017); as for other DCLs in Hibiscus rosa-sinensis participating in different-lengths vsiRNAs generation will be the future discussed topic. Furthermore, the distance of 5′ ends (overhangs) of 21-nt vsiRNAs duplexes were analyzed. The results suggested that 21-nt vsiRNA duplexes with 2-nt overhangs were the most abundant 21-nt duplexes, followed by 1-nt overhangs and then 0-nt overhangs 21-nt duplexes in HLFPV-Ch-infected Hibiscus rosa-sinensis (Fig. 3C), which was similar that of other viruses-plant and viruses-invertebrates systems (Liu et al., 2018; Niu et al., 2017). Thus, the results revealed that 21-nt vsiRNAs duplexes with 2-nt overhangs were the most efficient triggers of RNAi in Hibiscus rosa-sinensis.
Many researches have suggested that the 5′-terminal nucleotides of vsiRNAs have a important role in modulating the sorting of vsiRNAs to different AGO complexes in plants (Mi et al., 2008). In this study, our analysis showed that HLFPV-Ch unique vsiRNAs demonstrated a clear tendency to begin with uracil (U, 31%) and adenine (A, 28%) as compared with cytosine (C, 23%) and guanidine (G, 17%) (Fig. 3D), which was consistent with previous studies for diverse plant-virus systems (Donaire et al., 2008, 2009; Lan et al., 2019; Xu and Zhou, 2017). To obtain further understanding of HLFPV-Ch vsiRNA sorting, the complexity of vsiRNAs was analyzed for different sizes species. For 21-nt and 22-nt HLFPV-Ch vsiRNAs, a clear preference for vsiRNAs beginning with a 5′-U was observed (Fig. 3D). This is consistent with the function described for AGO1 in defending against RNA viruses in plant (Morel et al., 2002; Qu et al., 2008). However, for 23-nt, 24-nt, and 25-nt HLFPV-Ch vsiRNAs, a strong bias for vsiRNAs beginning with a 5′-A was observed (Fig. 3D), implying the high binding affinity of AGO2 and AGO4 for these vsiRNAs. The low proportion of vsiRNAs beginning with G was also observed and consistent with previous reports of pathogen-systems (Donaire et al., 2009; Li et al., 2016; Mi et al., 2008; Mitter et al., 2013). Thus, our finding suggested the involvement of different AGO proteins in binding of vsiRNAs in different plant species. In future, isolation and determination of AGO-vsiRNAs complexes from hibiscus plants, Hibiscus rosa-sinensis infected with HLFPV-Ch will provide us more functions information about vsiRNAs sorting in hibiscus plants.
To reveal the origin of vsiRNAs, we aligned vsiRNAs with positive and negative strands of HLFPV-Ch genome. Our results showed that HLFPV-Ch vsiRNAs were derived predominantly from the viral positive-strand RNA, accounting for about 86% (Fig. 3E). This is broadly contrasted to many ssRNA positive-strand viruses, including the cucumber green mottle mosaic virus of the same genus, vsiRNAs of which were produced nearly equally from the positive and the negative strands (Chen et al., 2018; Ho et al., 2007; Lan et al., 2020; Li et al., 2016; Xia et al., 2014; Yang et al., 2014). Additionally, single-base resolution maps of total unique vsiRNAs along with HLFPV-Ch genomes were created using Bowtie tools and in-house Perl scripts. Our results showed that the majority vsiRNAs were derived from the intragenic regions, not the 5′-or 3′-terminal regions of HLFPV-Ch genome (Fig. 2B). These vsiRNAs had a continuous but heterogeneous (Hot spot and Cold spot) distribution along the HLFPV-Ch genome (Fig. 2B). Taken together, our results implied that highly structured regions present in HLFPV-Ch genome may be the substrates for DCL enzymes to cutting into vsiRNAs. To confirm the speculation, we evaluated the secondary structures of HLFPV-Ch genome with RNAfold server. Our results showed that lots of stem loop secondary structures were formed in intragenic regions and corresponded to the hotspots (Fig. 2C and D). Taken together, these results supported that vsiRNAs should originate predominantly by direct DCL cleavage of imperfect duplexes in the most folded regions of the positive strand of HLFPV-Ch RNA molecular in Hibiscus rosa-sinensis. This result is similar to the speculation for vsiRNAs origin from plants infected with diverse ssRNA positive-strand viruses (Molnár et al., 2005).
Identification and characterization of target genes is very important to understand deeply the functions and implications of vsiRNAs in interaction with host. In this study, we used the small RNA target analysis server Patmatch_v1.2 to predict putative target genes of Hibiscus rosa-sinensis by HLFPV-Ch-derived vsiRNAs. Totally, 722 putative target genes were predicted by HLFPV-Ch-derived siRNAs (Supplementary Table 1). In BLAST2GO analysis, these predicted target genes are involved in abroad range of cellular component, molecular function and biological processes (Fig. 4, Supplementary Table 1). The three most highly represented GO terms were “intracellular membrane-bounded organelle”, “membrance” and “intracellular cytoplasm” under the cellular component category, comprising 90% of the annotated genes (Fig. 4A). The two most abundant GO terms under the molecular function category, “catalytic enzyme activity” and “binding”, represented 90% of the annotated genes (Fig. 4B). The terms “metabolic process”represented 73% of the classified targets under the biological process section (Fig. 4C). These results suggested that vsiRNAs may play vital roles due to their interactions with host by targeting their genes during HLFPV-Ch infection in hibiscus plants, Hibiscus rosa-sinensis.
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
This work was supported by the Nature Science Foundation of Fujian (2018J01465), the National Natural Science Foundation of China (grant no. 31601613), the Nature Science Foundation of Zhangzhou (grant no. ZZ2017J03) and the Outstanding Youth Research Talent Project for Fujian University (2017).
Fig. 1
Detection of HLFPV-Ch infection in Hibiscus rosasinensis sampled from Taiwan Farmers Entrepreneurship Park in Zhangzhou, Fujian. (A) Symptoms of Hibiscus rosa-sinensis leaves infected with HLFPV-Ch. (B) Agarose gel electrophoresis of RT-PCR products of HLFPV-Ch CP gene. M, DNA mareker D2000; 1 and 2, RT-PCR products of HLFPV-Ch CP gene from HLFPV-Ch-infected and healthy samples, respectively. (C) Electron microscopy of Hibiscus rosa-sinensis leaves infected with HLFPV-Ch showing virus particle morphology through the negative staining. Virion morphology is typical of recognized tobamoviruses. HLFPV, Hibiscus latent Fort Pierce virus; RT-PCR, reverse transcription polymerase chain reaction; CP, coat protein. Scale bar = 100 nm.

Fig. 2
Generation sites of vsiRNAs are associated with the putative secondary structures in the virus genomes RNA of HLFPV-Ch. (A) Genome organization of HLFPV-Ch. Boxes represent ORFs of replicase (128.5 kDa), polymerase (186.5 kDa), MP (31.1 kDa), and CP (17.6 kDa). The nucleotide numbers show the start and stop locations of ORFs. Dotted vertical line in the box indicates amber stop codon. Untranslated regions at the 5′-and 3′-terminals are represented by horizontal lines. Internal poly(A) is represented by black solid box. The data shown was the representative results of three replicates of sRNA-seq. (B) Distribution and hotspots for vsiRNAs along HLFPV-Ch genome. Green and red colors denoted vsiRNAs derived from positive and negative strands of HLFPV-Ch genome, respectively. (C) Secondary structures of HLFPV-Ch genome predicted with RNAfold server (http://rna.tbi.univie.ac.at/). Color bar with No. 0-1 denotes the possibilities of bases paring. (D) This is enlarged images of the boxed areas of the HLFPV sequence in panel C to show the 5′-and 3′-terminal regions. vsiRNA, virus-derived siRNA; HLFPV, Hibiscus latent Fort Pierce virus; ORF, open reading frame; MP, movement protein; CP, coat protein; UTR, untranslated region.
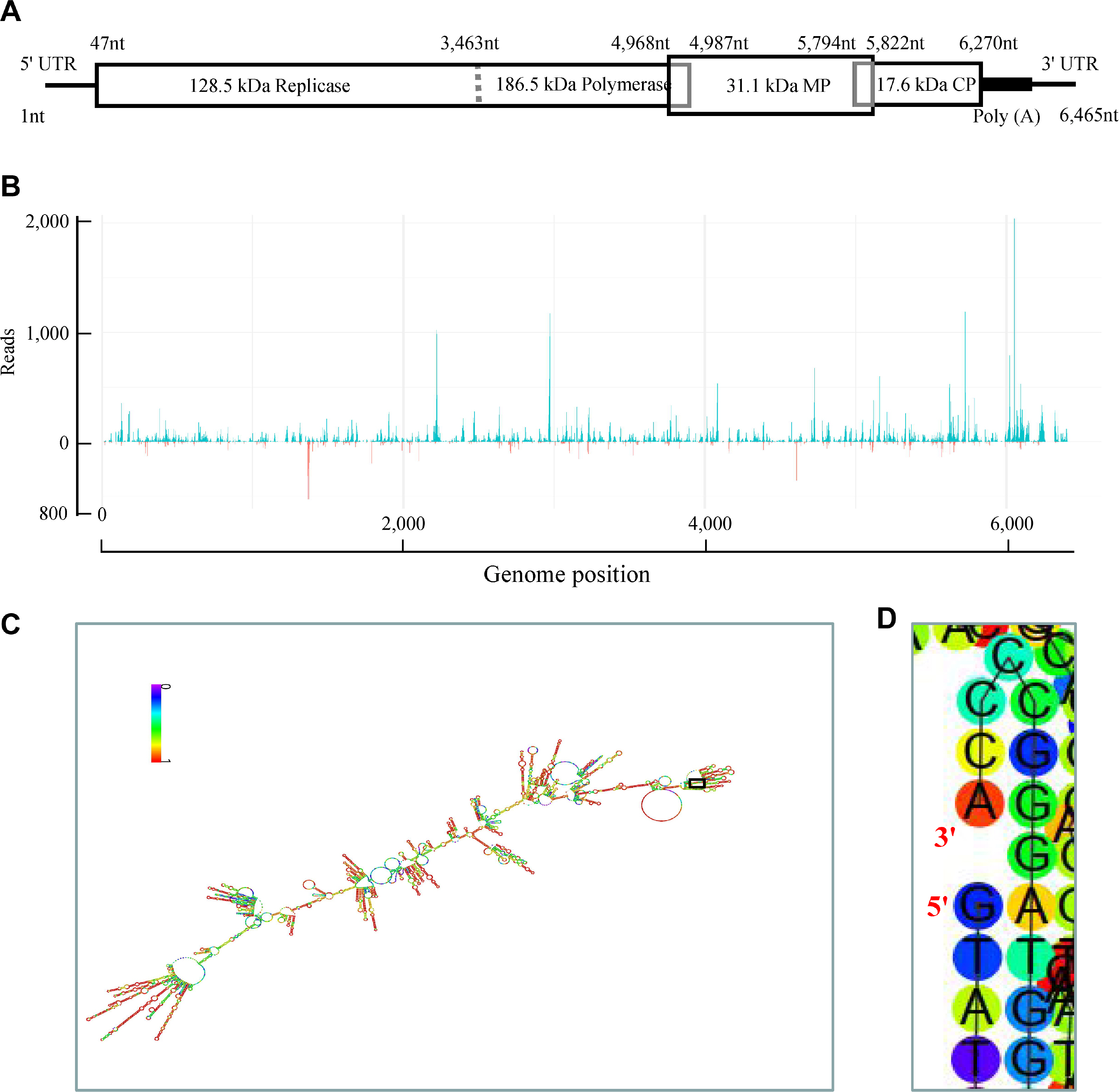
Fig. 3
Characterizations of vsiRNAs derived from HLFPV-Ch in Hibiscus rosa-sinensis. (A) Size distribution of total small RNAs (18-32 nt) in Hibiscus rosa-sinensis. Black and pink colors denoted the total small RNAs come from HLFPV-Ch-infected and healthy Hibiscus rosa-sinensis, respectively. (B) Size distribution of HLFPV-Ch-derived unique vsiRNAs (18-32 nt) in Hibiscus rosa-sinensis. (C) Reads of 21-nt vsiRNAs with 1-21 nt distance between 5′ ends in HLFPV-Ch-infected Hibiscus rosa-sinensis. (D) Percentages of 5′-terminal nucleotide of HLFPV-Ch-derived unique vsiRNAs (18-32 nt) in Hibiscus rosa-sinensis. (E) Percentages of unique vsiRNAs (18-32 nt) derived from positive and negative strands of HLFPV-Ch genome in Hibiscus rosa-sinensis. “+” and “-” indicate vsiRNAs derived from positive and negative genomic strands, respectively. The data shown was the representative results of three replicates of sRNA-seq. vsiRNA, virus-derived siRNA; HLFPV, Hibiscus latent Fort Pierce virus.
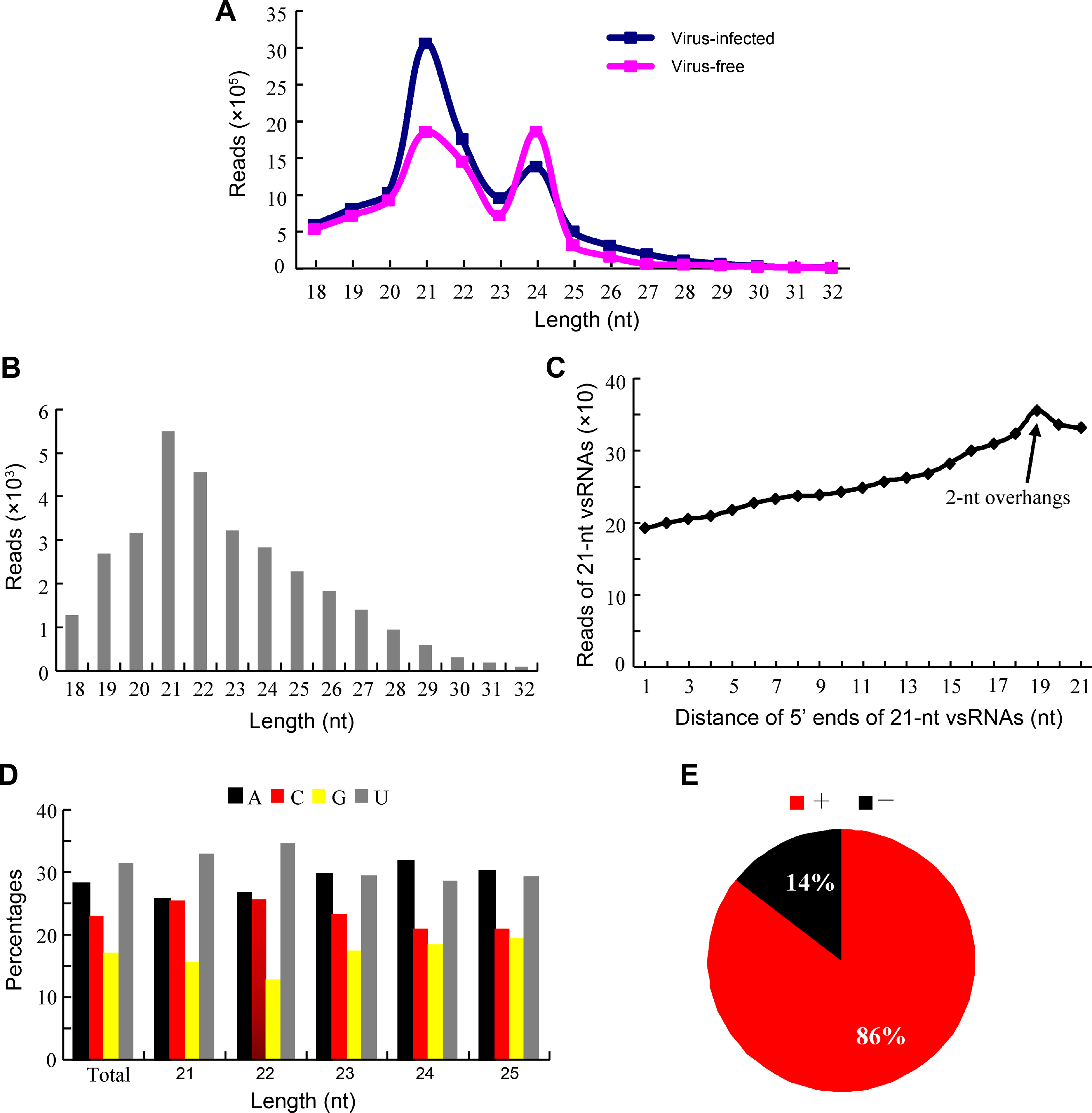
Fig. 4
Gene Ontology categories of predicted target genes of HLFPV-Ch-derived vsiRNAs in in Hibiscus rosasinensis. The predicted target genes were subject to BLAST alignment and Gene Ontology annotation using BLAST2GO software. These predicted target genes are involved in cellular component (A, red), molecular function (B, green) and biological processes (C, blue). HLFPV, Hibiscus latent Fort Pierce virus; vsiRNA, virus-derived siRNA.
Table 1
Percentages (%) of nucleotide and amino acid sequence identities of the UTRs and ORFs among HLFPV isolates
REFERENCES
Adams, M. J., Antoniw, J. F. and Kreuze, J. 2009. Virgaviridae: a new family of rod-shaped plant viruses. Arch. Virol. 154:1967-1972.



Adkins, S., Kamenova, I., Achor, D. and Lewandowski, D. J. 2003. Biological and molecular characterization of a novel tobamovirus with a unique host range. Plant Dis. 87:1190-1196.


Adkins, S., Kamenova, I., Chiemsombat, P., Baker, C. A. and Lewandowski, D. J. 2006. Tobamoviruses from hibiscus in Florida and beyond. Acta Hortic. 722:65-70.

Beauchemin, C. and Laliberté, J.-F. 2007. The poly(A) binding protein is internalized in virus-induced vesicles or redistributed to the nucleolus during turnip mosaic virus infection. J. Virol. 81:10905-10913.



Blevins, T., Rajeswaran, R., Shivaprasad, P. V., Beknazariants, D., Si-Ammour, A., Park, H.-S., Vazquez, F., Robertson, D., Meins, F. J, Hohn, T. and Pooggin, M. M. 2006. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34:6233-6246.



Chen, S., Yu, N., Yang, S., Zhong, B. and Lan, H. 2018. Identification of Telosma mosaic virus infection in Passiflora edulis and its impact on phytochemical contents. Virol. J. 15:168




Deleris, A., Gallego-Bartolome, J., Bao, J., Kasschau, K. D., Carrington, J. C. and Voinnet, O. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 313:68-71.


Donaire, L., Barajas, D., Martínez-García, B., Martínez-Priego, L., Pagán, I. and Llave, C. 2008. Structural and genetic requirements for the biogenesis of Tobacco rattle virus-derived small interfering RNAs. J. Virol. 82:5167-5177.



Donaire, L., Wang, Y., Gonzalez-Ibeas, D., Mayer, K. F., Aranda, M. A. and Llave, C. 2009. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology. 392:203-214.


Gao, R., Niu, S., Dai, W., Kitajima, E. and Wong, S.-M. 2016. Hibiscus latent Fort Pierce virus in Brazil and synthesis of its biologically active full-length cDNA clone. Virus Genes. 52:754-757.


Ho, T., Wang, H., Pallett, D. and Dalmay, T. 2007. Evidence for targeting common siRNA hotspots and GC preference by plant Dicer-like proteins. FEBS Lett. 581:3267-3272.


Kamenova, I. and Adkins, S. 2004. Transmission, in planta distribution, and management of Hibiscus latent Fort Pierce virus, a novel tobamovirus isolated from Florida Hibiscus. Plant Dis. 88:674-679.


Kreuze, J. F., Perez, A., Untiveros, M., Quispe, D., Fuentes, S., Barker, I. and Simon, R. 2009. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: a generic method for diagnosis, discovery and sequencing of viruses. Virology. 388:1-7.


Lan, H., Lai, B., Zhao, P., Dong, X., Wei, W., Ye, Y. and Wu, Z. 2020. Cucumber mosaic virus infection modulated the phytochemical contents of. Passiflora edulis. Microb. Pathog. 138:103828

Lan, H.-H., Wang, C.-M., Chen, S.-S. and Zheng, J.-Y. 2019. siRNAs derived from Cymbidium mosaic virus and Odontoglossum ringspot virus down-modulated the expression levels of endogenous genes in Phalaenopsis equestris. Plant Pathol. J. 35:508-520.



Lan, H., Chen, H., Liu, Y., Jiang, C., Mao, Q., Jia, D., Chen, Q. and Wei, T. 2015. Small interfering RNA pathway modulates initial viral infection in midgut epithelium of insect after ingestion of virus. J. Virol. 90:917-929.



Lan, H., Wang, H., Chen, Q., Chen, H., Jia, D., Mao, Q. and Wei, T. 2016. Small interfering RNA pathway modulates persistent infection of a plant virus in its insect vector. Sci. Rep. 6:20699



Li, Y., Deng, C., Shang, Q., Zhao, X., Liu, X. and Zhou, Q. 2016. Characterization of siRNAs derived from cucumber green mottle mosaic virus in infected cucumber plants. Arch. Virol. 161:455-458.


Liu, C., Chen, Z., Hu, Y., Ji, H., Yu, D., Shen, W., Li, S., Ruan, J., Bu, W. and Gao, S. 2018. Complemented palindromic small RNAs first discovered from SARS coronavirus. Genes (Basel). 9:442



Mandadi, K. K. and Scholthof, K.-B. G. 2015. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell. 27:71-85.



Matsui, Y., Adkins, S. and Natsuaki, K. T. 2005 Hibiscus latent Fort Pierce virus (HLFPV) from Hibiscus rosa-sinesis in Japan and Indonesia. Jpn. J. Phytopathol. 71:232-233 (in Japanese).
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., Wu, L., Li, S., Zhou, H., Long, C., Chen, S., Hannon, G. J. and Qi, Y. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’-terminal nucleotide. Cell. 133:116-127.



Mitter, N., Koundal, V., Williams, S. and Pappu, H. 2013. Differential expression of Tomato spotted wilt virus-derived viral small RNAs in infected commercial and experimental host plants. PLoS ONE. 8:e76276



Molnár, A., Csorba, T., Lakatos, L., Várallyay, E., Lacomme, C. and Burgyán, J. 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured singlestranded viral RNAs. J. Virol. 79:7812-7818.



Morel, J.-B., Godon, C., Mourrain, P., Béclin, C., Boutet, S., Feuerbach, F., Proux, F. and Vaucheret, H. 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in posttranscriptional gene silencing and virus resistance. Plant Cell. 14:629-639.



Nerva, L., Vallino, M., Turina, M. and Ciuffo, M. 2018. Identification and characterization of Hibiscus latent Fort Pierce virus in Italy. J. Plant Pathol. 100:145.

Niu, X., Sun, Y., Chen, Z., Li, R., Padmanabhan, C., Ruan, J., Kreuze, J. F., Ling, K., Fei, Z. and Gao, S. 2017. Using small RNA-seq data to detect siRNA duplexes induced by plant viruses. Genes (Basel). 8:163



Prabha, K., Baranwal, V. K. and Jain, R. K. 2013. Applications of next generation high throughput sequencing technologies in characterization, discovery and molecular interaction of plant viruses. Indian J. Virol. 24:157-165.



Qu, F., Ye, X. and Morris, T. J. 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U. S. A. 105:14732-14737.



Rubio, M., Rodríguez-Moreno, L., Ballester, A. R., de Moura, M. C., Bonghi, C., Candresse, T. and Martínez-Gómez, P. 2015. Analysis of gene expression changes in peach leaves in response to Plum pox virus infection using RNA-Seq. Mol. Plant Pathol. 16:164-176.


Sharma, N., Sahu, P. P., Puranik, S. and Prasad, M. 2013. Recent advances in plant-virus interaction with emphasis on small interfering RNAs (siRNAs). Mol. Biotechnol. 55:63-77.


Vaucheret, H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20:759-771.


Wang, A. 2015. Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 53:45-66.


Xia, Z., Peng, J., Li, Y., Chen, L., Li, S., Zhou, T. and Fan, Z. 2014. Characterization of small interfering RNAs derived from Sugarcane mosaic virus in infected maize plants by deep sequencing. PLoS ONE. 9:e97013



Xu, D. and Zhou, G. 2017. Characteristics of siRNAs derived from Southern rice black-streaked dwarf virus in infected rice and their potential role in host gene regulation. Virol. J. 14:27




Yan, F., Zhang, H., Adams, M. J., Yang, J., Peng, J., Antoniw, J. F., Zhou, Y. and Chen, J. 2010. Characterization of siRNAs derived from rice stripe virus in infected rice plants by deep sequencing. Arch. Virol. 155:935-940.


Yang, J., Zheng, S.-L., Zhang, H.-M., Liu, X.-Y., Li, J., Li, J.-M. and Chen, J.-P. 2014. Analysis of small RNAs derived from Chinese wheat mosaic virus. Arch. Virol. 159:3077-3082.


Yoshida, T., Kitazawa, Y., Komatsu, K., Neriya, Y., Ishikawa, K., Fujita, N., Hashimoto, M., Maejima, K., Yamaji, Y. and Namba, S. 2014. Complete nucleotide sequence and genome structure of a Japanese isolate of hibiscus latent Fort Pierce virus, a unique tobamovirus that contains an internal poly(A) region in its 3′ end. Arch. Virol. 159:3161-3165.


- TOOLS



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




