Antagonistic Activity of Bacteria Isolated from Apple in Different Fruit Development Stages against Blue Mold Caused by Penicillium expansum
Article information
Abstract
Blue mold caused by Penicillium expansum is one of the most significant postharvest diseases of apples. Some microorganisms associated with the surface of ripening apples possess the ability to inhibit the growth of P. expansum. However, the existing literature about their colonization in the stages before ripening is not explored in depth. This study aims to characterize the antagonistic capacity of bacterial populations from five fruit development stages of ‘Royal Gala’ apples. The results have shown that the density of the bacterial populations decreases throughout the ripening stages of fruit (from 1.0 × 105 to 1.1 × 101 cfu/cm2). A total of 25 bacterial morphotypes (corresponding to five genera identified by 16S RNA) were differentiated in which Bacillus stood out as a predominant genus. In the in vitro antagonism tests, 10 Bacillus strains (40%) inhibited the mycelial growth of P. expansum from 30.1% to 60.1%, while in fruit bioassays, the same strains reduced the fruit rot ranging from 12% to 66%. Moreover, the bacterial strains with antagonistic activity increased in the ripening fruit stage. B. subtilis subsp. spiziennii M24 obtained the highest antagonistic activity (66.9% of rot reduction). The matrix-assisted laser desorption ionization- time of flight mass spectrometry analysis revealed that bacteria with antagonistic activity produce antifungal lipopeptides from iturin and fengycin families.
Apples (Malus domestica) is one of the most valued and cultivated fruits in temperate areas of the world (Spadoni et al., 2015). However, this fruit crop suffers economic losses in the post-harvesting stages due to the decay caused by phytopathogenic fungi in which the blue mold caused by Penicillium expansum contributes to the major postharvest disease, causing yield losses of up to 20% (Barad et al., 2016). Chemical methods based on synthetic products which are used to control P. expansum have exhibited negative effects, such as the potential risk of the development of resistance in P. expansum, as well as toxicological problems related to human health and environmental impact (Dukare et al., 2019; Li and Xiao, 2008). Therefore, the demand for safe and effective strategies to control apple blue mold has increased in recent years (Spadoni et al., 2015). The use of microorganisms with antagonistic activity as biocontrol agents is a promising alternative as compared to the utilization of chemical fungicides (de Oliveira-Nascimento et al., 2016). Microorganisms, mainly bacteria, with biocontrol potential have been found naturally associated with the surfaces of plant organs, which are called epiphytic microorganisms (Droby and Wisniewski, 2018). Epiphytic bacteria strains belonging to the genera Pantoea, Pseudomonas, Enterobacter, and Serratia can reduce the flower rot caused by Erwinia amylovora, which have been isolated from pear blossoms (Sharifazizi et al., 2017). Antagonistic epiphytic bacteria with antifungal activity against Exserohilum turcicum, mainly those from the genus Bacillus, have been successfully isolated from leaves of maize (Sartori et al., 2015). Isolation of biocontrol epiphytic microorganisms has been also reported from fruit where bacteria and yeast having the ability to inhibit the growth of phytopathogenic fungi has been identified. Campos-Martínez et al. (2016) reported the isolation of Candida intermedia and Wickerhamomyces anomalus from avocados with the capacity to reduce the incidence and severity of fruit anthracnose caused by Colletotrichum gloeosporioides. Whereas Kasfi et al. (2018) documented the isolation of Candida, Meyerozyma, and Bacillus species from grape berries and grape leaves with the capacity to control the grape rot disease produced by Aspergillus spp.
Bacterial communities are the most abundant on plant epiphytic microbiota. It can be probably due to their capacity to grow under nutrient-limited and unfavorable environmental conditions (Methe et al., 2020). On apple fruit, as in other fruit crops, the studies of antagonistic bacteria strains naturally associated with fruit are limited, only reporting Bacillus amyloliquefaciens (Calvo et al., 2017), Bacillus subtilis (Sholberg et al., 1995), Gluconobacter oxydans (Bevardi et al., 2013), and Rahnella aquatilis (Calvo et al., 2007) against the four of the main pathogenic apple fungi which are: P. expansum, Botrytis cinerea, Botryosphaeria dothidea, and Monilinia fructicola (Chen et al., 2016; Leibinger et al., 1997). The fact that these bacteria are isolated from the apple fruit indicating that the native appleassociated bacterial populations are an excellent source of antagonists against post-harvested fruit decaying fungi. Furthermore, it has been shown that the fruit surface of the apple is predominantly colonized by bacteria from the stage of fruit set to being harvested (Teixidó et al., 1999). Nevertheless, the dynamics of the antagonistic bacterial strains which appear during the development of apple fruit have not been studied, which might be hampering the comprehension of these bacterial populations with antagonistic potential associated natively with the fruit surface of the apple (Juhneviča et al., 2011).
The objectives of this study are to (1) identify the cultivable bacteria associated with the apple fruit surface emerging from the early stage of development to maturity; (2) evaluate in vitro and in vivo, the antagonistic activity of the bacterial isolates against P. expansum; and (3) identify the biochemical traits related to the antagonistic activity of the antagonistic strains.
Materials and Methods
Fruit sampling
For the isolation of the bacterial populations, fruit samples were collected from three 10-year-old trees of the ‘Royal Gala’ variety grown in an orchard in the field of Amealco, Queretaro, Mexico (latitude 20.175, longitude ‒100.175, 2,279 m.a.s.l.). Three fruits in five stages: fruit set, 20 days after blooming (DAB) (S1); green fruit 1, 62 DAB (S2); green fruit 2, 95 DAB (S3); green fruit 3, 124 DAB (S4) and ripe fruit, 186 DAB (S5) (Janssen et al., 2008; Lopes et al., 2013); from February to August 2017 (Fig. 1) were sampled and placed aseptically in polypropylene bags and transported to the laboratory for the examination which is carried out on the same day (Nongkhlaw and Joshi, 2015).
Bacterial isolation
Sterile phosphate buffer (10 mM KH2PO4, at pH 7.5) (Burch et al., 2016) was added to cover the fruit in the polypropylene bags. Then, the samples were sonicated (135 W, 42 kHz; Branson 5510, Branson Ultrasonic Corporation, Danbury, CT, USA) and shaken for 5 min on a rotary shaker (120 rpm/min). The microbial pellet was recovered by centrifugation (4,000 ×g for 10 min) and re-suspended in 1 ml of saline solution (0.85% wt/v NaCl). Appropriate 10-fold dilutions were plated by triplicate on nutrient agar (NA; Dibico, Cuautitlan Izcalli, Mexico) (Lorenzini and Zapparoli, 2020; Rungjindamai, 2016) containing 100 mg/l of fluconazole (Pisa, Guadalajara, Mexico) (Janaki et al., 2016) and incubated for 7 days at room temperature (28°C). The resultant bacterial isolates were classified into morphotypes, considering that a morphotype is defined as a group of different types of bacterial isolates distinguishable from other bacterial isolates by morphological, biochemical, and Gram staining characteristics that may or may not indicate different species (Luziatelli et al., 2019). Each bacterial morphotype was purified and maintained on nutrient agar plates for the forthcoming analysis. All the morphotypes were preserved in 50% glycerol at –80°C.
Molecular identification of bacterial strains
A representative isolate of each morphotype was identified by 16S rRNA gene sequencing analysis (Fredriksson et al., 2013). For this, bacterial DNA was extracted using a Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA, USA). The 16S rRNA gene was PCR-amplified in a 25 μl reaction containing 12.5 μl (1×) Radiant 2× PCR Master Mix, 1 μl (5 ng/μl) gDNA as a template, and 1 μl of previously reported primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (each at 10 mM). The PCR amplicons were purified with DNA Clean and Concentrator TM-5 (Zymo Research), and sequencing was carried out at LANGEBIO, Mexico. The resulting sequences were compared against the non-redundant database of GenBank (Bethesda, MD, USA; https://www.ncbi.nlm.nih.gov/). Best hits were compared and the highest sequence score (≥99% of the sequence cover for the comparison with the database of NCBI) was used to identify bacterial species. The obtained partial sequences have been deposited in the NCBI GenBank database.
In vitro antagonistic assays against P. expansum
To evaluate the antifungal activity of the bacterial morphotypes, in vitro dual confrontations were performed according to the methodology previously proposed by (Chen et al., 2016), in three different culture media used to test antagonist activity against P. expansum (Ávalos et al., 2012; Wang et al., 2016; Yang et al., 2015). A mycelial disc (5 mm of diameter) of P. expansum Link strain CFNL2016, previously grown in potato dextrose agar (PDA; Dibico) for 10 days, was inoculated on the center of PDA, nutrient yeast glucose (NYDA; Dibico), and yeast extract peptone dextrose (YPD; Dibico) plates; finally incubated at 28°C for 3 days. One bacterial colony from a fresh culture was inoculated at a distance of 3 cm from the mycelial disc. The negative control consisted of PDA plates only inoculated with P. expansum, and as positive controls a commercial biological strain Bacillus spp. Q11 (Rodríguez-Chávez et al., 2019) and nystatin fungicide (Shehata et al., 2017) were used. All the plates were incubated at 28°C for 8 days and the mycelial growth diameter was measured, and the antagonistic activity was expressed as the percentage of inhibition of radial growth (PIRG): PIRG (%) = [(R1 ‒ R2)/R1] × 100, where R1 is the radial growth of P. expansum in negative control and R2 is the radial growth produced by the co-inoculation.
In vivo antagonistic assay of bacteria with antifungal activity
To evaluate the capacity to reduce the fruit rot infection caused by P. expansum of the bacterial morphotypes and bacteria consortia by stage of fruit development, in vivo tests were carried out on apple fruit of the Golden Delicious variety, which is susceptible to fungal infection. Bacterial inoculum was prepared by growing each isolate in nutrient broth (Dibico) at 28ºC for 24 h and constant agitation (180 rpm). Cells were recovered by centrifugation at 4,000 ×g for 10 min and washed twice using the saline solution (NaCl 0.85%). Finally, the cellular pellet was re-suspended in a minimum volume of saline solution to be counted in a Neubauer chamber, and then adjusted to a cellular density of 108 cells/ml. For the inoculum of P. expansum, conidia were harvested from 12-day-old culture using saline solution; mycelia were separated by filtration and the conidial suspension was quantified and adjusted to 105 conidia/ml. For the control infection assay, apple fruits were externally disinfected by immersion in a sodium hypochlorite solution (2%) for 2 min, rinsed three times with sterile water, and then air-dried. Then, four wounds (5 × 5 × 3 mm) were made in the equatorial zone of each fruit and were inoculated with 20 μl of the conidial suspension followed by 20 μl of bacterial and bacterial consortium inoculum. Controls were the same as those used in the in vitro antagonistic assays. All inoculated fruits were placed in closed plastic containers and incubated at room temperature (28°C) for 14 days (Wallace et al., 2017). The antagonistic activity was expressed as the percentage of reduction of fruit rot (PRFR): PRFR (%) = [(D1 ‒ D2)/D1] × 100, where D1 is the rot diameter of apples only with the conidial suspension of P. expansum and D2 is the rot diameter produced by co-inoculation.
Characterization of antifungal bacterial compounds
The bacterial strains that exhibited the highest antagonistic activity were subjected to analysis to determine the bacterial production of compounds with antifungal activity. The lipopeptide production related to the surfactant activity was tested on blood agar plates in triplicate (MCD LAB, Tlalnepantla de Baz, Mexico). A fresh inoculum of each bacteria was streaked on blood agar plates and afterward, it was incubated at 30°C for 3 days. The bacteria that produced clearing zones around colonial growth were considered positive for surfactant activity (Sarwar et al., 2018). To determine the nature of the produced lipopeptides, the bacterial supernatants were analyzed through the technique of the matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) and detected in a range of 900 to 1,600 of mass/charge (m/z) (Rodríguez-Chávez et al., 2019; Yang et al., 2015). Siderophore production was determined qualitatively using the Chrome azurol S (CAS) assay. For this, ten microliters of bacterial suspension (aforementioned details on the preparation) of each isolate were inoculated directly onto CAS medium plates in triplicate. The bacterial inoculum that caused a color change from blue to yellow on plates, after 10 days of incubation (at 28°C), were considered as positive for siderophore production (Schwyn and Neilands, 1987).
Analysis of culturable bacterial density
Bacterial densities were calculated as the colony-forming unit, transformed into logarithms (Log10) per square centimeter of fruit surface (cfu/cm2), and analyzed by the non-parametric Kruskal-Wallis test. The relative abundance percentage of each bacterial species was calculated by the following formula: RA (%) = di/dT × 100; where d is the density (cfu/cm2) of the species “i”, and dT represents the total cfu per square centimeter of the fruit’s surface. The diversity of the bacterial species in each development stage was estimated with the Shannon diversity (H’) index using the BIODIVERSITYR (Kindt and Code, 2005). All analyses were performed using the 3.6.2 version of R software (R Core Team, 2013).
Statistical analysis
Non-metric multidimensional scaling (NMDS), with Bray-Curtis as distance matrix, followed by permutational multivariate analysis of variance (PERMANOVA) (Anderson, 2008), were used to analyze the effect of the development stage of apple fruit in the structure of the complete bacterial populations and bacteria populations with antagonistic capacity. They were run in MASS (Venables and Ripley, 2002) and Vegan (Oksanen et al., 2015) packages. In vitro antagonist assays were replicated four times while in vivo assays were replicated eight times. The obtained data were analyzed with a one-way ANOVA, and the statistical differences were evaluated using a Tukey-Kramer test (0.05 confidence level). The percentages were transformed to angular degrees for the analysis and expressed in terms of the original variable. All statistical analyses were performed using the 3.6.2 version of R software (The R Foundation, 2020).
Results
Changes of bacterial populations during fruit development
The bacterial abundance of the surface of apple fruit showed changes throughout the five developmental stages that were examined (fruit set – ripe fruit). The highest bacterial density per square centimeter of fruit was observed on fruit set (S1) (1.0 × 105 cfu/cm2), followed by green fruit 3 (S4) (9.5 × 10 cfu/cm2), green fruit 2 (S3) (8.3 × 10 cfu/cm2), green fruit 1 (S2) (3.4 × 10 cfu/cm2), and ripe fruit S5 (1.1 × 10 cfu/cm2) (Kruskal-Wallis test, P ≤ 0.01) (Table 1). The calculated diversity values for the five stages increased as the fruit maturation progressed, being the fruit set (S1) the stage with the lowest diversity (H’ = 0.67 ± 0.02) while the highest diversity was found on green fruit 3 (H’ = 1.73 ± 0.19) and ripening fruit stage (H’ = 1.80 ± 0.10) (Table 1). These results have demonstrated that the density of the bacteria decreases but the number of different species (diversity) increases as the apple fruit matures.

Bacterial density (cfu/cm2), diversity (H’), and antagonistic activity estimated for the five different stages of apple development
From the total sampled fruit in the five developmental stages, 828 bacterial isolates were recovered, which were further classified into 25 morphotypes (bacteria isolates with differences in morphological and biochemical traits) (Supplementary Table 1). 16S rRNA analysis allowed their classification into five different genera: Arthrobacter, Bacillus, Curtobacterium, Microbacterium, and Plantibacter. Bacillus, the genera exhibited the highest number of isolates (57.6%), followed by Curtobacterium (20.8%), Microbacterium (13.5%), Anthrobacter (5.5%), and Plantibacter (2.3%). Bacillus was the only genus detected in all developmental stages of fruit and its relative abundance (% RA) increased with the ripening of the fruit, finding its pick in S4 (92.1% RA), followed by S5 (88.4% RA), S3 (38% RA), S2 (35.5%), and the lowest value in S1 (34.1% RA) (Table 1, Fig. 2). PERMANOVA analyses revealed that the bacterial population’s composition was mostly influenced by the development stage of apple fruit (pseudo-F = 3.6, R2 = 0.59, P ≤ 0.001). This factor explained 59% of the total variance, separating the bacterial populations found in stage 4 and 5 from the first three stages of fruit development (stage 1-stage 3) (Fig. 3A).
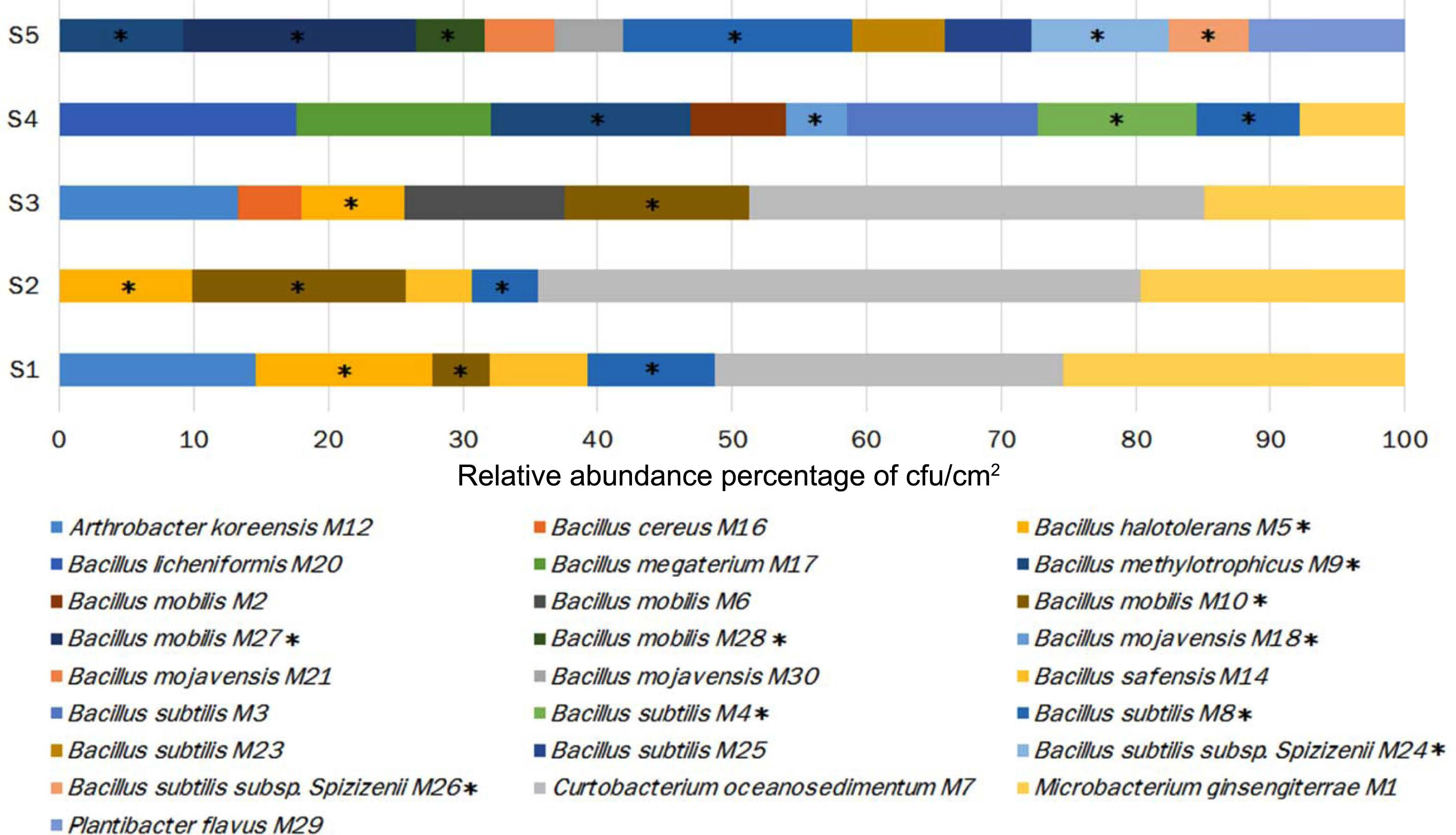
Changes in the distribution of the bacterial species and their relative abundance throughout the development of apple fruit. Bacteria with antagonistic potential against Penicillium expansum exhibited in each stage are shown with an asterisk (*).
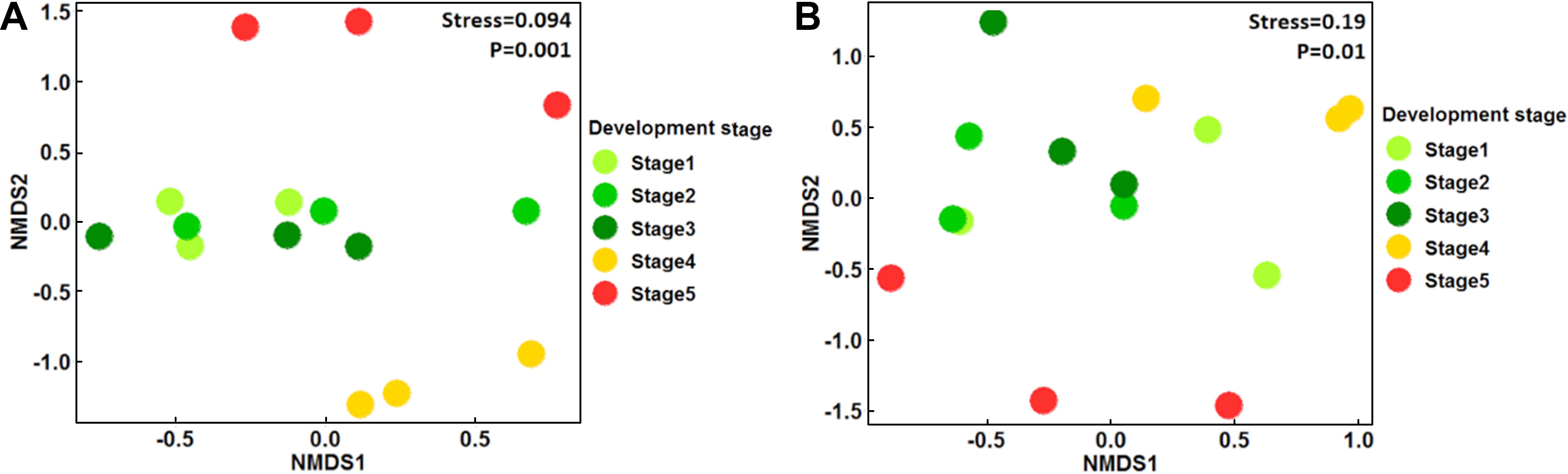
Non-metric multidimensional scaling (NMDS) clustering of total bacterial population (A) and the population of bacteria with antagonistic activity (B), associated in the five stages of fruit apple development. Stage: 1, fruit set; 2, green fruit 1; 3, green fruit 2; 4, green fruit 3; and 5, ripe fruit.
Antagonistic activity of bacterial morphotypes
The results of the in vitro assays revealed differences in the antifungal activity of the bacterial strains against P. expansum. Ten of the 25 bacterial morphotypes (40%) showed the highest PIGR on the three tested culture media (≥30% of PIGR) (Table 2, Supplementary Table 2, Supplementary Figs. 1 and 2), with values ranging from 30.1% to 60.1% of PIGR. These ten bacterial morphotypes belong to the Bacillus genus, where B. methylotrophicus M9 (PDA = 55.4 ± 1.4%, YPD = 53.1 ± 3.5%, and NYDA = 60.1 ± 0.5% of PIRG) and B. subtilis subsp. spizizenii M26 (PDA = 51.6 ± 1.7%, YPD = 48.2 ± 0.8%, and NYDA = 59.0 ± 1.4% of PIRG) showed the strongest inhibitory activity. Both strains inhibited more in the three media than the chemical fungicide (PDA = 46.1 ± 5.5%, YPD = 44.0 ± 5.5%, and NYDA = 44.0 ± 5.5% of PIRG) and equally or greater than the commercial biocontrol agent B. subtilis Q11 (PDA = 35.2 ± 1.9%, YPD = 60.1 ± 1.6%, and NYDA = 55.2 ± 2.2% of PIRG) (Table 2, Fig. 4). In fruit assays, the chemical fungicide showed the highest PRFR, followed by B. subtilis subsp. spizizenii M24 (66.9 ± 6.5% of PRFR), B. mojavensis M18 (49.3 ± 4.7% of PRFR), B. halotolerans M5 (48.0 ± 10.4% of PRFR), the commercial biocontrol agent B. subtilis Q11 (44.7 ± 10.0% of PRFR) and B. methylotrophicus M9 (40.3 ± 3.6% of PRFR) (Table 2, Fig. 5). The relative abundance of the bacterial morphotypes with antagonistic activity increased throughout the fruit development (Table 1, Fig. 2). The highest relative abundance was found during the ripening fruit stage (S5) (six bacterial morphotypes, 47.5% RA), followed by S4 (four bacterial morphotypes, 38.4% RA), S1 (three bacterial morphotypes, 22.6% RA), S2 (three bacterial morphotypes, 14.7% RA), and S3 (two bacterial morphotypes 7.6% RA) (Table 1). PERMANOVA analyses revealed that the composition of the bacterial populations with antagonistic activity was influenced by the stage of apple fruit development (pseudo-F = 1.9, R2 = 0.43, P ≤ 0.01), which explained 43% of the total variance. This pattern was also apparent in NMDS plot, where the bacterial population associated with the ripe fruit (stage 5) was different from those characterized in the other stages of development (Fig. 3B).
Characterization of the antagonistic activity of the Bacillus strains to Penicillium expansum, and compounds related to the antifungal activity
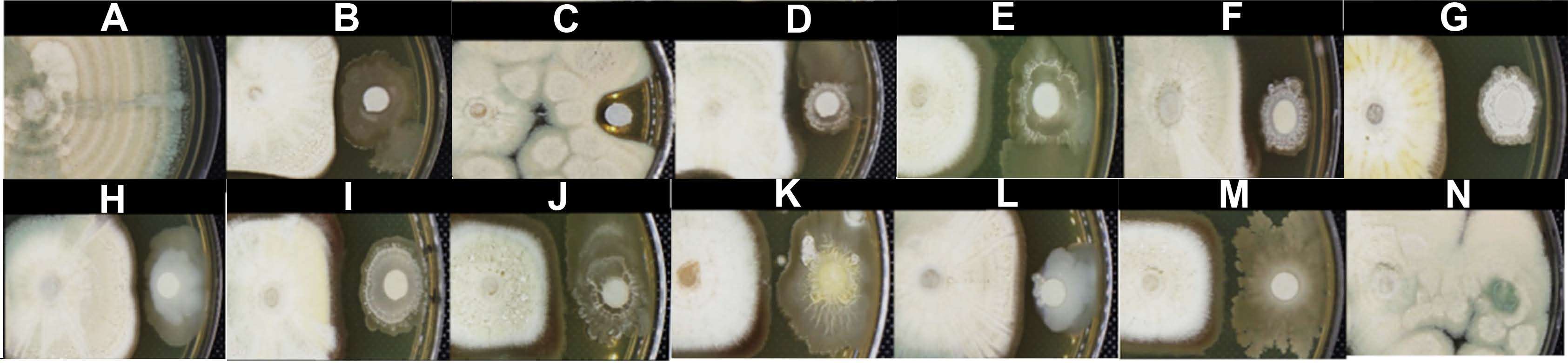
In vitro assay of bacterial strains with the highest antagonist activity against Penicillium expansum in yeast extract peptone dextrose agar. The normal mycelial growth of P. expansum (A). The antifungal activity of the biocontrol agent Bacillus spp. Q11 (B), fungicide nystatin (C), B. subtilis M4 (D), B. halotolerans M5 (E), B. subtilis M8 (F), B. methylotrophicus M9 (G), B. mobilis M10 (H), B. mojavensis M18 (I), B. subtilis subsp. spizizenii M24 (J), B. subtilis subsp. spizizenii M26 (K), B. mobilis M27 (L), B. mobilis M28 (M), and the comparison with the non-antagonist B. mojavensis M30 (N).
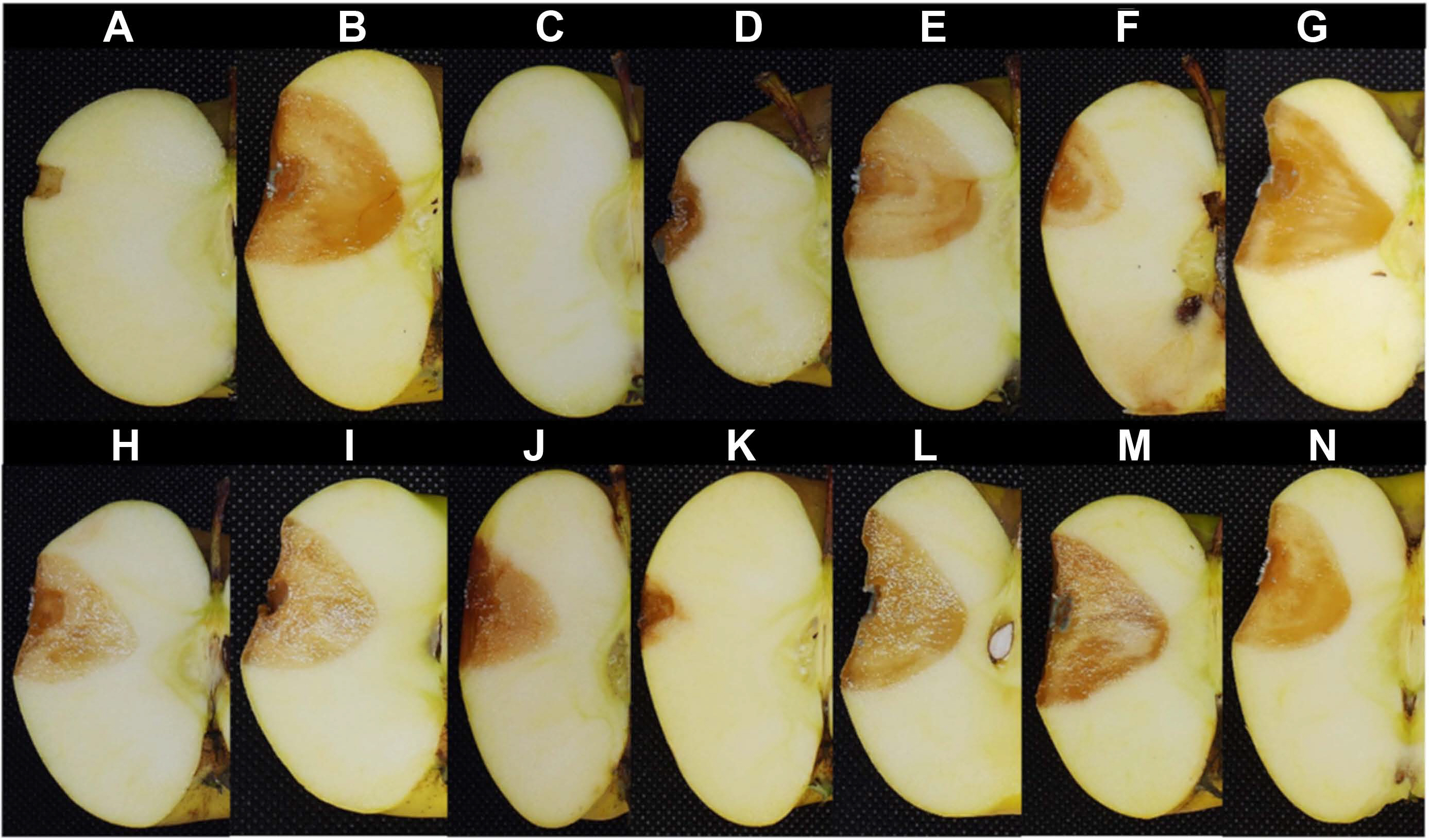
Antagonistic assay in the fruit of bacterial strains with the highest in vitro antagonist activity against Penicillium expansum. Fruit inoculated only with saline solution (A), fruit inoculated with P. expansum without treatment (B), fruit inoculated with P. expansum and fungicide nystatin (C), the biocontrol agent Bacillus spp. Q11 (D), and bacterial strains B. subtilis M4 (E), B. halotolerans M5 (F), B. subtilis M8 (G), B. methylotrophicus M9 (H), B. mobilis M10 (I), B. mojavensis M18 (J), B. subtilis subsp. spizizenii M24 (K), B. subtilis subsp. spizizenii M26 (L), B. mobilis M27 (M), and B. mobilis M28 (N).
In the fruit assays, from the antagonistic activity of the bacterial consortium inoculum formed by the different strains with in vitro antifungal activity found in the fivestage of development, the stage of green fruit 2 (S3 = 26.8 ± 8.6% of PRFR), green fruit 3 (S4 = 23.9 ± 3.6% of PRFR), and ripe fruit (S5 = 27.1 ± 6.6% of PRFR) showed the highest biocontrol activity (Table 1, Supplementary Fig. 3), but it was inferior to those found with individual strains.
Bacterial traits involved in the antagonistic activity
Bacterial strains that inhibited the growth of P. expansum exhibited biochemical properties related to the antifungal activity (Table 2). In CAS medium, 87.5% of them showed the capacity to produce siderophores. On the other hand, all the bacterial strains were positive for the surfactant activity on blood agar. MALDI-TOF-MS analysis of supernatants of strains with the best in vitro antagonistic and surfactant activity showed the production of a mixture of lipopeptides. The lipopeptides produced by each Bacillus strain showed differences in relative abundance (%) and/or mass/charge ratio. The cluster with the most abundant and diverse peaks of the set of ions of mass-to-charge (m/z) was detected in ratios between 1,440 and 1,560 suggesting protonated fengycin isoforms and their alkali metal adducts (Fig. 6, Supplementary Fig. 4). In the fengycin isoforms fraction produced by B. subtilis subsp. spiziennii M24, the isolate with the highest antagonistic activity showed the most intense peaks which were attributed to fengycin B C-17 and their adducts with sodium [M+Na]+. Whereas in B. mojavensis M18, the second-best antagonist, it was found that C-17 fengycin A/B and C-18 fengycin A were the most prominent peaks. In the remaining strains, the C-17 fengycin B showed the greatest abundance for the cases in which the antagonistic activity was similar. Another lipopeptide cluster was detected in ratios between 1,020 and 1,100 suggest the production of iturine-type compounds; the peaks related to this type of lipopeptides were few having a very small abundance.
Discussion
In recent years, the relevance of microbial ecology in the maintenance of agroecosystems has been recognized. The study of the beneficial interactions between bacteria and plant hosts, and the antagonistic interactions between bacteria and phytopathogenic fungi, have shown the important role that bacteria play in the health of the plant and to improve the crop productivity and quality (Fediala Abd El-Gleel Mosa et al., 2018). Almost all bacterial species that improve the health of fruit crops have been isolated from mature fruit and characterized by their ability to control postharvest fruit diseases caused by phytopathogenic fungi (Chen et al., 2016; Li et al., 2013). However, the knowledge about native bacterial populations with antifungal activity during fruit development is scarce.
In our study, bacteria antagonistic to P. expansum were found throughout apple fruit development (Royal Gala var.), this bacterial population was affected by the stage of fruit development whose relative abundance increased during the fruit maturation from 6.2% in a fruit set to 47% in ripe fruit. Additionally, an increase during the ripening of the total morphotypes with antagonistic activity was also noted. This could probably be due to an increase in the glucose and fructose content on the fruit exudates during the development of the fruit which also stimulates the growth of other microorganism groups, including phytopathogenic fungi, as reported by Wrona and Grabowski (2004) on Novamac apples. This could also explain the appearance of fungal infections (i.e., Botrytis) at the final stages of fruit ripening which has also been reported in plumb and nectarine (Fourie and Holz, 1998). The effects of plant exudates on bacterial populations have been widely described on rhizospheric bacteria (Bever et al., 2012), but it was unknown for the bacterial populations associated with the fruit.
The bacterial and fungi populations commonly coexist in all plant organs and possess a synergistic and antagonistic interaction between them (Frey-Klett et al., 2011, Janisiewicz et al., 2014). Owing to this reason, their isolation and characterization from all stages of plant growth are possible. In our results, the antagonistic bacterial population associated with apple fruit was mainly composed of Bacillus species. The predominance of Bacillus species in fruit surfaces of different crops, such as strawberries, grapes, eggplants, and tomatoes, has been documented (Kaur et al., 2017; Leff and Fierer, 2013). This could be due to their ability to form a biofilm, which improves the plant organs’ colonization. Additionally, this genus displays different physiological adaptations to survive in extreme environments, which also include the formation of endospores (Demoz and Korsten, 2006). On the other hand, its predominance over other bacterial genera, as it’s the case in our study, could be related to the same antagonistic activities that it exerts against phytopathogenic fungi to compete for space and nutrients on the plant surface (de Oliveira-Nascimento et al., 2016). Additionally, it has been shown that most epiphytic microorganisms, including the Bacillus genus, offer a horizontal transfer (environment). In our study, we found that some strains, such as B. subtilis M8 were not found in all the development stages. It could be due to the lower density of bacteria in the environment surrounding the apple fruit during some stages of fruit development making them not detectable within the bacterial population (Jackson and Denney, 2011; Shade et al., 2017).
In the present study, Bacillus isolates exhibited different antagonistic activity against P. expansum in vitro, being B. subtilis subsp. spizizenii M24 obtained from the final apple fruit maturation stages, which presented the higher activity. Bacillus subsp. spizizenii has been reported as epiphyte of alfalfa and rice, exhibiting antagonistic activity against Fusarium semitectum (Wen et al., 2015).
Our results have demonstrated that the stage of fruit development could influence the antagonistic bacterial population structure in apples. In some way, the fruit exudates excreted in these last stages could be conditioning for the establishment of the populations with the greater antagonistic activity. Rudrappa et al. (2008) reported that an increase in the secretion of L-malic acid from roots of Arabidopsis thaliana improved the rhizo-colonization of Bacillus subtilis strain FB17, a plant growth-promoting bacteria that induces the mechanisms of resistance to pathogens. L-malic acid is the major organic acid on the mesocarp of ripe apple fruit (Zhang et al., 2010), and this could be secreted through lenticels and micro-cracks, which also increased with the ripening of the fruit (Konarska, 2014; Tessmer et al., 2016).
The application of microbial consortia has been demonstrated to improve the efficacy of biocontrol activity against diverse plant pathogens (Krzyzanowska et al., 2019). These bacteria integrating the consortia must be compatible with each other and possess synergistic interactions to increase the biocontrol capacity against the phytopathogen (Xu and Jeger, 2013). Our results have revealed that the bacterial consortium as a factor combining by the stage of development did not show higher antagonistic activity than that produced by individual bacterial strains. Janisiewicz (1996) reported that the better form for combining the antagonist bacteria against postharvest diseases in apple is selecting the microorganisms that occupy different niches and carbon metabolism to be able to coexist and be more effective biocontrol consortia. Our results suggest that the native bacteria found in the same stage of development (same niche) are not amenable to increasing antagonistic activity either.
The production of antimicrobial metabolites by Bacillus is one of the most reported strategies in the antagonistic activity. In our study, the antagonistic strains to P. expansum produced siderophores and lipopeptides, two of the metabolites most reported in Bacillus strains with antifungal activity (Rodríguez-Chávez et al., 2019; Yu et al., 2011). Lipopeptides comprise mainly three families of non-ribosomally synthesized compounds: surfactin, iturin, and fengycin. This last family of compounds has turned out to be the most active in reported Bacillus species, with antifungal activity against Aspergillus, Fusarium, Pythium (Hanif et al., 2019; Malfanova et al., 2011), Botrytis (Tosco et al., 2015), and Penicilium (Arrebola et al., 2010; Yu et al., 2011) species.
Finally, the production of siderophores found on the strains of this study could be participating during the antagonist interaction. The microbial production of these molecules has been recognized as a strategy aimed at reducing the concentration of iron available in situ, thus limiting growth, establishment, and phytopathogenic activity (Yu et al., 2012). The antagonistic effect of microbial siderophores against P. expansum in apple fruit tests has already been described employing the yeast Metschnikowia pulcherrima (Saravanakumar et al., 2008). This kind of assays employing siderophores produced by Bacillus against P. expansum has not yet been reported.
In conclusion, the abundance and structure bacterial species antagonistic to P. expansum are modified throughout the development of apple fruit, being highest in the maturation stage. These isolates were dominated by Bacillus species producers of lipopeptides and siderophores, antifungal metabolites probably involved in the antagonistic activity. In a wholesome, our study provides the first ecological characterization of antagonistic bacteria associated with different stages of fruit development which could represent a promising vision for the isolation of new populations with biocontrol activity. Besides, data obtained in this work constitute the basis for determining if there are nutrients on the surface of apple fruit that are related to the changes in the observed microbial populations.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
JRPA is thankful to CONACYT for the postdoctoral and Ph.D. fellowships of JLRC and RCLG. The authors also acknowledge the technical support provided by Lucía del Carmen-Márquez and Lucero Ríos. The authors also express gratitude to Dr. Juan Campos-Guillén for the assistance provided at his laboratory.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.


