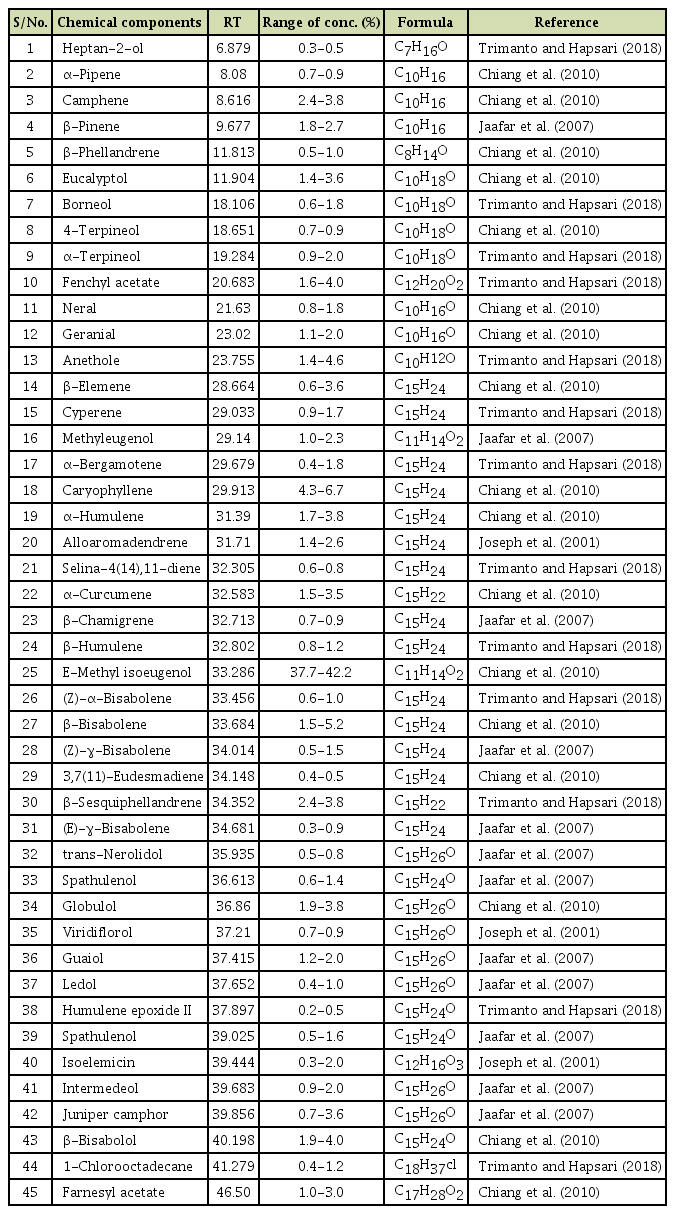
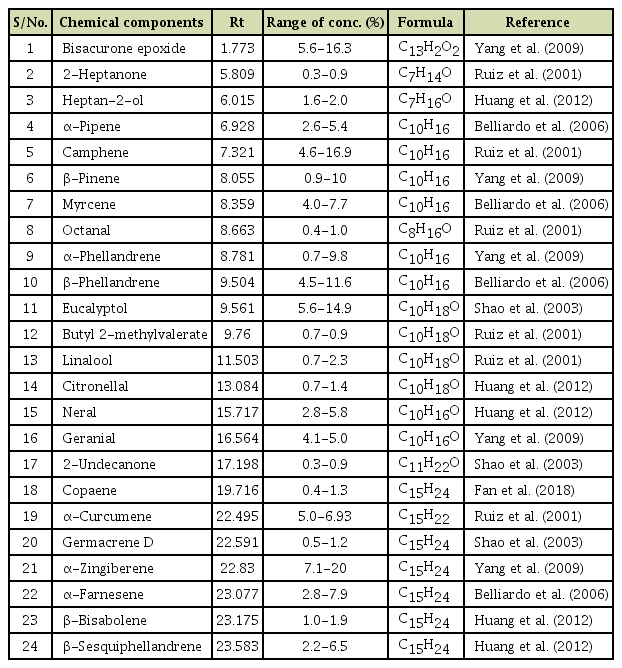
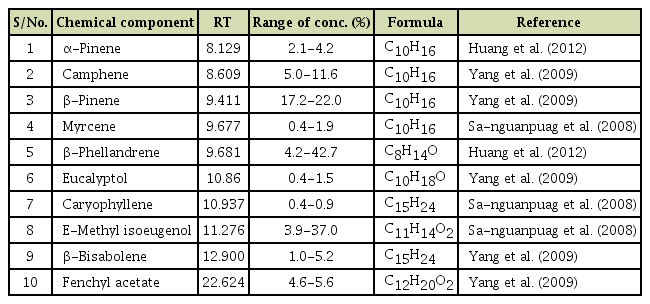
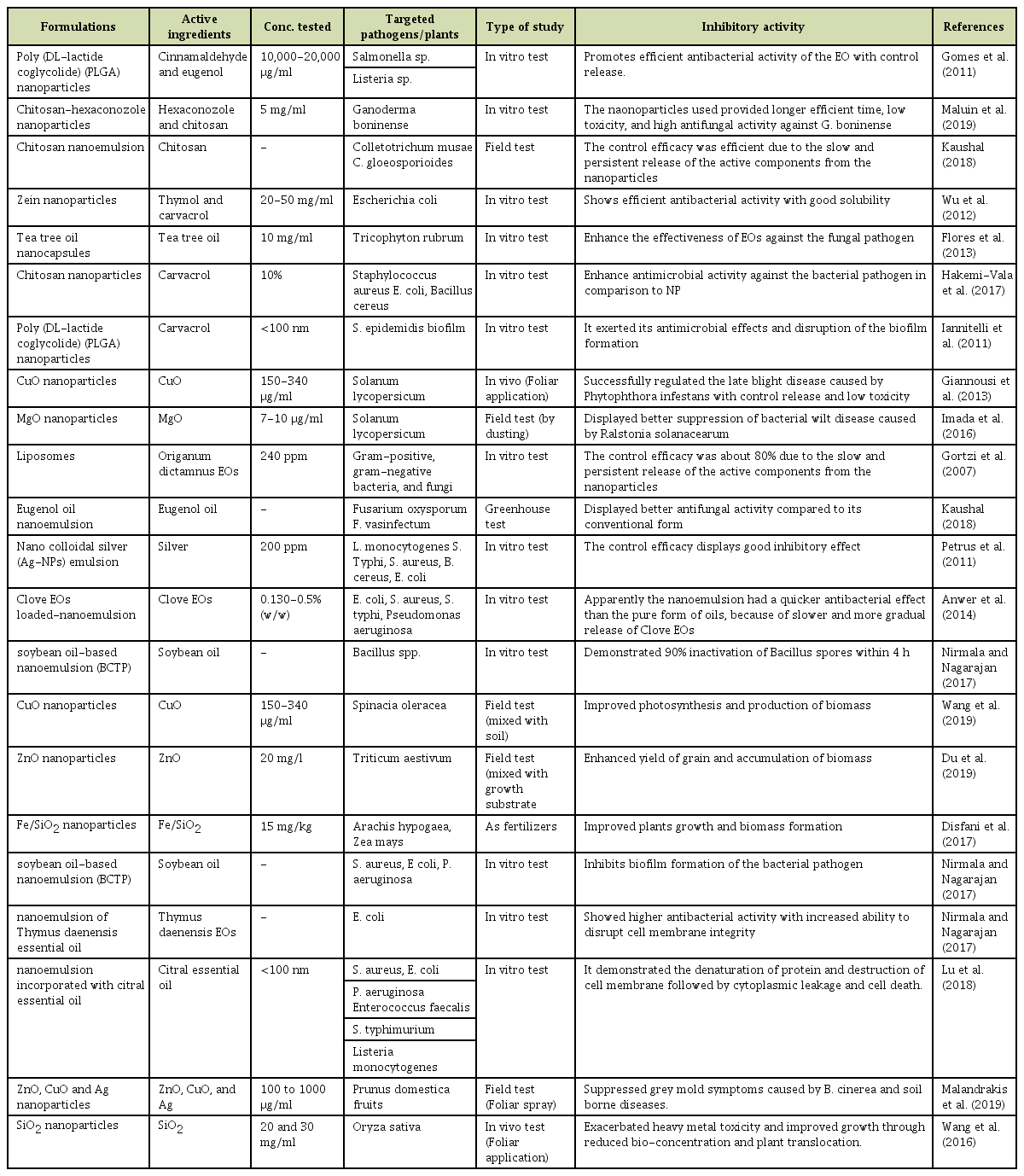
Variability in the Viral Protein Linked to the Genome of Turnip Mosaic Virus Influences Interactions with eIF(iso)4Es in Brassica rapa
Article information
Abstract
Plants protect against viruses through passive and active resistance mechanisms, and in most cases characterized thus far, natural recessive resistance to potyviruses has been mapped to mutations in the eukaryotic initiation factor eIF4E or eIF(iso)4E genes. Five eIF4E copies and three eIF(iso)4E copies were detected in Brassica rapa. The eIF4E and eIF(iso)4E genes could interact with turnip mosaic virus (TuMV) viral protein linked to the genome (VPg) to initiate virus translation. From the yeast two-hybrid system (Y2H) and bimolecular fluorescence complementation (BiFC) assays, the TuMV-CHN2/CHN3 VPgs could not interact with BraA.eIF4E.a/c or BraA.eIF(iso)4E.c, but they could interact with BraA.eIF(iso)4E.a in B. rapa. Further analysis indicated that the amino acid substitution L186F (nt T556C) in TuMV-UK1 VPg was important for the interaction networks between the TuMV VPg and eIF(iso)4E proteins. An interaction model of the BraA. eIF(iso)4E protein with TuMV VPg was constructed to infer the effect of the significant amino acids on the interaction of TuMV VPgs-eIF(iso)4Es, particularly whether the L186F in TuMV-UK1 VPg could change the structure of the TuMV-UK1 VPg protein, which may terminate the interaction of the BraA.eIF(iso)4E and TuMV VPg protein. This study provides new insights into the interactions between plant viruses and translation initiation factors to reveal the working of key amino acids.
Plants fight viruses through two resistance mechanisms, including passive and active resistance regulation. In previous studies, some silenced genes or resistance genes (R genes) could mediate certain dominant active resistance mechanisms. Some R genes have characteristic special domains, such as (CC)-NBS-LRR or NBS-LRR (Li et al., 2018). Generally, potyviruses need to infect host plants, as they depend on host plant factors to encode proteins for replication and translation (Carrington et al., 1996; Wang and Krishnaswamy, 2012). In recent years, a variety of resistance (R) genes have been identified and used to improve brassicaceous crop resistance against turnip mosaic virus (TuMV), which not only to ensure vegetable production, but also to provide approaches for the control of other viruses, such as cucumber mosaic virus (CMV) and tobacco mosaic virus (TMV). Many genes that demonstrate resistance to TuMV have been mapped in brassica crops, such as TuRB01, TuRB02, TuRB03, TuRB04, TuRB05, and ConTR01. Dominant resistance genes control resistance to a narrower spectrum of TuMV isolates than the recessive genes (Li et al., 2016). In other plants, the eukaryotic initiation factor (eIF) genes were also found to be resistant to viruses. There are many types of eIF genes, including eIF4E, eIF4G, eIF(iso)4E, and eIF(iso)4G, which play an important role in recessive resistance genes with broad-spectrum resistance to viral infection (Dinkova et al., 2016).
To date, most mutations of the eIF4E or eIFiso4E genes have been mapped to natural recessive resistance to potyvirus, and the long co-existence of plants and potyviruses has promoted the emergence of virulent isolates that have adapted to the recessive resistance in plants (Sanfaçon, 2015). Many resistance genes have been cloned, especially after the genomes of brassica crops were assembled (BRAD, http://brassicadb.org/brad/). Some genes have been shown to be eIF family genes, and there are various eIF4E family members in brassica crops, such as ConTR01 [eIF(iso)4E.c], retr01/retr02 [eIF(iso)4E.a], and retr03 (eIF2Bβ). The reported susceptibility/resistance associated with eIF4E or eIF(iso)4E is mostly based on the interaction between these proteins and the genome-linked viral protein (VPg) covalently attached at the 5′ end of the viral-positive RNA strand (Michon et al., 2006). However, the phenomenon of eIF4E-VPg interaction does not provide sufficient evidence to explain the observed results in TMV-eIF4E (Roudet-Tavert et al., 2007). Therefore, additional hosts and virus determinants may be required to determine the interaction between TuMV VPg and eIF genes, as well as their functions during TuMV infection. Several functions have been summarized for the interaction of eIFs-TuMV VPgs during viral infection. First, TuMV VPg imitates the 5′ cap of cellular mRNAs, which may be important for viral RNA translation and might accelerate protein translation through the interaction with eIF genes (Contreras-Paredes et al., 2013). Second, the interactions between eIF4E and virus VPgs might promote viral replication and facilitate virus movement to other tissues to result in the infection of the entire plant (Wang and Krishnaswamy, 2012). Third, during the viral infection cycle, the interactions of virus VPgs and eIFs might boost viral RNA translation through restraining entire or specific cellular translation (Léonard et al., 2000).
In Brassica rapa, three genes, namely, retr01, retr02, and trs, are associated with eIF(iso)4E genes for recessive resistance (Kim et al., 2014; Qian et al., 2013; Rusholme et al., 2007). Sequencing with a sequence-based genetic map of B. rapa confirmed that retr02 is retr01 rather than a different allele. Nellist et al. (2014) reported retr01/retr02 broad-spectrum resistance to TuMV due to a natural mechanism based on the mis-splicing of the eIF(iso)4E allele in B. rapa. Kim et al. (2014) proved that there were some key variations in the B. rapa eIF(iso)4E protein for the interactions of TuMV VPg and eIF(iso)4E using a yeast two-hybrid (Y2H) system. In addition, the TuMVC4/CDN1/UK1 VPg could not interact with eIF4E, but only with the eIF(iso)4E of B. rapa based on Y2H and bimolecular fluorescence complementation (BiFC) assays (Li et al., 2018). Additionally, some single nucleotide polymorphisms (SNPs) were identified that may influence the interaction between eIF(iso)4E and VPg, such as the SNP T106C in BraA.eIF(iso)4E.c and the SNP A154C in VPg (Li et al., 2017, 2018). In addition to the recessive genes retr01 and retr02, the other recessive gene retr03 was mapped and cloned in Brassica juncea and encodes the eukaryotic translation initiation factor 2B-beta (eIF2Bβ), representing a new class of virus resistance genes conferring resistance to pathogens (Shopan et al., 2017).
Some amino acids have a significant influence on the interactions between TuMV VPg and eIF(iso)4E in B. rapa. A Y2H assay and co-immunoprecipitation analysis demonstrated that the W95L, K150L, and W95L/K150E amino acid mutations in B. rapa eIF(iso)4E proteins interrupted their interactions with TuMV VPg (Kim et al., 2014). Meanwhile, TuMV VPg could interact with eIF(iso)4E, but not with eIF4E, in B. rapa, and some SNPs were identified that may affect the interactions between eIF(iso)4E and VPg, such as the SNP T106C in BraA.eIF(iso)4E.c and the SNP A154C in VPg (Li et al., 2018). Based on the findings of a previous study, the interactions between TuMV-CHN2/CHN3 isolates and eIF4E/eIF(iso)4E were identified in this study, and some key amino acids were observed. In addition, an interaction model was constructed to infer the key amino acids influencing the structure of protein complexes. This study may be helpful for mapping new eIF(iso)4E resistance loci and further revealing the resistance mechanisms of TuMV to eIF(iso)4E in B. rapa.
Materials and Methods
Plant materials and TuMV isolates
In this study, eight highly-inbred B. rapa accessions were used, including ‘80186’, ‘Chiifu’, ‘80122’, ‘2079’, ‘80425’, ‘BP058’, ‘80124’, and ‘R-o-18’. The eIF(iso)4E and eIF4E gene sequences from the eight lines above were cloned and deposited in the NCBI database (MH614206-MH614238). The three representative pathotypes included the TuMV isolates CHN2 (GenBank: BAD20366) and CHN3 (Gen-Bank: BAD20367) from China, and UK1 (GenBank: NC_002509) from the UK.
Y2H.
A Gal4-based Y2H system was used to conduct the protein interaction assays according to the manufacturer’s instructions. Plasmid vectors and yeast strains AH109 were obtained from Clontech (Mountain View, CA, USA). The bait plasmid pGBKT7 could combine the TuMV VPg with the DNA-binding domain. The prey plasmid pGADT7 could express the eIF family genes. The restriction enzyme sites were designed based on the gene-specific primers. The forward/reverse primers (Bio120213 [NdeI site] and Bio120214 [XmaI site]) were used to amplify the VPg sequences from the TuMV-CHN2/CHN3 genome. BraA.eIF4E and BraA.eIF(iso)4E from eight B. rapa lines were amplified as described in a previous study (Li et al., 2018). The forward/reverse primers (Bio120582 [EcoRI site] and Bio120583 [XhoI site]) were used to amplify the eIF(iso)4E sequences from Arabidopsis Col-0, which was used as a positive control in the Y2H assays. XhoI/EcoRI and XmaI/NdeI were used to digest the amplified fragments and construct the pGADT7 and pGBKT7 recombination vectors, respectively. All recombination vectors were confirmed by PCR and sequencing.
Based on the manufacturer’s protocols, the Matchmaker GAL4 Y2H system was used to create the pGBKT7:TuMV VPg and pGADT7:eIF4E constructs, which were used to transform the recombination plasmids. After yeast transformation, selective media lacking histidine, adenine, leucine, and tryptophan were used to screen the colonies. Plates were incubated at 30°C and were assessed after 3-5 days. Each assay had three repeats. In the assays, two empty vectors were used as negative controls, and the interaction relationship between the SV40 large T antigen and murine p53 was used as a positive control. In addition, the interactions of Arabidopsis eIF(iso)4E (LSP) were also used as a positive control.
BiFC
BiFC was performed as described in a previous study (Li et al., 2018). The primers containing two enzyme sites (XhoI and BamHI) were used to amplify the eIF family gene, and the eIF family genes PCR products and the pSPYNE empty vector were recombined into the eIF(iso)4E-pSPYNE recombinant vector using BamHI/XhoI enzymes and T4 ligase. Similarly, using specific primers containing two enzyme sites (ClaI and XhoI), the TuMV VPg genes were amplified, and the pSPYCE empty vector and TuMV VPg gene PCR products were digested using XhoI and ClaI. Using T4 DNA ligase, we constructed the recombinant vector TuMV VPg-pSPYCE. The PCR test and sequencing were used to confirm all the recombinant vectors. Each assay was performed three times. The combination of Arabidopsis eIF(iso)4E (LSP) and TuMV-VPg was the positive control, and the YCE-empty and YNE-empty vectors were the negative controls. Each empty vector was used as a blank control.
The B. rapa protoplasts were extracted as described in a previous study (Li et al., 2018). Fresh leaves at the four-leaf stage were obtained from Chinese cabbage plants. The reagents for the assays included 0.4% macerozyme R10, 1.5% cellulase R10, 20 mM KCl, 0.4 M D-mannitol, 10 mM CaCl2, 20 mM MES (pH 5.7), 5 mM β-mercaptoethanol, and 0.1% bovine serum albumin, and the assays were conducted according to the manufacturer’s instructions. The recombinant plasmids (TuMV VPg-pSPYCE, eIF(iso)4EpSPYNE, YCE-empty, and YNE-empty, constructed for the BiFC assays) were transformed into the Chinese cabbage protoplasts. The Chinese cabbage protoplasts were cultured in the dark firstly, and after 24 h, a laser confocal scanning microscope was used to scan the fluorescence signals and assess the transformation effects. Fluorophores were detected through the ZOE Fluorescent Cell Imaging System (Li et al., 2018).
Results
Identification of eIF4E and its isoform eIF(iso)4E in B. rapa
eIF4E and eIF(iso)4E share the same functional domain, IF4E, while eIF4G and eIF(iso)4G contain the same functional domain, MIFG (or MIF4G_like). nCBP, the novel cap-binding protein (currently termed 4EHP), has both functional domains, IF4E and MIFG. From the BRAD and TAIR database, three eIF4E genes (AT1G29550, AT1G29590, and AT4G18040), one eIF(iso)4E gene (AT5G35620), and one nCBP gene (AT5G18110) were identified in Arabidopsis thaliana. In B. rapa, five eIF4E (Bra030147, Bra032325, Bra013283, Bra012622, and Bra021026), three eIF(iso)4E (Bra035531, Bra039484, and Bra035393), and three nCBP (Bra002134, Bra006439, and Bra023664) genes were identified using BLASTN and BLASTP from the genomic sequence. A comparative analysis of homology between A. thaliana and B. rapa eIF genes is shown in Table 1, and the homologous gene similarities between A. thaliana and B. rapa were greater than 85%. Genome-wide association studies of the diploid B. rapa have indicated that B. rapa evolved from a hexaploid ancestor (Wang et al., 2011). Our results are consistent with this hypothesis. Compared to the eIF4E genes identified by Jenner et al. (2010), two additional eIF4E genes and three nCBP genes were identified in this study.
B. rapa eIF4E genes non-interacting with TuMV CHN2/CHN3 VPg
TuMV isolates were classified into 12 pa thotypes as determined in the B. napus lines. TuMV CHN2/3/4 belongs to pathotype 3, the UK1 isolate belongs to pathotype 1, and CDN1 belongs to pathotype 4. In our previous study, TuMV-CHN4/UK1/CDN1 VPg could not interact with eIF4E, and we inferred that the same pathotype TuMV isolate exhibited the same interaction relationships with eIF4E. To further assess the interactions between TuMV and eIF4E in B. rapa, the TuMV-CHN2/CHN3 isolates (pathotype 3) were used in the interaction analysis with eIF4E/eIF(iso)4E. There were two variant BraA.eIF4E.a alleles detected in B. rapa, i.e., BraA.eIF4E. a-1 and BraA.eIF4E.a-2, while four variant BraA.eIF4E. c alleles were detected, namely, BraA.eIF4E.c-1 to BraA. eIF4E.c-4. Two methods (Y2H and BiFC) were used to detect the interactions between BraA.eIF4E/BraA.eIF(iso)4E and TuMV-CHN2/CHN3 VPgs.
The Y2H assays suggested the Arabidopsis eIF(iso)4E LSP could interact with the TuMV-CHN2/CHN3 VPgs, which confirmed that the LSP gene was susceptible to TuMV, whereas BraA.eIF4E.c-1 -BraA.eIF4E.c-4, BraA. eIF4E.a-1, and BraA.eIF4E.a-2 could not interact with the TuMV-CHN2/CHN3 VPgs, which indicated that the eIF4E alleles in B. rapa were resistant to TuMV-CHN2/CHN3 (Fig. 1A). In addition, B. rapa protoplast cells were used in the BiFC assays to verify the interactions, and the results were consistent with those obtained by the Y2H assays (Fig. 1B). The blank controls (the empty vector pGADT7 and the empty vector pGBKT7) could not interact, which confirmed that the experimental processes were reliable.
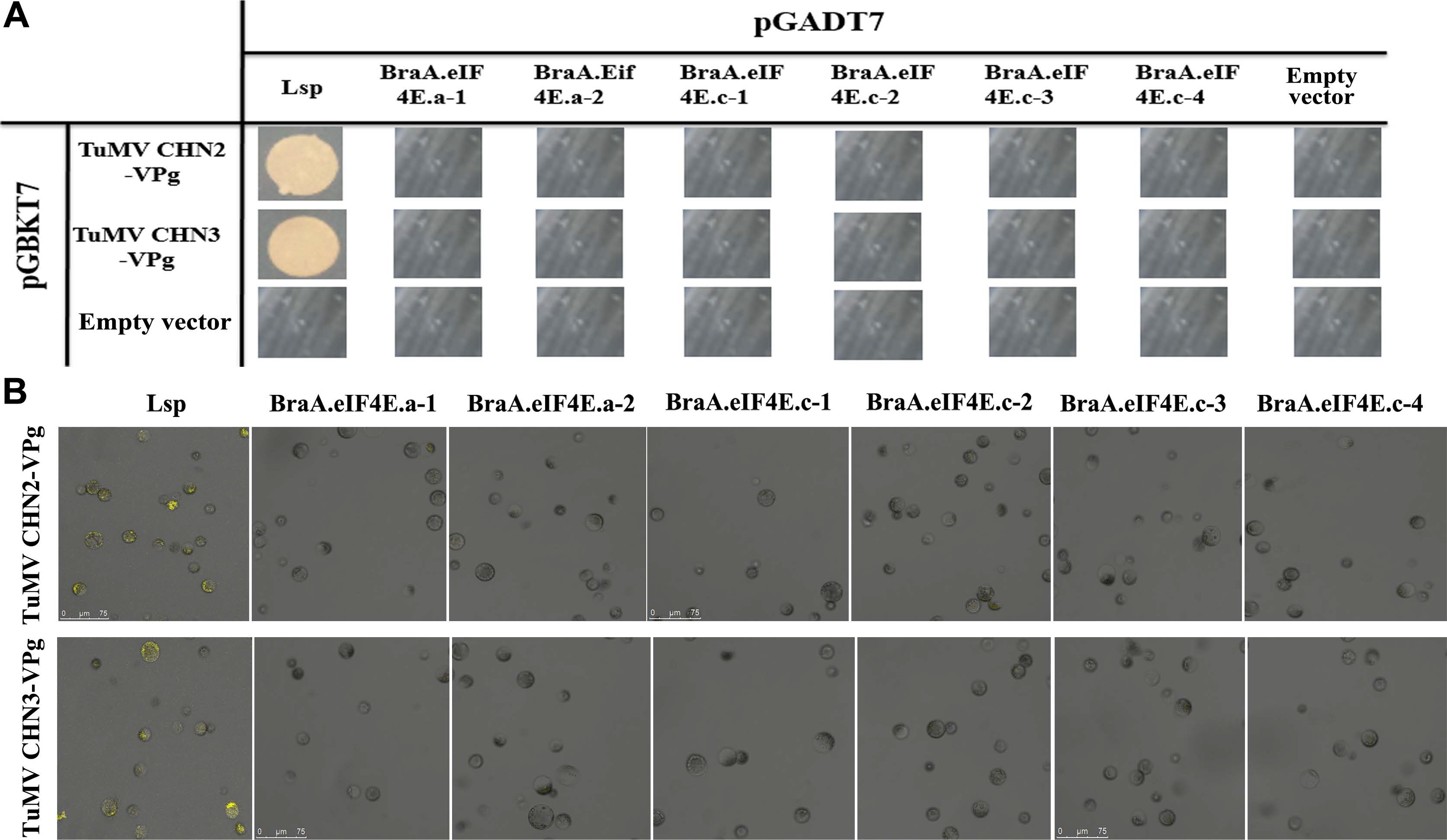
TuMV-CHN2/CHN3 VPgs do not interact with BraA.eIF4E.a or BraA.eIF4E.c. (A) The interaction was confirmed by Y2H assays. Negative control: the empty vectors pGADT7 and pGBKT7; positive controls: TuMV-VPg and Arabidopsis eIF(iso)4E (LSP); assay controls: each partner and empty vector. (B) The interactions were confirmed by BiFC. Positive controls: TuMV-VPg and Arabidopsis eIF(iso)4E (LSP); negative controls: YNE-empty and YCE-empty vectors; assay controls: each partner and empty vectors (data not shown). TuMV, turnip mosaic virus; VPg, viral protein linked to the genome; Y2H, yeast two-hybrid; BiFC, bimolecular fluorescence complementation.
TuMV-CHN2/CHN3 isolates interacting with different eIF(iso)4Es in B. rapa
There were four variant BraA. eIF(iso)4E.a alleles in the eight resistant/susceptible B. rapa lines, namely, BraA.eIF(iso)4E.a-1 to BraA.eIF(iso)4E. a-4, and there were two variant BraA.eIF(iso)4E.c alleles, i.e., BraA.eIF(iso)4E.c-1 and BraA.eIF(iso)4E.c-2. The eIF(iso)4E genotypes were used to verify the interaction relationship with TuMV-CHN2/CHN3 VPgs.
The Y2H and BiFC analyses suggested that TuMVCHN2/CHN3 VPg could not interact with BraA.eIF(iso)4E. a-1, BraA.eIF(iso)4E.a-2, BraA.eIF(iso)4E.c-1, or BraA. eIF(iso)4E.c-2, suggesting that the four above alleles were resistant to TuMV-CHN2/CHN3, whereas TuMV-CHN2/CHN3 VPg could interact with BraA.eIF(iso)4E.a-3 and BraA.eIF(iso)4E.a-4, which revealed that the two alleles were susceptible to TuMV-CHN2/CHN3 in B. rapa (Fig. 2A and B). In a previous study, TuMV-CHN4 obtained the consistent interaction results with TuMV-CHN2/CHN3.
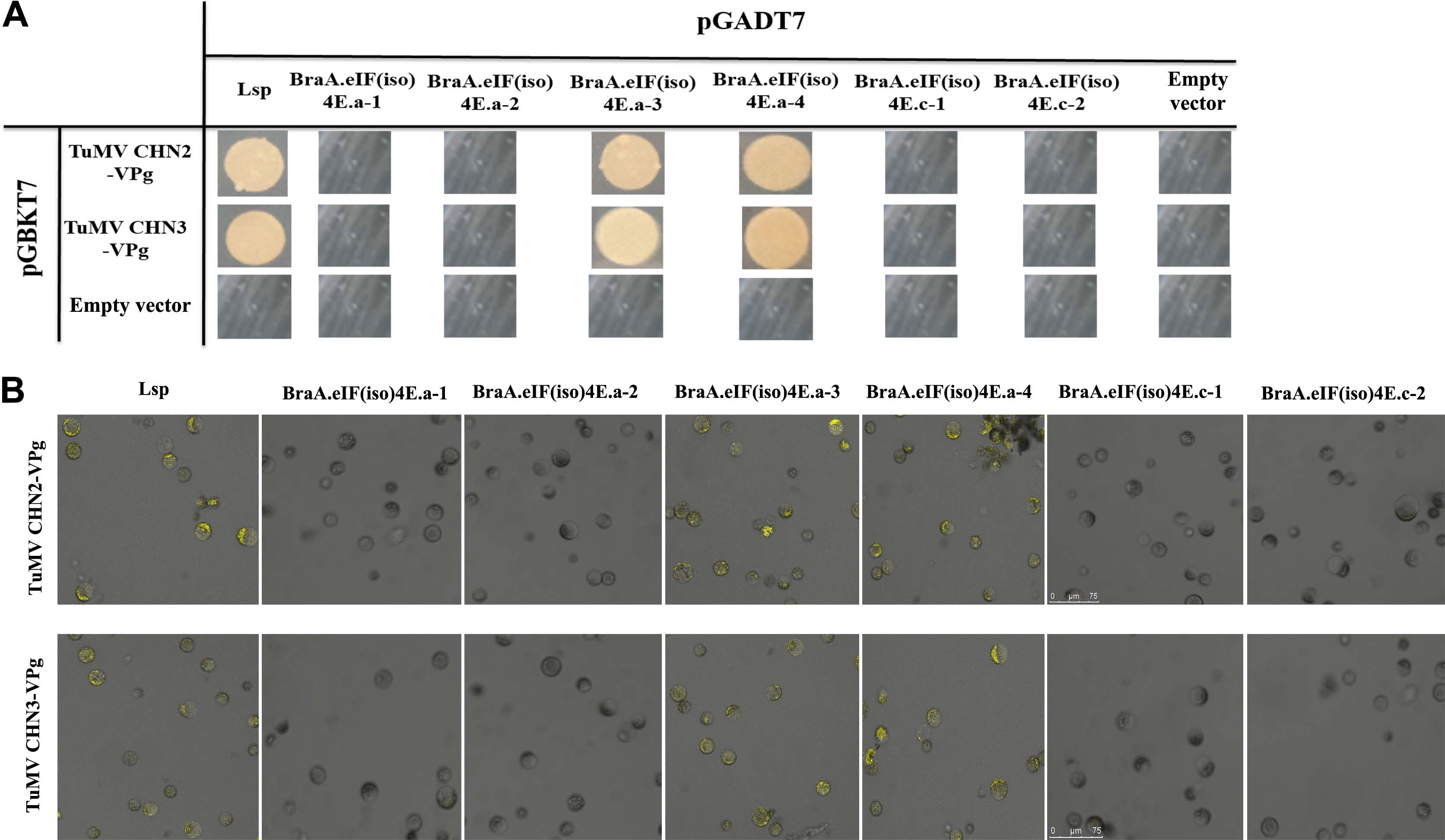
TuMV-CHN2/CHN3 VPgs interacts with BraA.eIF(iso)4E.a, but not with BraA.eIF(iso)4E.c. (A) The results are from the Y2H. Negative control: the empty vectors pGADT7 and pGBKT7; positive controls: TuMV-VPg and Arabidopsis eIF(iso)4E (LSP); assay controls: each partner and empty vector. (B) Verification of the results using BiFC. Positive controls: TuMV-VPg and Arabidopsis eIF(iso)4E (LSP); negative controls: YNE-empty and YCE-empty vectors; each partner and empty vector were used as controls (data not shown). TuMV, turnip mosaic virus; VPg, viral protein linked to the genome; Y2H, yeast two-hybrid; BiFC, bimolecular fluorescence complementation.
Furthermore, the TuMV-CHN2/CHN3/CHN4 isolates all belong to pathotype 3, which may indicate that the TuMV pathotype 3 isolates could not interact with BraA. eIF(iso)4E.c, but could interact with BraA.eIF(iso)4E.a in B. rapa. This suggested that the eIF(iso)4E.c gene in B. rapa was resistant to TuMV-CHN2/CHN3/CHN4, and the eIF(iso)4E.a gene in B. rapa was susceptible to the three TuMV isolates (TuMV pathotype 3).
Specific SNPs affecting the interaction between TuMV VPg and eIF(iso)4E
Some amino acid changes may play important roles in strain-specific interactions between eIF(iso)4E and TuMV VPg. The Y2H and BiFC analyses both indicated that BraA.eIF(iso)4E.a could interact with TuMV-CHN2/CHN3 VPgs, but not with TuMVUK1 VPg. Between TuMV-CHN2 VPg and TuMV-UK1 VPg, five differing bases (nt C154A, A313G, A323G, T556C, and T562C) were identified, which were predicted to result in five amino acid changes (I52L, D105N, G108E, L186F, and P188S) (Table 2). Therefore, primers were designed based on the five locus sequences, and site-directed mutagenesis (using TuMV-UK1 VPg as a template) was successfully implemented through overlap-extension PCR to assess which SNP was important for the TuMV-eIF interactions. The Y2H and BiFC methods indicated that the amino acid change L186F (nt T556C) in TuMV VPg was significant in the interaction between TuMV-CHN2 VPg and BraA. eIF(iso)4E. This indicated that the SNP site could influence infection in TuMV VPg, while the amino acids I52L (nt C154A), D105N (nt A313G), G108E (nt A323G), and P188S (nt T562C) had little influence, which suggested that these four sites did not influence infection in TuMV VPg (Fig. 3A and B). In our previous study, the interaction between TuMV-CHN2 VPg and BraA.eIF(iso)4E.a-3 was found to be regulated by the amino acid changes I52L, E97K, and N105D of the TuMV VPg protein, which indicated that some special amino acids in TuMV-CHN2 VPg were important for these interactions.
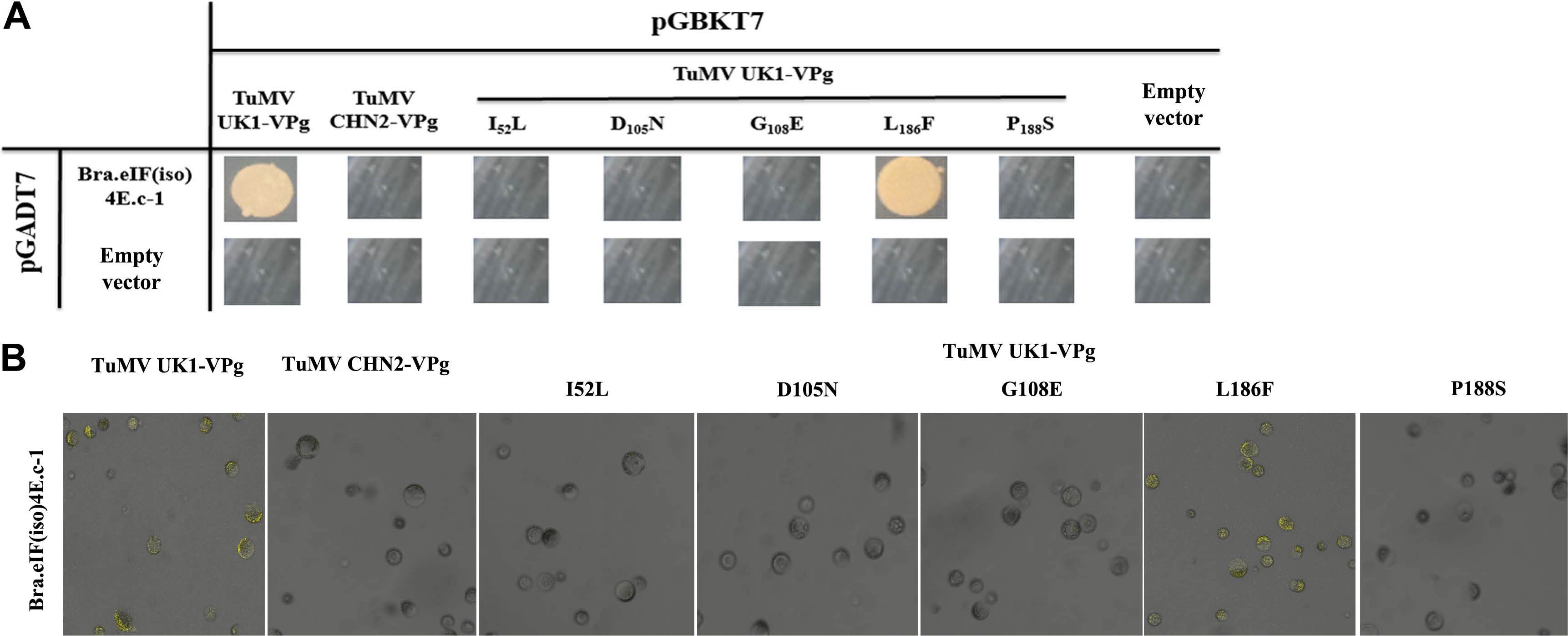
Specific SNPs affect the interaction between eIF(iso)4E and TuMV VPg. Variations between TuMVUK1/CHN2 VPgs could affect the interaction as indicated by Y2H (A) and by BiFC (B). Y2H: negative control, the empty vectors pGADT7 and pGBKT7; positive controls, TuMV-VPg and Arabidopsis eIF(iso)4E (LSP); assay controls, each partner and empty vector. BiFC: positive controls, TuMV-VPg and Arabidopsis eIF(iso)4E (LSP); negative controls, YNE-empty and YCE-empty vectors; assay controls, each partner and empty vectors (data not shown). SNP, single nucleotide polymorphism; TuMV, turnip mosaic virus; VPg, viral protein linked to the genome; BiFC, bimolecular fluorescence complementation; Y2H, yeast two-hybrid.
Constructing the interaction model of TuMV VPgeIF(iso)4E
In plants, the cap structure at the mRNA 5′ end can be recognized by the cap-binding protein eIF(iso)4E during translation initiation. However, interactions with eIF(iso)4G can enhance this to accelerate mRNA translation. When TuMV infects a host plant, its VPg acts like a cap and should interact with eIF4E or eIF(iso)4E to initiate translation. Some SNPs have been identified that may have affected the interaction between eIF(iso)4E and VPg, such as the SNP T106C in BraA.eIF(iso)4E.c and the SNP A154C in VPg (Li et al., 2018). In this study, we also discovered that the amino acid change L186F (nt T556C) in TuMV-UK1 VPg played an important role in the interaction between BraA.eIF(iso)4E and TuMV-UK1 VPg. Thus, the key amino acid variation could transform the three-dimensional structure of the eIF(iso)4E or TuMV VPg proteins, which could influence the interaction between TuMV and eIF(iso)4E (Tables 3-6). The wild-types of eIF(iso)4E and TuMV VPg could interact, which would be useful for reserving TuMV in the host plant; however, the amino acid variation in TuMV VPg (L186F) could alter the structure of the TuMV VPg protein, which would result in no interaction between eIF(iso)4E and TuMV VPg. Similarly, the amino acid variation in eIF(iso)4E (F36L) could also alter the structure of the eIF(iso)4E protein, which would result in no interaction between eIF(iso)4E and TuMV VPg, i.e., the virus could not proliferate and transmit in B. rapa plants.
Discussion
The Potyviridae family, which includes many economically important pathogens of cultivated crops especially members of the genus Potyvirus, is one of the largest families of plant viruses in the world (Gibbs and Ohshima, 2010). Crop species have co-evolved with potyviruses, resulting in the development of recessive resistance. To date, mutations in the eIF4E or eIFiso4E genes have been mapped for natural recessive resistance to potyviruses (Sanfaçon, 2015). Sixteen genes resistant to potyviruses have been detected based on eIF4Es, such as pvr1/pvr12 in Capsicum spp. (Arroyo et al., 1996; Kang et al., 2005); mo1 in Lactuca sativa (German-Retana et al., 2008; Nicaise et al., 2003); sbm1/wlv/cyv2 in Pisum sativum (Gao et al., 2004); bc-3 in Phaseolus vulgaris (Naderpour et al., 2010); pot-1 in Solanum habrochaites (Ruffel et al., 2005); rym4/rym5 in Hordeum vulgare (Kanyuka et al., 2005); SNP-mapping in Citrullus lanatus (Ling et al., 2009); several quantitative trait loci in Prunus armeniaca (Marandel et al., 2009); cum1-1 in A. thaliana (Sato et al., 2005); eIF4E RNAi in Cucumis melo (Rodríguez-Hernández et al., 2012); Sle-IF4E1-G1485A (Piron et al., 2010), eIF4E-1 and eIF4E-2 (Mazier et al., 2011) in Solanum lycopersicum; and pvr12 (Cavatorta et al., 2011), eIF4E (Cavatorta et al., 2011), and Eva1 (Duan et al., 2012) in Solanum tuberosum. In addition, seven resistance genes to potyviruses were detected based on eIF(iso)4Es, such as retr01 (Rusholme et al., 2007), retr02 (Qian et al., 2013), and trs (Kim et al., 2013), overexpression mutagenesis (Kim et al., 2014) in B. rapa; EMS mutagenesis (Lellis et al., 2002), Spm (Jenner et al., 2010) in A. thaliana, and RNAi mutagenesis in Prunus domestica (Wang et al., 2013).
TuMV VPg, and eIF4E/eIF(iso)4E have co-evolved in plants. The eIF4E and eIF(iso)4E proteins shared 50% amino acid sequence identity, and interestingly, eIF(iso)4E and eIF4E were functionally redundant following knockout of the eIF(iso)4E gene, which was compensated for by the increased expression of eIF4E (Sanfaçon, 2015). There are several copies of eIF4E and eIF(iso)4E in plants, and so if eIF4E or eIF(iso)4E mutate, the other copies can compensate their function to interact with TuMV. Additionally, there are at least 124 isolates of TuMVs (Jenner and Walsh, 1996) that could threaten all Cruciferae vegetables, including horticultural brassica crops (calabrese, cauliflower, broccoli, brussels sprout, cabbage, swede, kale, kohlrabi, and Chinese cabbage), arable brassica crops (oilseed rape, B. napus, and B. rapa), edible horticultural non-brassica crops (peas, watercress, radish, rhubarb, chicory, spinach, chrysanthemum, tobacco, and lettuce), ornamentals (Abutilon, stocks, and wallflowers), and 14 weed plants from various families (Jenner and Walsh, 1996; Provvidenti, 1982; Shattuck, 1992). If one TuMV isolate mutates, the other isolates substitute it to infect plants.
Some amino acid positions that are relevant for potyvirus resistance have been analyzed. The G107R mutation in the eIF(iso)4E protein 3D ribbon, which could affect both VPg and cap binding, is associated with virus resistance in pea, tomato, and pepper, while L79R, which is located in an external loop, could affect VPg but not cap binding (Kang et al., 2005; Nicaise et al., 2003; Ruffel et al., 2005). Some amino acid changes in eIF genes located in the central domain could be associated with eIF4E-mediated resistance (Ruffel et al., 2005, 2007). Twenty-two eIF4E sequences have been obtained from monocot and dicot plant species, which suggests that there are four SNP positions for natural and positive selection (Sanfaçon, 2015). Furthermore, the eIF4E/eIF(iso)4E protein 3D ribbon was constructed based on the wheat eIF4E protein structure (Li et al., 2018). Therefore, the assessment of eIF family gene resistance spectra and genetic backgrounds would be helpful for eIF4E-mediated resistance mechanism analysis (Dinkova et al., 2016; Li et al., 2019).
In our previous study, we used TuMV-C4 VPg as a template and found that the amino acid change I52L was important for the interaction between TuMV VPg and eIF(iso)4E (Li et al., 2018). However, in this study, the TuMV-UK1 VPg was used as a template, and the amino acid substitution I52L was non-essential for the interaction between TuMV-CHN2/CHN3 VPgs and eIF(iso)4E, which indicated that it may not be just one amino acid that could influence the interaction between TuMV VPg and eIF(iso)4E, but may rather be several loci working together, with one or two sites playing a key role. Of course, we obtained sitedirected mutagenesis using different isolates as the templates (TuMV-UK1/C4 VPg), which have great differences in genetic background.
Natural recessive virus resistance has provided new insights into the interactions between translation factors and plant viruses (Sanfaçon, 2015). In addition, the systematic identification of the interactions between translation factors and viral RNAs or proteins has established new potential targets for antiviral resistance. With the development of plant genome sequencing, our understanding of eIF family gene evolution, as well as other translation factors in plants, has expanded. However, it is unclear whether viruses possess special functions in regulating the interactions between plants and viruses (Dinkova et al., 2016). For instance, when the eIF family genes knocked-out or subjected to other abiotic stresses such as drought, salinity, and so forth, how do plants survive? Genetics, biotechnology, bioinformatics, and biochemistry approaches should be combined to address these challenges in agriculturally relevant species.
Acknowledgments
This work was funded by State Key Laboratory of North China Crop Improvement and Regulation, and the Natural Science Foundation of Beijing (6212030), and the National Natural Science Foundation of China (31772302). This work was performed at the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, Beijing, China.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.