 |
 |
| Plant Pathol J > Volume 37(2); 2021 > Article |
|
Abstract
To establish an infection, fungal pathogens must recognize diverse signals from host surfaces. The rice blast fungus, Magnaporthe oryzae, is one of the best models studying host-pathogen interactions. This fungus recognizes physical or chemical signals from the host surfaces and initiates the development of an infection structure called appressorium. Here, we found that protein MoAfo1(appressorium formation, MGG_10422) was involved in sensing signal molecules such as cutin monomers and long chain primary alcohols required for appressorium formation. The knockout mutant (ΔMoafo1) formed a few abnormal appressoria on the onion and rice sheath surfaces. However, it produced normal appressoria on the surface of rice leaves. MoAfo1 localized to the membranes of the cytoplasm and vacuole-like organelles in conidia and appressoria. Additionally, the ΔMoafo1 mutant showed defects in appressorium morphology, appressorium penetration, invasive growth, and pathogenicity. These multiple defects might be partially due to failure to respond properly to oxidative stress. These findings broaden our understanding of the fungal mechanisms at play in the recognition of the host surface during rice blast infection.
Plant pathogens encounter a wide variety of surfaces during their life cycle, including soil particles, water, and nonhost plants; none of which favors pathogens’ infection. Pathogens sense several signals from the host plant and need to respond timely for successful disease establishment to occur. Therefore, identifying sensed host signals is crucial for pathogen adaptation to the host. The plant pathogenic fungus Magnaporthe oryzae (synonym: Pyricularia oryzae) is the causal agent of rice blast disease and one of the best models for studying plant-microbe interactions (Ebbole, 2007). The interactions between rice and this fungus causes severe loss in rice world production which is the third largest in agriculture, and a major contributor to human nutrition. Recently, global wheat production has been threatened by the outbreak of wheat blast in Bangladesh (Islam et al., 2019).
During infection, M. oryzae produces a specialized infection structure called appressorium which generates a high turgor pressure to penetrate epidermis of rice leaves. Then, the penetration peg from the appressorium differentiates into infectious hyphae for multicellular infection. The formation of an appressorium consists of two developmental phases: recognition and maturation. In the recognition phase, the growing germ tube ceases to elongate and exhibits morphological changes such as a swelling and bending—called hooking—at the tip (Bourett and Howard, 1990). In the maturation stage, appressorium increase its volume and the cell wall accumulates melanin. This melanized cell wall results in the generation of a high turgor pressure due to increased intracellular glycerol. Proper surface signal sensing by the fungus stimulates the morphological changes during the recognition phase. Thus, several signals are known to induce appressorium formation in M. oryzae. For example, an artificial hydrophobic surface is sufficient for appressorium formation (Lee and Dean, 1993). The spores of M. oryzae extend a germ tube g without forming an appressorium on a hydrophilic surface like a glass slide; however, on a hydrophobic surface the spores form short germ tubes and melanized appressoria within 8 h. Indeed, chemical components in the cuticle or wax layers induce appressorium formation even on a hydrophilic surface. Furthermore, several primary fatty alcohols are known to induce appressorium formation (Gilbert et al., 1996). These alcohols are also called cutin monomers due to the structural similarity of cutin. Thus, 1,16-hexadecanediol (C16) and cis-9-octadecen-1-ol (C18) have effects on the induction of appressorium formation, and C16 has been widely used in the M. oryzae research. In addition, longer primary alcohols such as 1-octacosanol (C28) and 1-triacontanol (C30), which are one of the main components of the plant wax layer, induce appressorium formation in the rice blast fungus (Liu et al., 2011). As these chemicals have long hydrocarbon chains in common, the hydrogen bonds of these alcohols made by the alkyl group (−OH) are easily broken, leading to insolubility in water.
These physical and chemical signals are recognized by germ tubes and subsequently transduced into cells via intracellular 3′,5′-cyclic adenosine monophosphate (cAMP). Treatment with a high concentration of exogenous cAMP bypasses signal-sensing and directly induces appressorium formation even on a hydrophilic surface (Lee and Dean, 1993). This signal transduction mechanism is mediated by the heterotrimeric G protein, which consists of three subunits, Gα, Gβ, and Gγ. When Gα is dissociated from Gβγ, free Gα binds to adenylate cyclase which transforms adenosine triphosphate to cAMP. The Δmagb mutant, in which the Gα gene has been deleted, exhibited no sign of swelling or hooking (Liu and Dean, 1997). However, the Δmac1 mutant, which shows defects in the adenylate cyclase gene, still showed swelling and hooking (Adachi and Hamer, 1998; Choi and Dean, 1997). These results indicate that swelling and hooking of the rice blast fungus is related to the Gα subunit but not the production of cAMP.
Three genes have been characterized as sensing-related proteins for appressorium formation in the rice blast fungus: the G protein coupled receptor (GPCR)-like protein (Pth11, MGG_ 05871) (DeZwaan et al., 1999; Kou et al., 2017), the chitin-binding protein (Cbp1, MGG_01868) (Kamakura et al., 2002), and the surface mucin protein (Msb2, MGG_6033) (Liu et al., 2011). The pth11 mutant exhibited long germ-tubes on an inductive surface and showed delayed appressorium formation (Kou et al., 2017). Because the defect is compensated for by exogenous cAMP, and as Pth11 has multiple transmembrane domains like human GPCRs (DeZwaan et al., 1999; Kulkarni et al., 2005), Pth11 is predicted to function as GPCR for activation of the cAMP-mediated protein kinase A pathway. However, Pth11 is different from the other sensing-related proteins because the Δpth11 mutant exhibits swelling and hooking on hydrophobic surfaces (DeZwaan et al., 1999). The ΔMocbp1 mutant (Cbp1−) showed no sign of hooking or swelling at the germ tube tips on the inductive surface (Kamakura et al., 2002). Addition of C16 effectively corrected the ΔMocbp1 mutant defect, indicating that the mutant can recognize C16 as a signal to form appressorium. Consistently, both ΔMocbp1 and its wildtype showed similar virulence on plant leaves, suggesting that Cbp1 is a specific sensor protein for the physical signal hydrophobicity (Kamakura et al., 2002). The ΔMomsb2 mutant showed similar defects in appressorium formation (Liu et al., 2011). However, neither C16 nor C18 induced appressorium formation in the ΔMomsb2 mutant. Instead, long-chain primary alcohols, such as C28 and C30, enabled ΔMomsb2 to produce appressorium. Therefore, the function of MoMsb2 protein seemingly involves sensing both hydrophobicity and cutin monomers.
Although the appressorium formed is usually observed at the tip of a germ tube in M. oryzae, an appressorium-like structure (ALS) can also be produced at the tip of hyphae in the laboratory conditions. ALS formation was initially observed by Lee and Dean (1993) and analyzed again to test the ability of appressorium formation in the ΔMohox2 mutant, which could not generate a conidium at all (Kim et al., 2009). Recently, the regulation of ALS formation has been extensively examined and compared to normal appressorium formation, indicating that the two processes were not identical (Kong et al., 2013). For example, ALS formation was not induced by the addition of cAMP, a well-known inducer of appressorium formation; however, it was effectively induced by C16. The Δpmk1 mutant failed to form both appressorium and ALS, whereas the Δchs7 mutant did form an ALS although it was defective in appressorium formation on the germ tube (Kong et al., 2013).
In the process of mutant screening, we identified a deletion mutant that was defective in appressorium formation. The deleted gene (MGG_10422) was namedMoAFO1 due to its phenotypic defects in appressorium formation. This mutant failed to form an appressorium on artificial hydrophobic surfaces despite treatment with some of the known appressorium-inducing chemicals. We further characterized this mutant to understand the role of this gene in host surface-sensing mechanisms and pathogenicity.
The MoAFO1 (MGG_10422) protein sequence was retrieved from the online database Comparative Fungal Genomics Platform (CFGP, http://snu.ac.kr) (Choi et al., 2013). Homologous genes were found in the BLASTMatrix tool built in CFGP using “Fungal Genome Gold Standard release 1.3.” Alignment of sequences was done by the ClustalW algorithm (Thompson et al., 1994). The phylogenetic tree was inferred using the Neighbor-Joining method implemented in the MEGA 6.0 software and tested by bootstrapping with 5000 replications (Tamura et al., 2013). Online InterProScan database was used to obtain the MoAFO1 gene structure (http://www.ebi.ac.uk/).
M. oryzae wild-type strain KJ201 and ΔMohox7 were collected from the Center of Fungal Genetic Resources (CFGR; http://cfgr.snu.ac.kr). All strains were grown on V8 agar (V8; 8% V8 juice (v/v), 1.5% agar (w/v), adjusted to pH 6.0 using NaOH) or complete agar media (CM; 0.6% yeast extract (w/v), 0.6% casamino acids (w/v), 1% sucrose (w/v), and 1.5% agar (w/v)) at 25ºC (Goh et al., 2017). Strains were cultured on V8 juice agar media for 7 days for seven days at 25°C under the constant light to promote conidial production. Five stages of samples were used for RNA extraction: mycelia (liquid CM for 8 days), conidia (V8 agar for 7 days), germinated conidia (2 h after inoculation of a slide glass), appressoria (6 h after inoculation of a cover glass), and infectious hyphae stages in rice leaves (72 h after inoculation on rice leaves).
For the PCR-based screening of transformants, genetic DNA was extracted through previously established method (Chi et al., 2009). In the purpose of Southern hybridization, genomic DNA was extracted from the mycelia according to a standard protocol (Sambrook et al., 1989). Genomic DNA was digested with BamH1 and blot was probed with 0.5 kb 5′-flanking sequences (Supplementary Fig. 1A). The RNA (5 μg) were extracted from the five-stage samples by using Easy-Spin Total RNA extraction kit (Intron, Seongnam, Korea). For expression analysis through quantitative Real Time-PCR, cDNA synthesis was done with 5 μg of total RNA using the oligo dT primer with the ImProm-II Reverse Transcription System kit (Promega, Madison, WI, USA) following the manufacturer’s instruction.
Knock-out construct of MoAFO1 was generated by double-joint PCR where ~1 kb sequences of this gene were amplified and fused to hygromycin resistant gene (HPH). Then the construct was transferred to wildtype protoplasts and resulting transformants were primarily selected through PCR-based screening using specific primer pair MoAFO1_SF and MoAFO1_SR (Supplementary Table 1). Knock-out mutant was confirmed by Southern blot analysis using 5′-flanking sequence as a probe (Supplementary Fig. 1B). Complemented strain for ΔMoafo1 was generated by reintroducing ORF of MoAFO1 (2.8 kb) and complementation was confirmed through PCR-based screening with specific primers MoAFO1_NF and MoAFO1_NR (Supplementary Table 1). Complemented strain was indicated as Moafo1c. Monomeric red fluorescent protein (RFP) was tagged to the 3′ end of the MoAFO1 gene using Gateway cloning with the pFPL-Rh vector (Gong et al., 2015). The native promoter of MoAFO1 was replaced with that of the EF1α gene in Fusarium verticillioides for strong expression. The vector was transformed into wildtype protoplasts using the PEG-mediated transformation method.
Vegetative mycelial growth was measured at 7 days with three replications. All strains were grown on both complete agar media [modified CM; 1% glucose (w/v), 0.2% peptone (w/v), 0.1% yeast extract (w/v), 1% casamino acid (w/v), 0.1% trace element, 0.1% vitamin supplement, 0.6% NaNO3 (w/v), 0.05% KCl (w/v), 0.05% MgSO4 (w/ v), 0.15% KH2PO4 (w/v)] and on minimal agar media [MM; 1% glucose (w/v), 0.1% trace element, 0.1% vitamin supplement, 0.6% NaNO3 (w/v), 0.05% KCl (w/v), 0.05% MgSO4 (w/v), 0.15% KH2PO4 (w/v), pH 6.5]. For conidiation, 7-day-old V8 agar plates were used to collect conidia with 5 ml sterilized distilled water. Conidiation was determined by counting the numbers of conidia with a hemocytometer under a microscope. During conidial germination and appressorium formation assays, conidia were harvested from 7-day-old V8 juice agar plates and conidial concentration was adjusted to 2 × 104 conidia/ml. And then, 40 μl conidial suspension was dropped onto coverslip and on onion epidermis. At 4 and 8 h after incubation, conidial germination rate and appressorium formation was determined by counting at least a hundred of conidia per replicate under a microscope. These processes were performed with three replicates in three independent experiments. For coating glass surfaces, ethyl alcohol and chloroform were used for making 10 mM final concentration of cis-9-octadecen-1-ol (C18, oleyl alcohol) and 1,16-hexadecanediol (C16), respectively. Long chain primary alcohol 1-octacosanol (C28) and 1-triacontanol (C30) were dissolved to 4 mg/ml in chloroform. Fifty μl of chemical solutions were dropped on microscopic glass slides (Marienfeld, Lauda-Königshofen, Germany) and evaporated inside the gas hood for 30 min.
Ten ml of conidial suspensions (1 × 105 conidia/ml) containing Tween 20 (250 ppm final concentration) were sprayed onto the 3-week-old rice seedlings (Oryza sativa cv. Nakdongbyeo). Sprayed rice seedlings were placed in a dew chamber for 24 h under the dark condition at 25°C and then, transferred to the rice growth incubator maintained 25°C, 80% humidity and with photoperiod of 16 h with fluorescent lights. For microscopic observation of infectious growth on rice tissue, excised rice sheath of Nakdongbyeo were prepared with an established method (Koga et al., 2004). Conidia suspension (2 × 104 conidia/ ml) was injected with or without cAMP in excised rice sheaths and incubated in moistened box for 24 and 48 h at room temperature. After incubation, the infected rice sheaths were trimmed to remove chlorophyll enriched plant parts. Left epidermal layers of mid vein (three to four cell layers thick) were used for microscopic experiment. To observe the lesion development through drop inoculation with conidial suspension, 30 μl of conidial suspension (1 × 105 conidia/ ml) were dropped onto punctured or intact detached rice leaves and incubate in a humid plastic box chamber at room temperature.
ROS sensitivity was tested using both solid and liquid media. Sevenday old culture on V8 agar media was used as inoculum. In solid assays, CM was supplemented with 2.5 mM and 5 mM H2O2 and 0.2 mM 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonate) (ABTS) for peroxidase and laccase activities, respectively. Pictures were taken at 7 dpi (days post inoculation) and 4 dpi for peroxidase and laccase activities, respectively. In liquid assays, 3-day-old culture in the liquid CM were filtered through Whatman filter paper no. 42. For laccase enzyme assay, 150 μl of 9.1 mM ABTS solution and 250 μl of culture filtrates were mixed and 1.1 ml of 100 mM phosphate buffer was added. For peroxidase enzyme assay, 50 μl of 0.3% peroxide solution and 50 μl of culture filtrates were mixed and 1.45 ml of 9.1 mM ABTS solution was added. Absorbance at 405 nm was measured. All experiments were performed with three biological replicates.
The homologous proteins of MoAfo1 (MGG_10422) were queried using BLASTMatrix, a bioinformatics tool, available in the CFGP (Choi et al., 2013). Twelve orthologs of MoAfo1 were found among 29 fungal species with an e-value cut-off of 1e−5 (Supplementary Fig. 2). The orthologs were conserved specifically in subphylum Pezizomycotina of phylum Ascomycota (Fig. 1A). InterPro Scan analysis predicted that all orthologs had both the fungal transcription factor (IPR021858, green box in Fig. 1A) and Zn(II)2Cys6 fungal-type DNA-binding (IPR001138, red box in Fig. 1A) domains. The Phobius program calculating features of transmembrane domains and signal peptides predicted that many of them had transmembrane domains near their C-terminal ends (Käll et al., 2007) (Fig. 1B). Based on this prediction, most MoAfo1 might be located outside of the membrane (Fig. 1C). The Phobius tool also predicted signal peptides at the N-terminal proteins of Fusarium oxysporum and M. oryzae (yellow circle in Fig. 1). However, there is a conflict between these predicted results because transcription factors usually locate at nucleus. In addition, the PSORT II program predicted that these orthologs should localize to the cytoplasm (~26%) and nucleus (17%-22%) in the cell.
To confirm the localization of MoAfo1, the RFP gene was introduced to the C-terminal end of the wildtype strain (Fig. 2). In conidia, MoAfo1-RFP signals were weak and most signals were detected at plasma membrane or in some spots in the cytoplasm (0 h in Fig. 2). When germ tubes were formed (1 to 4 h), signals were stronger in conidia and several spots were observed in the germ tubes (4 h). However, no signal was observed in the membranes of germ tubes. During appressorium formation (5 to 8 h), the RFP localization was also observed in the plasma membranes and vacuole-like organelles of the appressoria. After 24 h of incubation, most signals were observed exclusively in the appressoria (data not shown). The numbers and sizes of cytoplasmic spots were different from cell to cell. The trend in the RFP signals during germination and appressorium formation was similar to that of the mRNA transcripts (Supplementary Fig. 3).
The gene deletion mutant and its complement strains were generated (Fig. 3). We compared the disease cycle of mutants with that of the wildtype strain to test whether MoAFO1 is a pathogenicity specific gene. The vegetative growth of the ΔMoafo1 mutant was similar to that of the wildtype (Table 1). Conidia production in the mutant was reduced by ~85% of the wildtype level (P < 0.05, one-way ANOVA). Wild-type conidia germinated and started to produce appressoria on the hydrophobic surface within 8 h. Conidial germination of the mutant was similar to the wildtype, while the ΔMoafo1 mutant failed to produce any appressorium on the cover slip (Table 1, Fig. 3). Incubation up to 72 h could not failed to correct the defect in appressorium formation of the mutant, suggesting that ΔMoafo1 did not delay the appressorium formation. Furthermore, we used the ΔMohox7 mutant as a positive control for hooking and swelling (data not shown) because this mutant showed hooking and swelling without developing any appressorium on the inductive surface (Kim et al., 2009). Conversely, the ΔMoafo1 mutant did not show any sign of hooking or swelling as ΔMohox7 did, indicating that it was defective in recognizing physical hydrophobicity (Fig. 3). In addition, functionality of MoAfo1-RFP was examined by the introduction of the MoAFO1-RFP construct to ΔMoafo1 (Supplementary Fig. 4). Appressorium formation was restored in the complement strain, indicating that the construct was functional.
We applied cAMP exogenously to wildtype and mutant strains on hydrophilic and hydrophobic surfaces to verify the lack of formation of an appressorium in the ΔMoafo1 mutant (Fig. 4). On both surfaces, 5 mM cAMP increased the appressorium forming rate of the mutant from zero to 65-70%, suggesting that the mutant failed to accumulate cAMP in response to the signals. This result highlights the involvement of MoAfo1 in the activation of cAMP production during early appressorium development. Interestingly, most cAMP-induced appressoria were abnormal in morphology when compared to those of wildtype (Fig. 4). The M. oryzae wildtype usually has dome-shaped appressoria shown as circles under a microscope, whereas rod- or kidney-shaped appressoria were observed in the ΔMoafo1 mutant (Fig. 4). On the other hand, dark colored appressoria provided evidence that melanization, one of the other maturation processes, was in progress.
M. oryzae produces appressorium-like structures (ALSs) from hyphal tips when a mycelial agar block is placed on a hydrophobic surface. We estimated the rate of ALS formation in the ΔMoafo1 mutant. Contrary to our expectation, the mutant produced ALSs at a rate of 15.3 ± 4.5% at 72 hour post inoculation (hpi), while wildtype did at a rate of 66.3 ± 2.3% at 72 hpi (P < 0.0001). This result shows that the mechanism of ALS formation is different from that of conidial appressorium formation, although the exact mechanisms are still unknown. Interestingly, the morphology of ALSs was deformed like the cAMP-treated conidia of the ΔMoafo1 mutant (Supplementary Fig. 5).
Pathogenicity assays showed that the wildtype strain caused many of lesions on the leaves at 7 dpi (Fig. 5A), whereas the ΔMoafo1 mutant developed very few lesions with a typical size and shape. These results suggest that the ΔMoafo1 mutant still made enough functional appressoria to infect rice leaves. However, as the ΔMoafo1 mutant could not develop appressorium on the artificial surface, we also tested the appressorium-forming ability of the mutant using rice sheath and onion epidermis (Fig. 5B and C). Long germ tubes were formed from most conidia of the ΔMoafo1 mutant on both plant surfaces, indicating that the mutant did not fully recognize surface signals. Unlike the artificial surface, however, 5% to 8% of the germ tubes produced appressoria at the tips of the long germ tubes (Fig. 5B and C). We also tested appressorium formation directly on the surface of rice. Surprisingly, approximately half of the observed conidia in the ΔMoafo1 mutant produced appressoria on the surface of rice leaves (Fig. 5D). These results indicated that the ΔMoafo1 mutant still sensed some signals from the rice leaf surface. In contrast to artificial or non-host surfaces, something in the surface of rice leaves induced the mutant to activate the downstream pathways for appressorium formation. In addition, the appressoria of the mutant appeared deformed on onion skins and rice sheaths (Fig. 5B and C), while those on the surface of rice leaves seemed to be normal in germ tube length and appressorium morphology (Fig. 5D).
Restoration of defects in appressorium development in the ΔMoafo1 mutant on rice leaves pointed at the need to identify the chemicals that induce appressorium formation. Thus, we tested cutin monomers and long chain primary alcohols (C16, C18, C28, and C30) (Gilbert et al., 1996; Liu et al., 2011) as signal molecules inducing appressorium formation. These chemicals had been known to restore the defects in appressorium formation of the sensor mutants (ΔMocbp1 and ΔMomsb2) (Kamakura et al., 2002; Liu et al., 2011). All tested chemicals induced appressorium formation by the wildtype fungal strain on a hydrophilic surface, but they failed to influence that of the ΔMoafo1 mutant (Fig. 6). Thus, MoAfo1 might play a role in recognizing a wide range of signal chemicals.
Because appressorium formation of the ΔMoafo1 mutant was partially restored on the onion and rice sheath surfaces, we examined the penetration and invasive growth of the mutant appressoria. After 48 h of incubation of the wildtype conidia, 89.2% of the counted appressoria showed invasive hypha or penetration peg (Fig. 7). However, no penetration was observed in the appressoria of the mutant, indicating that this mutant has a defect in penetration (Fig. 7). In addition, we investigated whether cAMP might compensate for such defect or not. The results showed that the rate of appressorium formation increased slightly in both wildtype and mutant strains with the existence of 5 mM cAMP. However, the penetration rate of the mutant increased to 77.6%, whereas that of the wildtype remained constant (~85%).
During penetration, we also observed that invasive growth was significantly delayed or defective in the ΔMoafo1 mutant (Fig. 7). All the invasive hyphae of the ΔMoafo1 mutant were limited to the first infected cells with dark brown granules, whereas those of wildtype expanded to neighboring cells (Fig. 7). In addition, we examined the invasive growth of the mutant using the punctured rice leaves (Fig. 8A and B). The conidial suspension was dropped on both intact and wounded rice leaves. The mutant showed a delay in disease development at 4 dpi, compared to the wildtype and complement strains under both conditions (Fig. 8A and B). These results confirmed that the ΔMoafo1 mutant is defective in invasive growth. Based on these findings on penetration and invasive growth, we examined the response of the mutant to oxidative stress because accumulation of dark brown granules is regarded as a sign of an oxidative burst during the host defense response (Chi et al., 2009). The ΔMoafo1 mutant showed a significant decrease in response to ABTS with and without H2O2 (Fig. 8D). Consistently, the mutant showed a weak color change in response to ABTS in the solid media assay (Fig. 8C). Thus, the growth defects within host cells might be caused by reduced oxidation of molecules of ROS in the ΔMoafo1 mutant.
The recognition of diverse host signals by the rice blast fungus is essential for the initiation of a series of developmental changes that facilitate penetration and invasive growth. In this study, we found that the membrane-bound protein MoAfo1 is involved in sensing well-known signals required for the induction of appressorium formation by the fungus. MoAfo1 plays a role in sensing multiple signals such as physical hydrophobicity of artificial surfaces, cutin monomers (C16 and C18), and long chain primary alcohols (C28 and C30). In addition, the mutant shows multiple defects in appressorium morphology, penetration, invasive growth, and pathogenicity. These findings revealed that MoAfo1-mediated host signal sensing is important not only for early appressorium development but for the subsequent events during infection development, such as penetration and invasive growth.
We observed that ΔMoafo1 continued to extend its germ tubes without any sign of hooking or swelling on an artificial hydrophobic surface (Fig. 3), thus suggesting that it failed to respond to the cues from inductive surfaces. In contrast, the Δpmk1 and ΔMohox7 mutants, which are defective in appressorium formation, exhibit hooking and swelling at several points in the germ tubes in response to hydrophobic signals (Kim et al., 2009; Xu and Hamer, 1996). ΔMoafo1 is not the first sensing-defective mutant in which, hydrophobicity was not sufficient to induce appressorium formation; similar observation were made in the mutants, ΔMocbp1 and ΔMomsb2. Treatment with cutin monomer (C16) activated appressorium development in ΔMocbp1, suggesting that Cbp1 is a sensor protein for physical signals such as hydrophobicity (Kamakura et al., 2002). Interestingly, although appressorium formation in ΔMomsb2 was not induced by treatment with cutin monomers (C16 or C18), induction did follow upon treatment with long chain primary alcohols (C28 and C30) (Liu et al., 2011). This means that MoMsb2 is a sensor for hydrophobicity and cutin monomers. However, ΔMoafo1 failed to respond to all known signals, such as hydrophobicity, C16, C18, C28, and C30, indicating that MoAfo1 is responsible for sensing a broad range of signal molecules (Fig. 6). Consistently, our results regarding protein localization lend support to the possibility that MoAfo1 is a sensing-related protein. MoAfo1-RFP signals were constantly observed in the plasma membranes and vacuole-like structures in both conidia and appressoria (Fig. 2). In turn, MoMsb2 and Pth11 were reportedly localized in the vacuole-like organelles in conidia or appressoria as well (Kou et al., 2017; Liu et al., 2011). These proteins were predicted to have signal peptides at the N-terminal, although their peptide sequences were not homologous to each other (Fig. 1). In the case of MoMsb2, removing the signal peptides from the gene resulted in the failure of vacuole localization (Liu et al., 2011). Thus, MoAfo1 is very likely involved in sensing surface signals.
On the onion surface, the mutants showed deformed appressoria defective in penetration (Fig. 7). When we complemented MoAFO1 with cAMP, this defect was compensated for, thus allowing the mutant to perform as the wildtype in effectively developing infection hyphae in this case. Therefore, the expression of MoAFO1 contributes to the accumulation of cAMP, which is required for successful penetration of the plant epidermal surface. Furthermore, this mutant showed limited growth, and dark brown granules accumulated around its infection hyphae (Fig. 7). This observation is important because these characteristic phenotypes were reminiscent of those of the Δdes1 and ΔMoap1 mutants (Defense Suppressor 1 and AP1-like transcription factor, respectively) (Chi et al., 2009; Guo et al., 2011), which consistently showed defects while dealing with oxidative stress generated as part of the plant defense response to the fungal infection. Similarly, the ΔMoafo1 mutant was sensitive to an in vitro oxidative stress (Fig. 8C) and exhibited significant delay in invasive growth (Fig. 8A and B). Virulence of the mutant on rice leaves was much reduced (2.1%), compared to the wildtype (15.3%) (Fig. 5A). Interestingly, MoAFO1 was predicted to have the MoAp1-binding motif at the promoter region (1,145 bp upstream from ATG) (Guo et al., 2011). Therefore, such defects in ROS-related responses might explain the reduced virulence of the mutant.
Recent findings indicate that endogenous ROS are more important to fungal development than the exogenous ROS mentioned above (Samalova et al., 2014). Endogenous ROS were produced during appressorial differentiation in the rice blast fungus. Another recent report revealed reduced levels of endogenous ROS in the Δpth11 mutant, indicating that endogenous ROS are responsible for the delay of appressorium formation in the mutant (Kou et al., 2017). This indicates that Pth11, a GPCR for the cAMP signaling pathway (Ramanujam et al., 2013), may be involved in ROS accumulation during appressorium formation. Pth11 and MoAfo1 proteins are involved in both sensing signals and balancing ROS. In addition, they showed a similar pattern in cellular localization. Although the exact relationships among ROS balancing, signal sensing, and the cAMP signaling pathway warrants further research, the phenotypic similarity does suggest a probable connection between two proteins.
Understanding the mechanisms by which microorganisms recognize surfaces is important in controlling plant pathogenic fungi. The ΔMoafo1 mutant exhibited surfacespecific responses, suggesting that the MoAFO1 gene plays a key role in detecting physical and chemical signals for appressorium formation. This gene is linked to the ROS signaling pathway, which is important for successful fungal growth during early development of the rice blast disease.
Acknowledgments
This work was supported by the Incheon National University Research Grant in 2015. We acknowledged the technical assistance of Bonjun Ku in confocal imaging. All authors have no conflict of interest to declare.
Electronic Supplementary Material
Supplementary materials are available at Plant Pathology Journal website (http://www.ppjonline.org/).
Supplementary Fig. 1.
Knock-out mutant generation. (A) Schematic diagram of knockout of MoAFO1 gene. The wildtype MoAFO1 gene was replaced with hygromycin resistance (HPH) gene cassette. B indicates the restriction site of BamHI. (B) Confirmation of mutants by Southern hybridization (on left) and cDNA amplification (on right).
Supplementary Fig. 2.
Screenshot of the BLAST Matrix result on MGG_10422 on the CFGP web page. “Fungal Genome Gold Standard release 1.3” was used for the reference dataset. E-value was limited under “1e-10”. “Class” in the “Taxonomical option” was applied for this view.
Supplementary Fig. 3.
Transcriptional expression of the MoAFO1 gene in five development stages of M. oryzae wildtype strain KJ201. MY, mycelia; CO, conidia; GE, germinating conidia; AP, appressoria; In72, early infection stage at 72 hpi.
Supplementary Fig. 4.
Appressorium formation of ΔMoafo1 (A) and its complement strain with the MoAFO1-RFP fused construct (B). Conidia placed on hydrophobic surface were stored in the humid chamber for 24 h.
Supplementary Fig. 5.
Appressorium-like structure (ALS) assay in the ΔMoafo1 mutant. Hyphal blocks of KJ201 and ΔMoafo1 were placed on plastic coverslips leaving the agar blocks on upper ward, and then incubated in a moist chamber box in the dark condition at 25ºC. Photographs were taken at 3 days after inoculation. Scale bars = 20 μm.
Fig. 1
Evolutionary relationships of the orthologs of MoAfo1 (MGG_10422). (A) The phylogenetic tree was inferred using the Neighbor-Joining method implemented in the MEGA 6.0 software, and tested by bootstrapping with 5,000 replications. The percentages of replicate trees in the bootstrap test are shown next to the branches. In the domain architecture, red and green boxes indicate Zn(II)2Cys6 fungal-type DNA-binding (IPR001138) and fungal transcription factor (IPR021858) domains, respectively. The yellow box and the violet circle indicate signal peptides and transmembrane domain, respectively. Pale violet color in transmembrane domains indicates lower probability in prediction. Fungal species are shown in italic and their abbreviations are as follows: Af, Aspergillus fumigatus; An, Aspergillus nidulans; Bc, Botrytis cinerea; Ci, Coccidioides immitis; Cg, Collectotrichum graminicola; Fg, Fusarium graminearum; Fo, Fusarium oxysporum f. sp. lycopersici; Hc, Histoplasma capsulatum; Mo, Magnaporthe oryzae; Mg, Mycosphaerella graminicola; Nc, Neurospora crassa; Pa, Podospora anserine. (B) Prediction of signal peptide and transmembrane by Phobius program. In the plot, y-axis is the probabilities of cytoplasmic, non-cytoplasmic, TM helix, and signal peptide. (C) A diagram based on predicted domain and topology. Colors are consistent with those in (A) and (B).

Fig. 2
Localization of MoAfo1-RFP during appressorium development in Magnaporthe oryzae. The photographs were taken using the same laser intensity of confocal microscopy. DIC, differential interference contrast; RFP, red fluorescent protein. Scale bars = 10 μm.
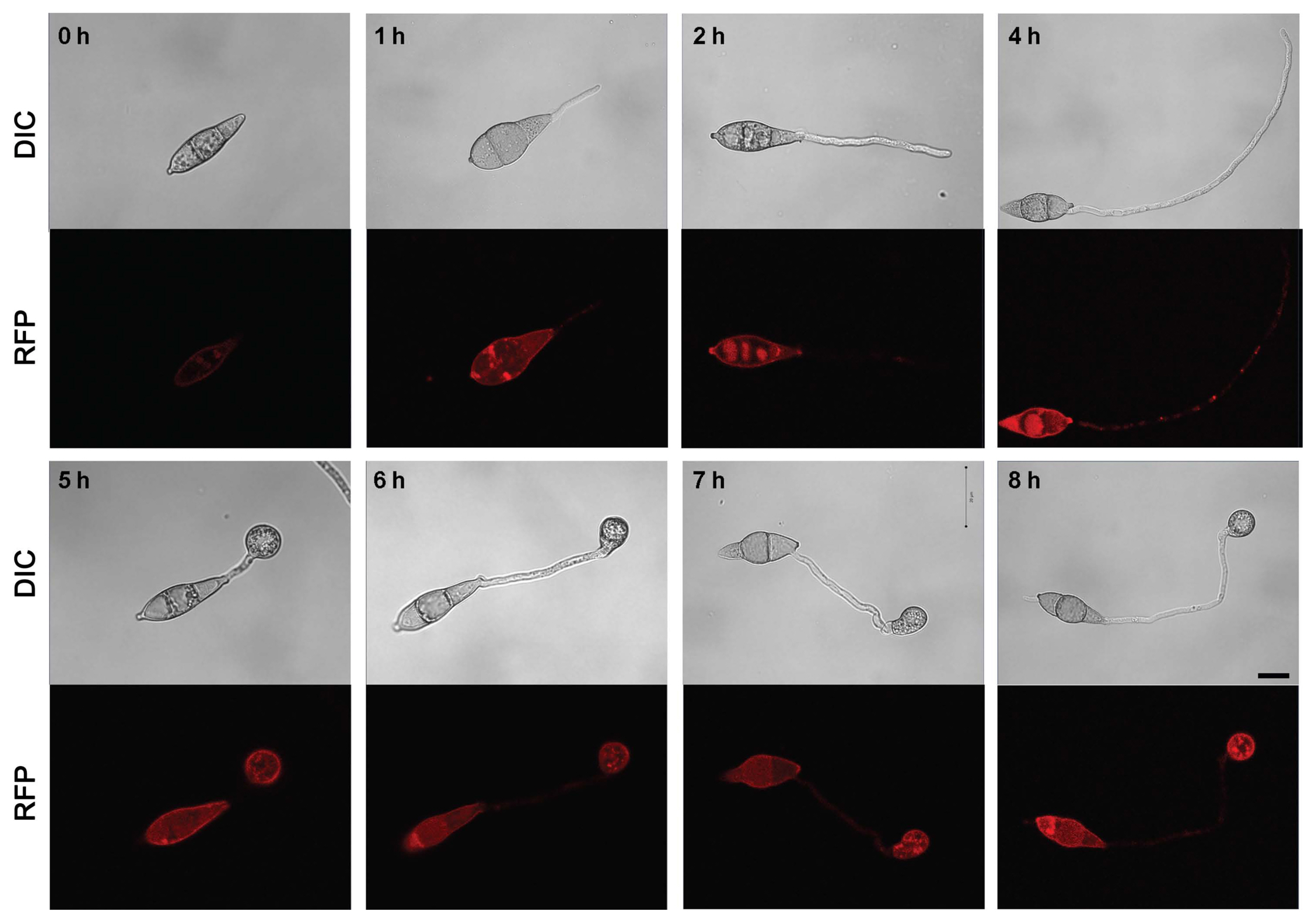
Fig. 3
Defects in appressorium formation in the ΔMoafo1 mutant strain. Conidial suspensions were dropped onto a plastic coverslip and pictures were taken at 8 hpi. KJ201, wildtype; ΔMoafo1, null mutant; Moafo1c, complement strain. Scale bars = 10 μm.
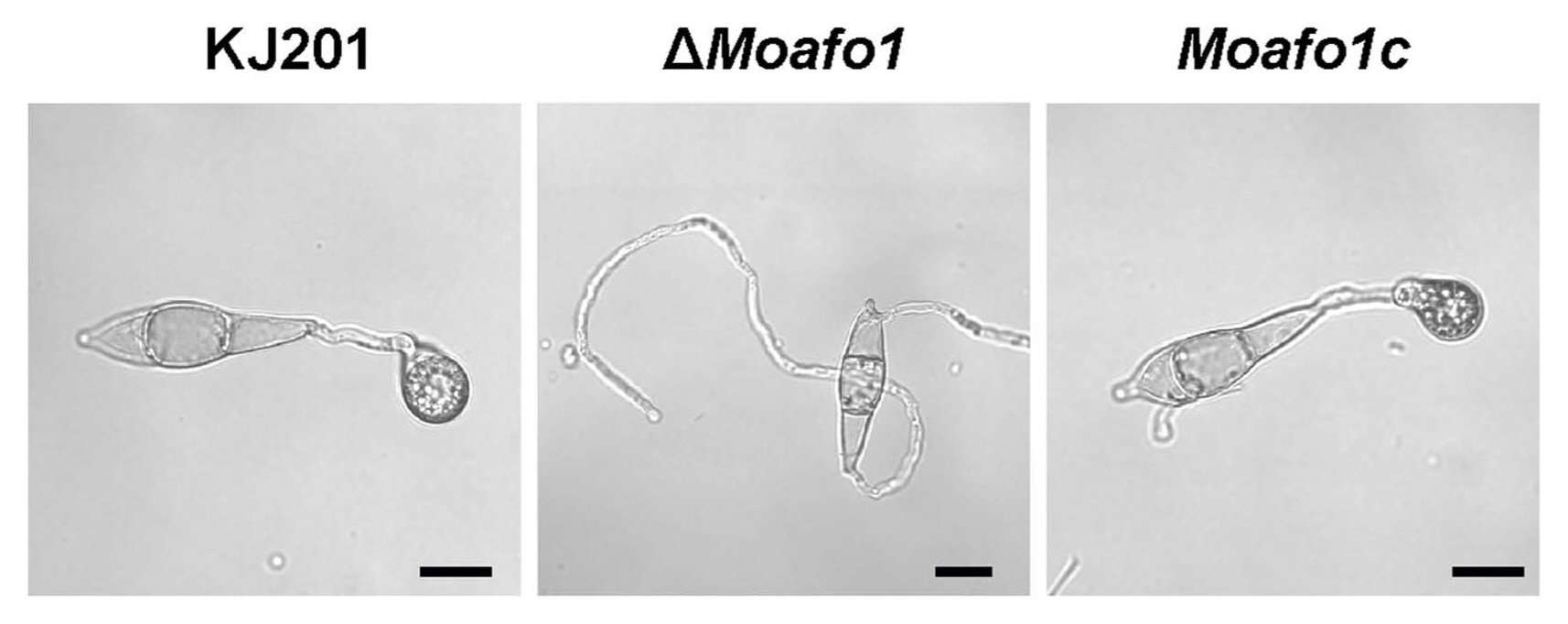
Fig. 4
The cAMP treatment restored appressorium formation in the ΔMoafo1 strain. Appressorium formation in ΔMoafo1 on both hydrophobic (upper) and hydrophilic (lower) surfaces with or without 5 mM cAMP. Photographs were taken at 24 hpi. Scale bars = 10 μm.

Fig. 5
Pathogenicity assay and appressorium formation on different plant surfaces. (A) Disease development after spraying a conidial suspension onto rice leaves. Conidia were sprayed onto the 3-week-old rice leaves and incubated in the dark. Pictures were taken at 7 dpi. (B, C) Appressorium formation was observed on onion epidermis (B) and in rice sheath (C). The conidial suspension was dropped on onion epidermis or injected in rice sheath with 2 × 104 conidia/ml and observed under a microscope at 48 hpi. Scale bar = 10 μm. (D) Appressorium formation assay on intact rice leaves. Forty μl of conidial suspensions (2 × 104 conidia/ml, 10 mg/ml Calcofluor white) were dropped onto the rice leaves and photographs were taken at 24 hpi. Arrowheads indicate formed appressoria.
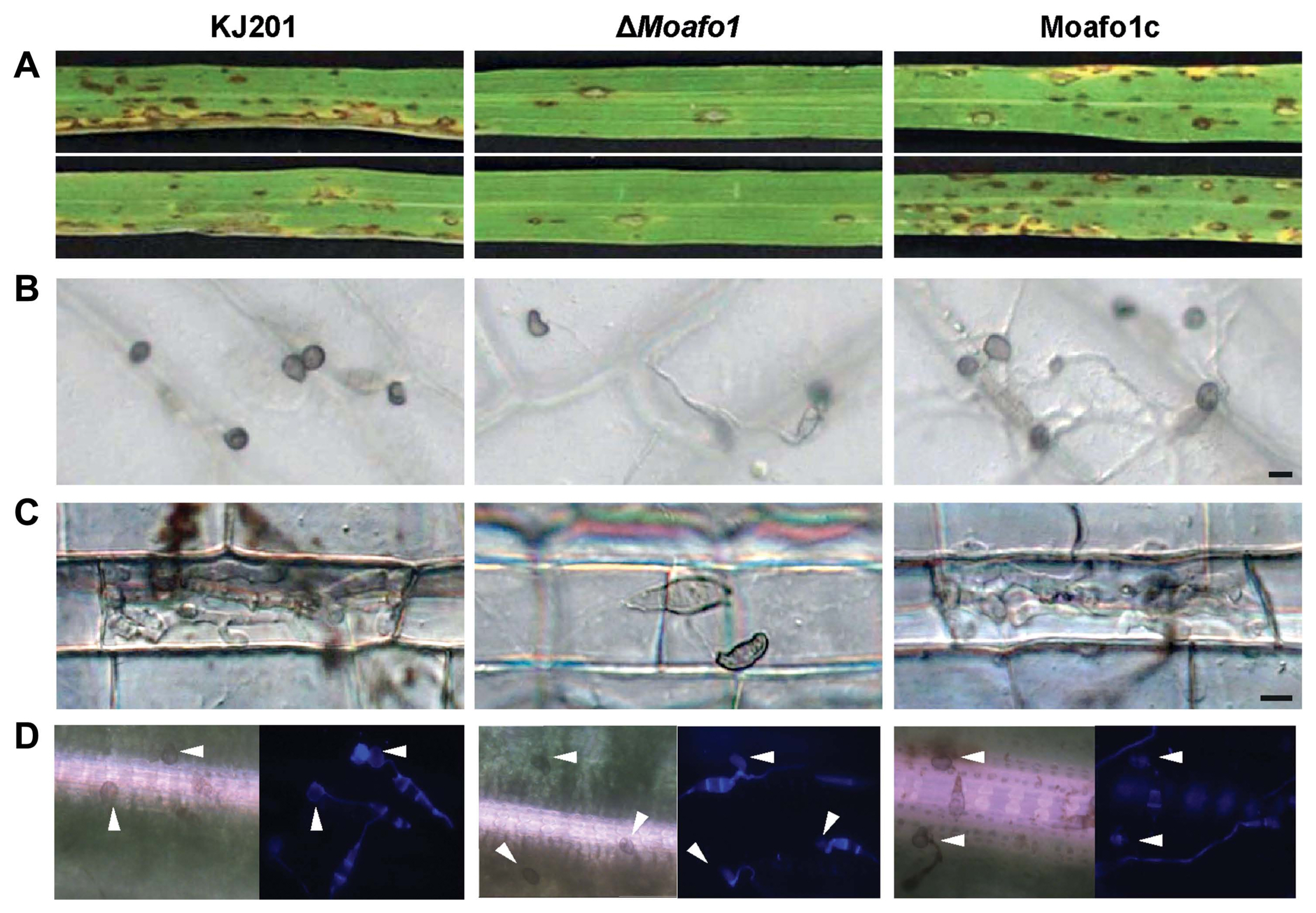
Fig. 6
Induction of appressorium formation by different signal molecules. Conidia of the wildtype and the ΔMoafo1 strains were placed on microscopic glass slides coated with different chemicals. Photographs were taken at 24 hpi. Scale bars = 10 μm.

Fig. 7
Leaf sheath assay with or without cAMP (5 mM). Thirty μl of conidial suspension (1 × 105 conidia/ml) was dropped onto punctured detached rice leaves and photographs were taken 4 dpi. Scale bars = 20 μm.

Fig. 8
Assays for pathogenicity (invasive growth) and reactive oxygen species sensitivity. The pathogenicity assay was performed using intact (A) and wounded (B) rice leaves. The rates of invasive growth were compared between them. Conidial suspensions (1 × 105 conidia/ ml) were dropped on the rice leaves and photographs were taken at 4 dpi. Lesion area was measured (right panels) using the Image J software. (C) Extracellular peroxidase and laccase activities were measured on solid CM. Photographs of peroxidase and laccase activities were taken at 7 and 4 dpi, respectively. (D) Extracellular peroxidase and laccase activities were measured using 3-day-old culture filtrates. Star marks in the graphs indicate statistical significance (t-test, P < 0.05).

Table 1
The phenotypes of ΔMoafo1 in different developmental stages
| Strains | Growth (mm)a | Conidiation (104/ml)b | Germination Rate (%)c | Appressorium formation (%)d,e |
|---|---|---|---|---|
| Wildtype | 35.6 ± 1.3 A | 22.3 ± 1.5 A | 98.6 ± 0.5 A | 95.0 ± 1.7 A |
| ΔMoafo1 | 35.3 ± 0.5 A | 18.0 ± 1.0 B | 97.0 ± 2.0 A | 0 B |
| Moafo1c | 36.0 ± 0.8 A | 23.6 ± 0.5 A | 97.3 ± 0.5 A | 94.0 ± 1.5 A |
References
Adachi, K. and Hamer, J. E. 1998. Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell. 10:1361-1374.



Bourett, T. M. and Howard, R. J. 1990. In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea. Can. J. Bot. 68:329-342.

Chi, M.-H., Park, S.-Y., Kim, S. and Lee, Y.-H. 2009. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 5:e1000401.



Chi, M.-H., Park, S.-Y. and Lee, Y.-H. 2009. A quick and safe method for fungal DNA extraction. Plant Pathol. J. 25:108-111.

Choi, J., Cheong, K., Jung, K., Jeon, J., Lee, G.-W., Kang, S., Kim, S., Lee, Y.-W. and Lee, Y.-H. 2013 CFGP 2.0: a versatile web-based platform for supporting comparative and evolutionary genomics of fungi and oomycete. Nucleic Acids Res. 41:D714-D719.


Choi, W. and Dean, R. A. 1997. The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell. 9:1973-1983.



DeZwaan, T. M., Carroll, A. M., Valent, B. and Sweigard, J. A. 1999. Magnaporthe grisea Pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell. 11:2013-2030.



Ebbole, D. J. 2007. Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45:437-456.
Gilbert, R. D., Johnson, A. M. and Dean, R. A. 1996. Chemical signals responsible for appressorium formation in the rice blast fungus Magnaporthe grisea. Physiol. Mol. Plant Pathol. 48:335-346.

Goh, J., Jeon, J. and Lee, Y. H. 2017. ER retention receptor, MoERR1 is required for fungal development and pathogenicity in the rice blast fungus, Magnaporthe oryzae. Sci. Rep. 7:1259.




Gong, X., Hurtado, O., Wang, B., Wu, C., Yi, M., Giraldo, M., Valent, B., Goodin, M. and Farman, M. 2015. pFPL vectors for high-throughput protein localization in fungi: detecting cytoplasmic accumulation of putative effector proteins. Mol. Plant-Microbe Interact. 28:107-121.


Guo, M., Chen, Y., Du, Y., Dong, Y., Guo, W., Zhai, S., Zhang, H., Dong, S., Zhang, Z., Wang, Y., Wang, P. and Zheng, X. 2011. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 7:e1001302.



Islam, M. T., Kim, K.-H. and Choi, J. 2019. Wheat blast in Bangladesh: the current situation and future impacts. Plant Pathol. J. 35:1-10.



Käll, L., Krogh, A. and Sonnhammer, E. L. 2007. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 35:W429-W432.



Kamakura, T., Yamaguchi, S., Saitoh, K.-I., Teraoka, T. and Yamaguchi, I. 2002. A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol. Plant-Microbe Interact. 15:437-444.


Kim, S., Park, S.-Y., Kim, K. S., Rho, H.-S., Chi, M.-H., Choi, J., Park, J., Kong, S., Park, J., Goh, J. and Lee, Y.-H. 2009. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 5:e1000757.



Koga, H., Dohi, K., Nakayachi, O. and Mori, M. 2004. A novel inoculation method of Magnaporthe grisea for cytological observation of the infection process using intact leaf sheaths of rice plants. Physiol. Mol. Plant Pathol. 64:67-72.

Kong, L.-A., Li, G.-T., Liu, Y., Liu, M.-G., Zhang, S.-J., Yang, J., Zhou, X.-Y., Peng, Y.-L. and Xu, J.-R. 2013. Differences between appressoria formed by germ tubes and appressoriumlike structures developed by hyphal tips in Magnaporthe oryzae. Fungal Genet. Biol. 56:33-41.


Kou, Y., Tan, Y. H., Ramanujam, R. and Naqvi, N. I. 2017. Structure-function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 214:330-342.


Kulkarni, R. D., Thon, M. R., Pan, H. and Dean, R. A. 2005. Novel G-protein-coupled receptor-like proteins in the plant pathogenic fungus Magnaporthe grisea. Genome Biol. 6:R24.



Lee, Y.-H. and Dean, R. A. 1993. cAMP regulates infection structure formation in the plant pathogenic fungus Magnaporthe grisea. Plant Cell. 5:693-700.



Liu, S. and Dean, R. A. 1997. G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 10:1075-1086.


Liu, W., Zhou, X., Li, G., Li, L., Kong, L., Wang, C., Zhang, H. and Xu, J.-R. 2011. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 7:e1001261.



Ramanujam, R., Calvert, M. E., Selvaraj, P. and Naqvi, N. I. 2013. The late endosomal HOPS complex anchors active Gprotein signaling essential for pathogenesis in Magnaporthe oryzae. PLoS Pathog. 9:e1003527.



Samalova, M., Meyer, A. J., Gurr, S. J. and Fricker, M. D. 2014. Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol. 201:556-573.


Sambrook, J., Fritsch, E. F. and Maniatis, T. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA. 1626 pp.
Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30:2725-2729.



- TOOLS



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print




