 |
 |
| Plant Pathol J > Volume 37(4); 2021 > Article |
|
Abstract
Alternaria leaf blight is one of the most common diseases in watermelon worldwide. In Korea, however, the Alternaria species causing the watermelon leaf blight have not been investigated thoroughly. A total of 16 Alternaria isolates was recovered from diseased watermelon leaves with leaf blight symptoms, which were collected from 14 fields in Korea. Analysis of internal transcribed spacer (ITS) region, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and RNA polymerase II second largest subunit (RPB2) were not competent to differentiate the Alternaria isolates. On the contrary, analysis of amplicon size of the histone H3 (HIS3) gene successfully differentiated the isolates into three Alternaria subgroups, and further sequence analysis of them identified three Alternaria spp. Alternaria tenuissima, A. gaisen, and A. alternata. Representative Alternaria isolates from three species induced dark brown leaf spot lesions on detached watermelon leaves, indicating that A. tenuissima, A. gaisen, and A. alternata are all causal agents of Alternaria leaf blight. Our results indicate that the Alternaria species associated watermelon leaf blight in Korea is more complex than reported previously. This is the first report regarding the population structure of Alternaria species causing watermelon leaf blight in Korea.
Watermelon (Citrullus lanatus Thunb.), a member of the Cucurbitaceae, is an economically important fruit crop and its annual production in 2018 was estimated at 103 million tons worldwide (Food and Agriculture Organization of the United Nations, 2018). It is widely grown in the most parts of Africa, the Caribbean, the southern United States, and Southeast Asia (Ma et al., 2021).
Dramatic problems in watermelon production have been caused by various pathogens, such as bacterial fruit blotch by Acidovorax citrulli, Fusarium wilt by Fusarium oxysporum, powdery mildew by Podosphaera xanthii, gummy stem blight by Didymella bryoniae, anthracnose by Colletotrichum orbiculare, and leaf spot disease by Nigrospora sphaerica (Burdman and Walcott, 2012; Ismail and Abd Razak, 2021; Noh et al., 2014). On the top of that, watermelon leaf blight caused by the genus Alternaria is an epidemic disease worldwide, resulting in reduced fruit size, quality, and yield (Chopra et al., 1974; Kim et al., 1994; Zhao et al., 2016b). Alternaria cucumerina was initially considered to be the causal agent of leaf blight in watermelons and other Cucurbitaceae crops (Jackson and Weber, 1959). Recently, however, various other Alternaria spp. including A. cucumerina, A. alternata, A. tenuissima, A. gaisen, and A. infectoria, have also been identified from watermelons in China, Serbia, and the United States (Blagojević et al., 2020; Ma et al., 2021; Zhao et al., 2016b; Zhou and Everts, 2008). In Korea, after A. cucumerina was first reported as a causal agent of watermelon leaf blight (Kim et al., 1994), additional studies concerning leaf blight have been rarely reported. Therefore, there is no further information about the presence of additional Alternaria spp. on watermelon in Korea. Although the survey conducted in Korea during 2008 to 2012 to examine different watermelon diseases reported Alternaria spp. to be one of 18 fungal pathogens, its species has not been identified (Noh et al., 2014).
Identification of the Alternaria species in watermelons is essential for setting up disease-management programs. The taxonomy of species within Alternaria have been previously based on morphological and physiological characteristics and differences in their host plants (Simmons, 2007). However, even in a single host, their morphological variation caused by environmental conditions, prolonged sub-culturing, and cultivation medium complicated species recognition. Molecular methods, therefore, have been more recently used to complement morphology-based approaches (Kang et al., 2001; Nishikawa and Nakashima, 2019). In the current study, we investigated what kinds of Alternaria species are associated with watermelon leaf blight in Korea and how to distinguish their species by comparing nucleotide sequences of key loci, internal transcribed spacer (ITS) region, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), RNA polymerase II second largest subunit (RPB2), and histone H3 (HIS3), commonly used for Alternaria species separation. Sequence analysis of the HIS3 gene successfully differentiated causal agents of Alternaria leaf blight into three different species, A. tenuissima, A. alternata, and A. gaisen. In addition to A. cucumerina, a previously reported causal agent, at least four different Alternaria spp. are now associated with watermelon leaf blight in Korea, which should be considered to set up effective management strategies.
In 2019 and 2020, a total of 14 fields from nine cities among the major watermelon production areas in Korea were selected for Alternaria spp. survey. Watermelon leaves and stems showing Alternaria-induced leaf blight lesions were harvested. Small pieces of tissue (10 × 10 mm) were removed from the margins between the healthy and symptomatic tissues, and their surfaces were disinfected with 70% ethanol for 1 min and rinsed with sterilized dH2O three times. The pieces were then placed on potato dextrose agar (PDA) (MBcell, Seoul, Korea) plates and incubated at 28°C in light for seven days. Multiple fungal colonies were obtained from each symptomatic tissue, and only those with Alternaria morphologies were selected as isolates. The isolates were sub-cultured onto new PDA plates for single spore purification.
Genomic DNA was extracted following a previously described protocol (Chen and Ronald, 1999) For the extraction, Alternaria mycelia were scrapped from the surface of the seven-day cultures grown on PDA and placed in a 2 ml microcentrifuge tube containing 700 μl cetyltrimethl-ammonium bromide (CTAB) extraction buffer (3% (w/v) CTAB, 20 mM EDTA pH 8.0, 1.42 M NaCl, 100 mM Tris-HCl pH 8.0, 2% (w/v) polyvinylptrrolidone, and 0.2% β-mercaptoethanol). After 10 min incubation at 65°C, the addition of 570 μl of chloroform:isoamyl alcohol (24:1) to the tube, and 10 min in a centrifuge at 13,000 rpm, genomic DNA was precipitated with 0.7 volume isopropanol. After centrifuging for 10 min at 13,000 rpm, the genomic DNA pellet was washed with 75% ethanol and then dried at room temperature. The genomic DNA was dissolved in 20 μl sterilized dH2O and stored at −20°C.
The ITS region of ribosomal DNA, GAPDH gene, RPB2 gene, and HIS3 gene were amplified using ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′)/ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990), gpd1 (5′-CAACGGCTTCGGTCGCATTG-3′)/gpd2 (5′-GCCAAGCAGTTGGTTGTGC-3′) (Berbee et al., 1999), RPB2-5F2 (5′-GGGGWGAYCAGAAGAAGGC-3′)/ fRPB2-7cR (5′-CCCATRGCTTGTYYRCCCAT-3′) (Liu et al., 1999), and H3-1a (5′-ACTAAGCAGACCGCCCGCAGG-3′)/H3-1b (5′-GCGGGCGAGCTGGATGTCCTT-3′) (Glass and Donaldson, 1995) primers, respectively. Polymerase chain reaction (PCR) was performed in a 20 μl reaction mixture containing 0.1 μl FIREPol DNA Polymerase (0.5 units) (Solisbiodyne, Tartu, Estonia), 1 μl each primer (10 μM), 0.2 μl dNTP mix (200 μM), 2 μl 10× buffer, 1 μl template (approximately 100 ng), 4 μl 5× Q-solution, and 10.7 μl sterilized dH2O. Amplifications were performed with an initial denaturation step at 94°C for 3 min followed by 35 cycles of 15 s at 94°C, 15 s at 53°C and 60 s at 72°C for ITS region, 35 cycles of 15 s at 94°C, 15 s at 57°C and 60 s at 72°C for GAPDH, 35 cycles of 15 s at 94°C, 15 s at 56°C and 2 min at 72°C for RPB2, 35 cycles of 15 s at 94°C, 15 s at 67°C and 60 s at 72°C for HIS3, and final extension at 72°C for 7 min. Amplified fragments were loaded on an agarose gel (0.8% w/v or 1.5% w/v) stained with RedSafe Nucleic Acid Staining Solution (20,000×) (Intronbio, Seongnam, Korea) and visualized under UV light.
The morphology of Alternaria isolates was examined as previously described (Andersen et al., 2001; Ma et al., 2021) with minor modification. For colony type, color, margin, and diameter (Andersen et al., 2001), Alternaria isolates were incubated on PDA and V8 juice agar (200 ml/l Campbell V8 juice, 3 g CaCO3, 15 g agar, and 800 ml dH2O) plates at 28°C dark condition for seven days. For the pattern of sporulation (Ma et al., 2021), the isolates were cultured at 22°C on PDA and V8 juice agar plates under UV light with light/dark cycle of 12 h photoperiod for seven days. The sporulation patterns and spore were examined using an inverted microscope Eclipse Ti (Nikon, Tokyo, Japan) fitted with an objective (200×).
The PCR amplification products of ITS region, HIS3, GAPDH, and RPB2 were purified with FavorPrep GEL/PCR Purification Kit (Favorgen, Ping-Tung, Taiwan) and sequenced by Sanger sequencing (Macrogen, Seoul, Korea). Sequence similarity searches were performed using the nucleotide BLAST at National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi). DNA sequences of ITS region, HIS3, and concatenated (GAPDH and RPB2) gene were aligned through ClustalW and refined manually using MEGA v.7.0 (Kumar et al., 2016). Maximum likelihood based on the Tamura-Nei model (Tamura and Nei, 1993) analyses of ITS region, HIS3, and concatenated (GAPDH and RPB2) gene datasets were performed using MEGA v.7.0 with 500 bootstrap replicates. Branches corresponding to partitions were reproduced in less than 50%. The phylogenetic trees were viewed in MEGA v.7.0. All analyzed DNA sequences, ITS region, HIS3, GAPDH, and RPB2, from isolates were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and their accession numbers are listed in Tables 1 and 2.
Alternaria inoculation was performed according to the previously described detached-leaf inoculation method (Pryor and Michailides, 2002) with minor modification. Briefly, to prepare the inoculum, Alternaria isolates were pure-cultured on PDA plates at 28°C in the dark for seven days and each mycelial plug was cut from the actively growing margin of the fungal colonies. Inoculations were conducted on detached leaves of watermelon cultivar, ‘Charleston gray’ (Wu et al., 2019). Fully expanded leaves were collected from six-to seven-week-old watermelon plants grown in a growth chamber. For the inoculation of each Alternaria isolate, six leaves from ‘Charleston gray’ were placed on papers soaked with distilled water in plastic boxes (25 × 19 × 5 cm [length × width × height]) and three mycelial plugs per leaf were placed on the detached leaves. The plastic boxes were individually covered with transparent plastics to maintain 100% humidity and incubated at 25°C with a 12 h photoperiod per day for seven days. The disease index was calculated after each disease area was divided by the whole leaf area using ImageJ. Alternaria isolates were re-isolated consistently from all the inoculated leaf to fulfill Koch’s postulates
Watermelon leaves and stems showing Alternaria-induced leaf blight lesions were collected from 14 fields in nine cities of Korea for Alternaria spp. survey (Table 1, Fig. 1A). Multiple fungal colonies were obtained from each symptomatic tissue (Fig. 1B), and only those with Alternaria morphologies were selected as isolates. A total of 16 Alternaria isolates were obtained and cultured to homogeneity. All isolates developed loosely cottony and light brown to grey colonies on PDA after incubation at 28°C for seven days in the dark. (Supplementary Fig. 1).
It has been reported that the amplicon lengths of the ITS regions varied among the Alternaria genus (Blagojević et al., 2020; Woudenberg et al., 2013). The ITS regions of Alternaria isolates were amplified with primers, ITS1 and ITS4. However, there was no length variation and sequence diversity in the ITS region throughout all isolates, except for one single-nucleotide substitution (T to C) in isolate CY202-29 (Supplementary Fig. 2A and B). Phylogenetic analyses based on the ITS region was able to divide seven references retrieved from GenBank into two different clades, A. cucumerina and A. infectoria. However, our isolates and many other references were not successful in dividing into clades and all were grouped into one clade containing A. alternata, A. tenuissima, and A. gaisen (Supplementary Fig. 3). These results indicate that the ITS region cannot be used for assaying genetic diversity in the Alternaria spp. causing leaf blight on watermelons in Korea.
Partial coding sequence of HIS3 was successfully used to divide Alternaria into subgroups of based on size analysis of PCR amplicons (Zhao et al., 2016a, 2016b; Zheng et al., 2015). After PCR analysis using a primer set, H3-1a and H3-1b, all 16 Alternaria isolates were successfully divided into three subgroups (Sub I, II, and III) based on their sizes (Table 2, Fig. 2). The largest amplicon with 546 bp was produced by Sub I, consisting of CY202-11, CY202-16, CY202-25, GC191-1, GC191-14, GC192-7, GJ192-1, GJ192-2, IC191-1, and YI201-2. The second largest one with 486 bp was amplified by Sub II GC192-13. Sub III producing the smallest amplicon with 440 bp was the dominant isolates, consisting of CJ191-1, CJ191-3, CY202-18, CY202-29, and IC191-2. Two isolates from dominant Sub I (CY202-11 and GC191-1) and one isolate from each Sub II (GC192-13) and Sub III (CJ191-1) were randomly selected for further analysis.
Four representative isolates, CY202-11, GC191-1, GC192-13, and CJ191-1, from three subgroups were subjected to morphological analysis on PDA and V8 juice agar plates. On the V8 juice agar plate, all isolates produced loosely cottony and dark grey colonies after incubation at 28°C in dark condition for seven days. On PDA plates, three isolates, CY202-11, GC191-1, and GC192-13, in Sub I and Sub II developed cotton light brown colonies, while CJ191-1 from Sub III produced a cotton dark grey colony (Table 3, Fig. 3A). In all isolates, conidiophores were short and arising singly (Fig. 3B). The conidia of them were mainly short ovoid to obclavate with a rounded apical cell, with no significant difference among the four representative isolates. A similar number of transverse septa and longitudinal septa of their conidia were also observed from 1 to 6 and from 0 to 2, respectively. All isolates had a similar size of conidiophores but slightly differed in sporulation on PDA and V8 juice agar (Table 4).
To examine whether the three subgroups can be identified into the Alternaria species, their HIS3 sequences were compared (Fig. 4, Supplementary Fig. 4). As a reference for sequence comparison, the HIS3 gene (accession no. XM_018526023) of an A. alternata isolate was used. Its full-length cDNA is 411 bp in length and the cDNA fragment amplified with primers H3-1a and H3-1b is 378 bp. In the analysis of Sub I isolates, CY202- 11 and GC191-1, the HIS3 gDNA amplicon was 546 bp in length and contained three introns 54 bp (Int A), 62 bp (Int B), and 52 bp (Int C) (Fig. 3). The HIS3 gDNA amplicon from GC192-13 isolate in Sub II was 484 bp with two introns 54 bp (Int A) and 52 bp (Int C). CJ191-1 isolate in Sub III was 440 bp in length with only one intron 62 bp (Int B).
Phylogenetic analyses based on HIS3 was performed together with available reference sequences retrieved from GenBank (eight from A. alternata. eight from A. tenuissima, one from A. gaisen, four from A. infectoria, three from A. cucumerina, two from Alternaria sp., and one outgroup Fusarium proliferatum) (Fig. 5). Sub I isolates, CY202-11 and GC191-1, were located in one clade with the known A. tenuissima from GenBank. Sub II GC192-13 isolate was together with A. gaisen and Sub III CJ191-1isolate was located in the A. alternata clade. Occurrence rates of each Alternaria sp. were relatively high with quite wide range depending on where the isolates were harvested (Supplementary Table 1). Including A. infectoria references and A. cucumerina references, the entire species-group within the Alternaria spp. was successfully split into five clades, suggesting that HIS3 can be used for classifying Alternaria spp. causing leaf blight on watermelon.
The amplicon lengths of GAPDH and RPB2 varied among some of Alternaria spp. associated with leaf spot and leaf blight disease in Solanaceae and Brassica (Bessadat et al., 2020; Blagojević et al., 2020; Lawrence et al., 2016). Although the sequences of GAPDH and RPB2 were not identical among four isolates and references retrieved from GenBank, none of the variations was specific for the Alternaria spp. (Supplementary Fig. 5). Therefore, four isolates and the reference Alternaria spp. were not in separate clades base on the phylogenetic tree of GAPDH and RPB2.
The pathogenicity of the four representatives isolates from three Alternaria species, A. tenuissima, A. gaisen, and A. alternata, was evaluated on a watermelon cultivar, ‘Charleston gray’ using the detached-leaf inoculation method (Pryor and Michailides, 2002). About seven days after inoculation, dark brown lesions around agar disks of the Alternaria spp. appeared on the detached leaves (Fig. 6), similar to those observed on naturally infected leaf samples. No symptoms were observed in the control detached leaves treated with agar disk only. The average disease incidence and average disease index caused by A. tenuissima, A. gaisen, and A. alternata ranged from 42.9% to 100% and 39.4% to 68.7%, respectively (Table 5). The Alternaria spp. were re-isolated consistently from all the leaf lesions to fulfil Koch’s postulates, while no Alternaria isolates were re-isolated from the control leaves.
In this study, we describe the occurrence and identification of the Alternaria spp. obtained from watermelon plants displaying leaf blight lesions. The identified species were A. tenuissima, A. gaisen, and A. alternata, and all of them displayed pathogenicity on a watermelon cultivar, ‘Charleston gray’. Although A. cucumerina has been reported as the prevalent species causing leaf blight on Cucurbitaceae including watermelon and muskmelon in Korea and China (Jackson and Weber, 1959; Kim et al., 1994; Liu et al., 2010; Ma et al., 2021; Zhao et al., 2016a, 2016b), this was not obtained in our study. Similarly, no A. cucumerina isolates were recovered from watermelon and muskmelon in the Beijing municipality of China (Zhao et al., 2016a, 2016b). Instead, A. tenuissima and A. alternata were reported as the predominant species of Alternaria leaf blight of watermelon in China (Ma et al., 2021; Zhao et al., 2016a, 2016b). These previous reports are consistent with our observation that 10 and five among the 16 Alternaria isolates were identified as A. tenuissima and A. alternata, respectively. These results suggest that the composition of the Alternaria species may have changed and become more complex, probably due to recent breeding programs for disease resistant cultivars.
Recently, A. gaisen was reported as new Alternaria sp. causing watermelon leaf blight in China (Ma et al., 2021). In our study, although there was only one isolate, A. gaisen was also obtained from Gochang city. These results suggest a possibility that A. gaisen is becoming one of casual agents of watermelon leaf blight worldwide. A. gaisen had been reported as a causal agent of black spot disease in Japanese pears (Pyrus pyrifolia) previously discovered only in Japan, Korea, and China (Simmons and Roberts, 1993). However, now, it has been discovered in various hosts-wintersweet (Chimonanthus praecox), rice (Oryzae sativa), gerbera daisy (Gerbera jamesonii), and watermelon-of many countries including France, Australia, China, Pakistan, and the US (Akhtar et al., 2014; Baudry et al., 1993; Ma et al., 2021; Perveen et al., 2018; Tian et al., 2020). Additional study will be necessary to determine whether A. gaisen can be another predominant species of Alternaria leaf blight of watermelon in Korea.
Information on the species of plant pathogens is necessary for disease management because different species often display different degrees of resistance to fungicides (Iacomi-Vasilescu et al., 2004). Morphological trait analysis has been the common method to identify and differentiate Alternaria species (Simmons and Roberts, 1993). However, previous studies indicate that morphological characters were often uninformative and insufficient to accurately distinguish Alternaria species (Pryor and Michailides, 2002; Zheng et al., 2015). To complement morphological trait analysis, phylogenetic analysis after combining DNA sequences has been suggested (Kang et al., 2001). Previously, the Alternaria species had been successfully differentiated using the ITS region, GAPDH, and RPB2 (Bessadat et al., 2020; Blagojević et al., 2020; Lawrence et al., 2016; Woudenberg et al., 2013). However, our results suggested that each of them and the combination of their sequences were not competent to differentiate the Alternaria spp. causing watermelon leaf blight in Korea. On the other hand, PCR amplification and sequence analysis of the HIS3 gene successfully differentiated Alternaria isolates. The PCR products amplified from A. tenuissima, A. gaisen, and A. alternata differed in size, depending on the presence and type of introns. Therefore, we recommend the analysis of the sequences of HIS3 rather than the ITS region, GAPDH, and RPB2 to identify Alternaria species causing leaf blight on watermelons in Korea.
Acknowledgments
This study was carried out with the support of the “Cooperative Research Program for Agriculture Science & Technology Development [Project No. PJ01421303 (C.J.P) and No. PJ01421302 (G.P.L)]” Rural Development Administration, Republic of Korea.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Watermelon farm locations and representative leaf blight symptoms on leaves. (A) Map showing nine cities in Korea. (B) Typical Alternaria leaf blight displaying discolored and necrotic symptoms on watermelon leaves.
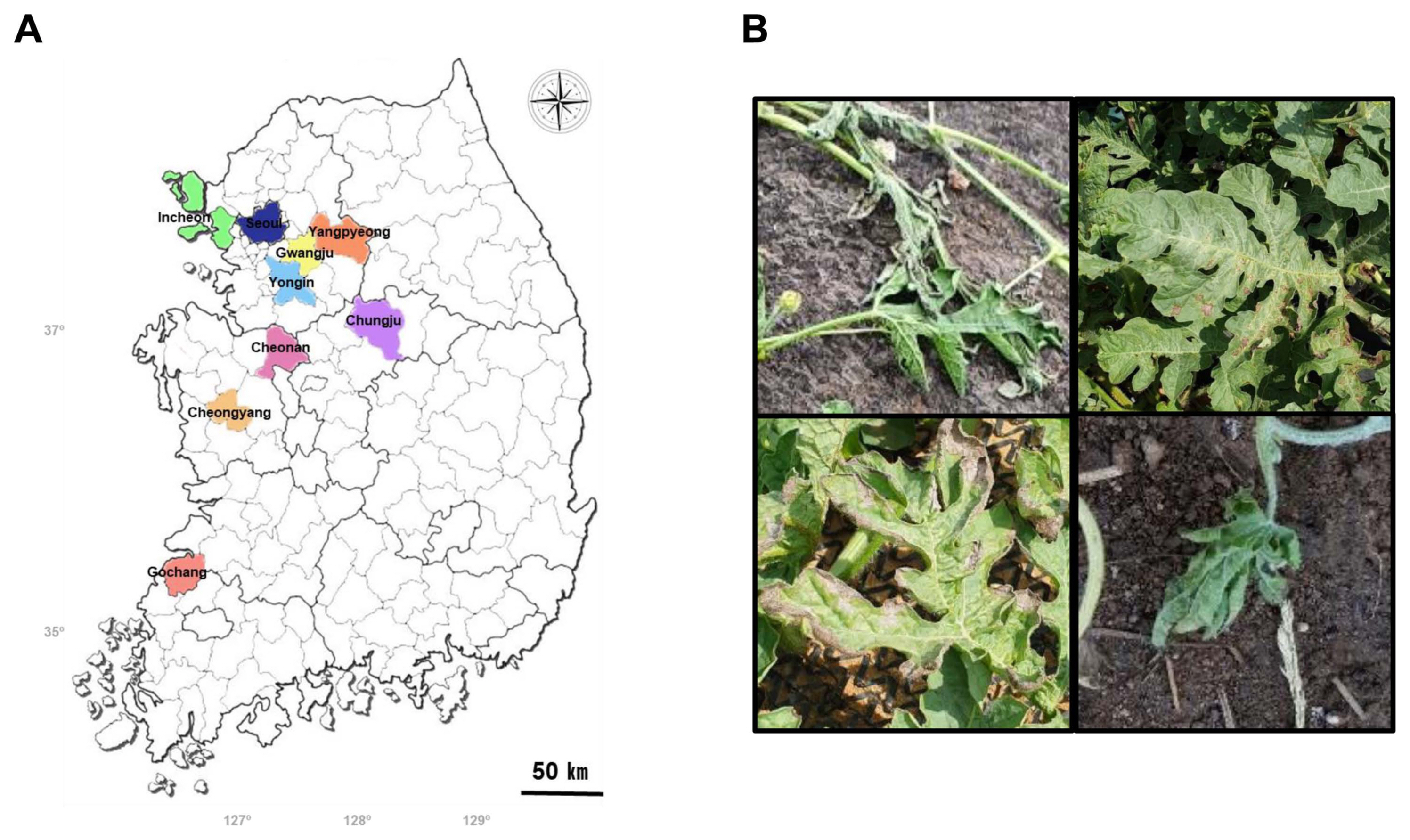
Fig. 2
Analysis of histone H3 gene amplicons from 16 Alternaria isolates. Polymerase chain reaction (PCR) amplicons with primers H3-1a and H3-1b were analyzed by gel electrophoresis. All the isolates were divided into three subgroups based on the PCR amplicon sizes. Subgroup I: CY202-11, CY202-16, CY202-25, GC191-1, GC191-14, GC192-7, GJ192-1, GJ192-2, IC191-1, and YI201-2; Subgroup II: GC192-13; Subgroup III: CJ191-1, CJ191-3, CY202-18, CY202-29, and IC191-2.
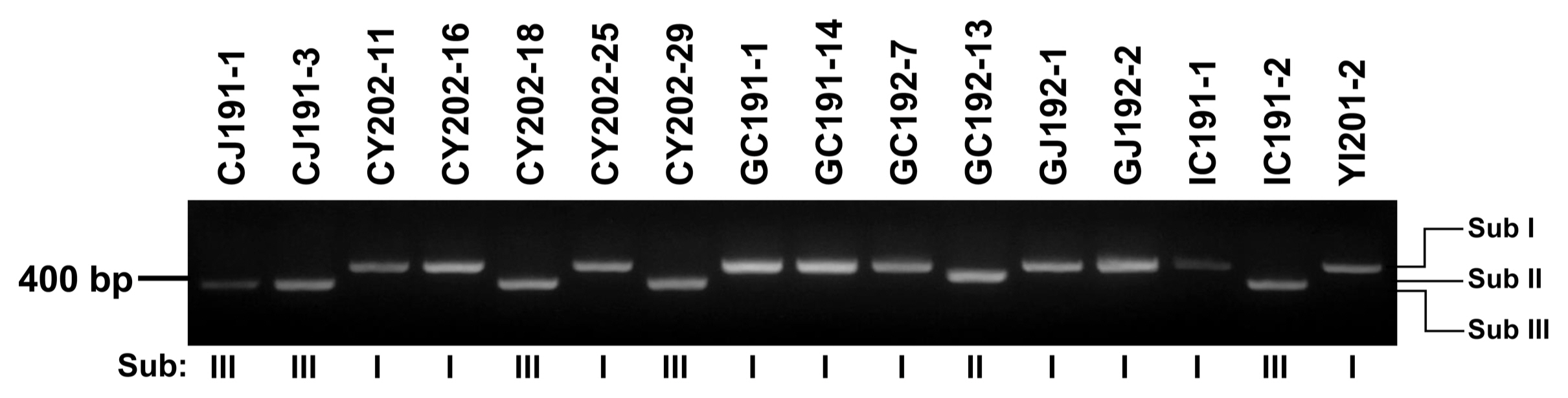
Fig. 3
Colonies of four Alternaria isolates. (A) Morphological characteristics of CY202-11, GC191-1, GC192-13, and CJ191-1 observed on different media, potato dextrose agar (PDA) and V8 juice agar (front and revers). (B) Conidial morphology of CY202-11, GC191-1, GC192-13, and CJ191-1 on PDA media. Scale bars = 50 μm.
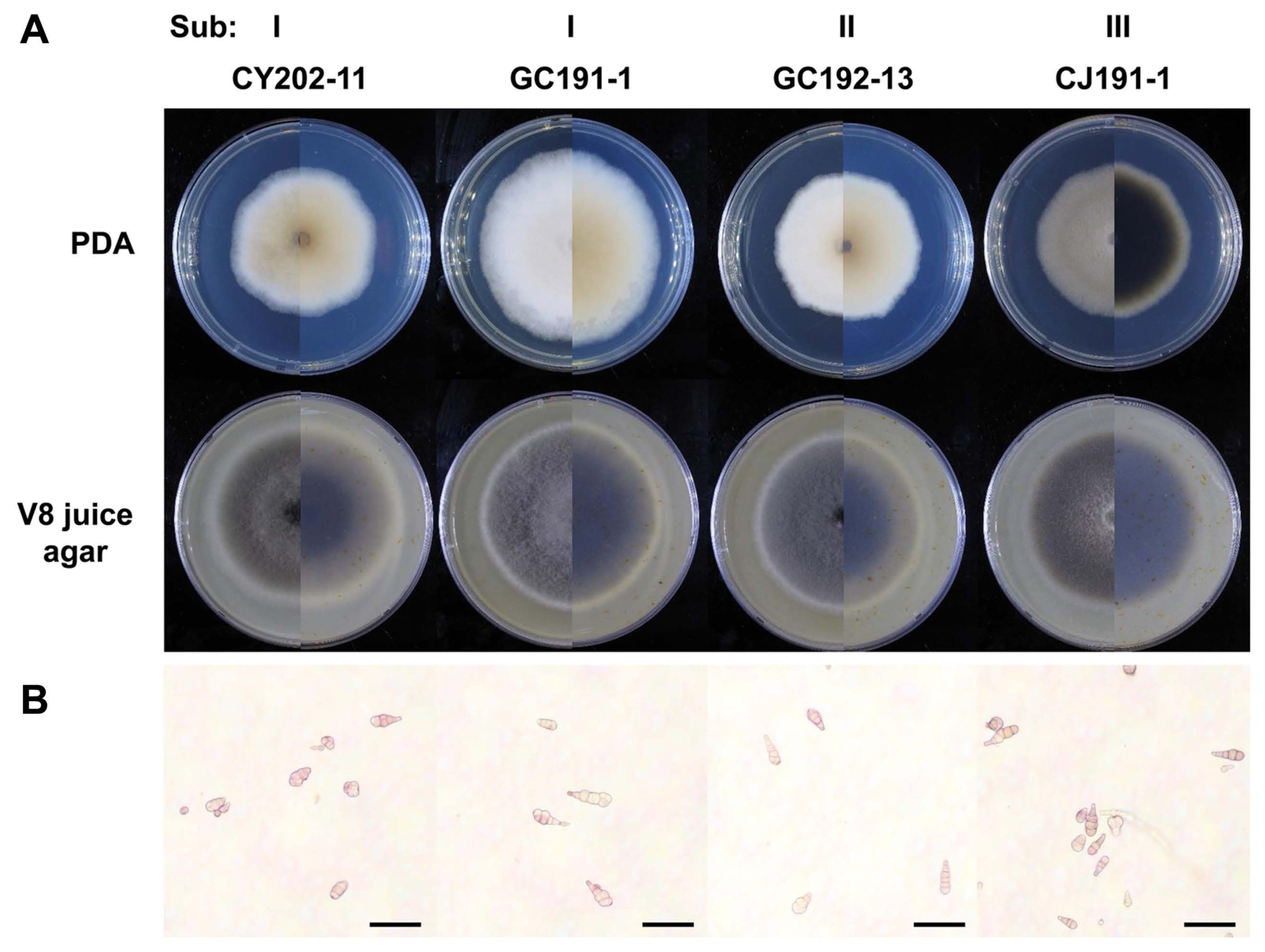
Fig. 4
Schematic of histone H3 gene in in Subgroup I, II, and III. Solid lines indicate introns. Intron (Int) A, B, and C are 54 bp, 62 bp, and 52 bp in size, respectively. Gray and white rectangles indicate cloned and noncloned histone H3 exons, respectively. Primers, H3-1a and H3-1b, used for the cloning of the exons were indicated as arrows in Alternaria Subgroup I, II, and III. Coding region sequence of histone H3 inferred from Alternaria alternata (GenBank accession no. XM_018526023) was used as a reference for this comparison.
Fig. 5
Maximum likelihood tree constructed based on histone H3 of four Alternaria isolates and 27 reference sequences retrieved from GenBank. Subgroup I (CY202-11 and GC191-1), II (GC192-13), and III (CJ191-1) were grouped into A. tenuissima, A. gaisen, and A. alternata, respectively. Red dots indicate Alternaria spp. isolated in this study. Bootstrap values are shown below branches. The bar indicates nucleotide substitutions per site.

Fig. 6
Pathogenicity of four representative isolates of three Alternaria species on detached watermelon leaves. Three agar disks growing each Alternaria species were placed on the detached of ‘Charleston gray’. Images were obtained seven days after inoculation. (A) Control with agar disks only. (B) A. tenuissima (CY202-11). (C) A. tenuissima (GC191-1). (D) A. gaisen (GC192-13). (E) A. alternata (CJ191-1). Scale bars = 2 cm.

Table 1
Origins of Alternaria isolates and GenBank accession numbers of ITS region
Table 2
Subgroups of Alternaria isolates and GenBank accession numbers of HIS3, GAPDH, and RPB2
Table 3
Phenotypic characteristics of four isolates on different media, PDA and V8 juice agar
Table 4
Spore size and sporulation of four Alternaria isolates
| Sub-group | Isolate | Sporulation (per ml)a | Spore size | ||
|---|---|---|---|---|---|
|
|
|||||
| PDA | V8 juice agar | Length (μm) | Width (μm) | ||
| I | CY202-11 | +++ | +++ | 12.5-32.4 (23.3 ± 6.7) | 7.2-14.4 (10.8 ± 2.4) |
| I | GC191-1 | + | ++ | 15.7-48.4 (27.4 ± 9.7) | 7.9-16.2 (10.0 ± 3.5) |
| II | GC192-13 | +++ | +++ | 16.2-43.4 (27.7 ± 7.9) | 6.9-18.1 (11.0 ± 3.1) |
| III | CJ191-1 | + | ++ | 14.6-35.1 (24.0 ± 5.9) | 4.8-15.4 (10.1 ± 2.9) |
Table 5
Disease incidence and disease index of the four Alternaria isolates on detached watermelon leaves
References
Akhtar, N, Hafeez, R and Awan, ZA 2014. First report of rice leaf spot by Alternaria gaisen from Pakistan. Plant Dis. 98:1440.

Andersen, B, Krøger, E and Roberts, RG 2001. Chemical and morphological segregation of Alternaria alternata, A. gaisen and A. longipes
. Mycol Res. 105:291-299.

Baudry, A, Morzières, JP and Larue, P 1993. First report of Japanese pear black spot caused by Alternaria kikuchiana in France. Plant Dis. 77:428.

Berbee, ML, Pirseyedi, M and Hubbard, S 1999.
Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia. 91:964-977.

Bessadat, N, Hamon, B, Bataille-Simoneau, N, Mabrouk, K and Simoneau, P 2020.
Alternaria telliensis sp. nov., a new species isolated from Solanaceae in Algeria. Phytotaxa. 440:89-100.

Blagojević, JD, Vukojević, JB and Ivanović, ŽS 2020. Occurrence and characterization of Alternaria species associated with leaf spot disease in rapeseed in Serbia. Plant Pathol. 69:883-900.

Burdman, S and Walcott, R 2012.
Acidovorax citrulli: generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol Plant Pathol. 13:805-815.



Chen, D-H and Ronald, PC 1999. A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol Biol Rep. 17:53-57.
Chopra, BL, Jhooty, JS and Bajaj, KL 1974. Biochemical differences between two varieties of watermelon resistant and susceptible to Alternaria cucumerina
. J Phytopathol. 79:47-52.

Food and Agriculture Organization of the United Nations. 2018 Food and Agriculture Organization of the United Nations Online Database. URL http://www.fao.org/faostat/en/#data [5 February 2021 [.
Glass, NL and Donaldson, GC 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 61:1323-1330.



Iacomi-Vasilescu, B, Avenot, H, Bataillé-Simoneau, N, Laurent, E, Guénard, M and Simoneau, P 2004. In vitro fungicide sensitivity of Alternaria species pathogenic to crucifers and identification of Alternaria brassicicola field isolates highly resistant to both dicarboximides and phenylpyrroles. Crop Prot. 23:481-488.

Ismail, SI and Abd Razak, NF 2021. First report of Nigrospora sphaerica causing leaf spot on watermelon (Citrullus lanatus) in Malaysia. Plant Dis. 105:488.

Jackson, CR and Weber, GF 1959. Morphology and taxonomy of alternaria cucumerina. Mycologia. 51:401-408.

Kang, J-C, Crous, PW and Schoch, CL 2001. Species concepts in the Cylindrocladium floridanum and Cy. spathiphylli complexes (Hypocreaceae) based on multi-allelic sequence data, sexual compatibility and morphology. Syst Appl Microbiol. 24:206-217.


Kim, WG, Cho, WD, Lee, YH and Yu, SH 1994. Leaf blight of watermelon caused by Alternaria cucumerina
. Korean J Plant Pathol. 10:245-248.
Kumar, S, Stecher, G and Tamura, K 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870-1874.



Lawrence, DP, Rotondo, F and Gannibal, PB 2016. Biodiversity and taxonomy of the pleomorphic genus Alternaria
. Mycol Prog. 15:3.


Liu, YJ, Whelen, S and Hall, BD 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol. 16:1799-1808.


Liu, ZH, LüBin Zhao, T-C, Yang, H, Liu, F, Sun, J and Han, XY 2010. Biological characteristics of the pathogenic fungus causing leaf spot disease of watermelon. J Shenyang Agric Univ. 41:161-164.
Ma, G, Bao, S, Zhao, J, Sui, Y and Wu, X 2021. Morphological and molecular characterization of Alternaria species causing leaf blight on watermelon in China. Plant Dis. 105:60-70.


Nishikawa, J and Nakashima, C 2019. Morphological and molecular characterization of the strawberry black leaf spot pathogen referred to as the strawberry pathotype of Alternaria alternata
. Mycoscience. 60:1-9.

Noh, J, Kim, J-H, Lim, JH, Kim, TB, Seong, MH, Jung, GT, Kim, JM, Cheong, S-S, Oh, NK and Lee, W-H 2014. Occurrence of diseases and case of clinical diagnosis on watermelon in South Korea,2008-2012. Res Plant Dis. 20:8-14.

Perveen, S, Akhtar, N and Nayab, M 2018.
Alternaria gaisen: a new pathogen causing leaf spot of Gerbera jamesonii from Pakistan. Mycopath. 16:7-10.
Pryor, BM and Michailides, TJ 2002. Morphological, pathogenic, and molecular characterization of Alternaria isolates associated with alternaria late blight of pistachio. Phytopathology. 92:406-416.


Simmons, EG 2007.
Alternaria: an identification manual. CBS Biodiversity Series 6. CBS Fungal Biodiversity Centre, Utrecht, The Netherlands. 775.
Simmons, EG and Roberts, RG 1993.
Alternaria themes and variations (73). Mycotaxon. 48:109-140.
Tamura, K and Nei, M 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512-526.

Tian, Y, Qiu, CD, Zhang, YY and Liu, ZY 2020. First report of Alternaria gaisen causing leaf blight on wintersweet (Chimonanthus praecox) in China. Plant Dis. 104:977.

White, TJ, Bruns, T, Lee, S and Taylor, J 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols a guide to methods and applications, eds. by MA Innis, DH Gelfand, JJ Sninsky and TJ White, 315-322. Academic Press, San Diego, CA, USA.

Woudenberg, JHC, Groenewald, JZ, Binder, M and Crous, PW 2013.
Alternaria redefined. Stud Mycol. 75:171-212.



Wu, S, Wang, X, Reddy, U, Sun, H, Bao, K, Gao, L, Mao, L, Patel, T, Ortiz, C, Abburi, VL, Nimmakayala, P, Branham, S, Wechter, P, Massey, L, Ling, K-S, Kousik, C, Hammar, SA, Tadmor, Y, Portnoy, V, Gur, A, Katzir, N, Guner, N, Davis, A, Hernandez, AG, Wright, CL, McGregor, C, Jarret, R, Zhang, X, Xu, Y, Wehner, TC, Grumet, R, Levi, A and Fei, Z 2019. Genome of ‘Charleston Gray’, the principal American watermelon cultivar, and genetic characterization of 1,365 accessions in the U.S. National Plant Germplasm System watermelon collection. Plant Biotechnol J. 17:2246-2258.



Zhao, J, Bao, S, Ma, G and Wu, X 2016a. Characterization of Alternaria species associated with muskmelon foliar diseases in Beijing municipality of China. J Gen Plant Pathol. 82:29-32.

Zhao, J, Bao, SW, Ma, GP and Wu, XH 2016b. Characterization of Alternaria species associated with watermelon leaf blight in Beijing municipality of China. J Plant Pathol. 98:135-138.



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




