 |
 |
| Plant Pathol J > Volume 37(6); 2021 > Article |
|
Abstract
Bacteriophages infecting Acidovorax citrulli, the causal agent of bacterial fruit blotch, have been proven to be effective for the prevention and control of this disease. However, the occurrence of bacteriophage-resistant bacteria is one of hurdles in phage biocontrol and the understanding of phage resistance in this bacterium is an essential step. In this study, we aim to investigate possible phage resistance of A. citrulli and relationship between phage resistance and pathogenicity, and to isolate and characterize the genes involved in these phenomena. A phage-resistant and less-virulent mutant named as AC-17-G1 was isolated among 3,264 A. citrulli Tn5 mutants through serial spot assays and plaque assays followed by pathogenicity test using seed coating method. The mutant has the integrated Tn5 in the middle of a cupin protein gene. This mutant recovered its pathogenicity and phage sensitivity by complementation with corresponding wild-type gene. Site-directed mutation of this gene from wild-type by CRISPR/Cas9 system resulted in the loss of pathogenicity and acquisition of phage resistance. The growth of AC-17-G1 in KingŌĆÖs B medium was much less than the wild-type, but the growth turned into normal in the medium supplemented with D-mannose 6-phosphate or D-fructose 6-phosphate indicating the cupin protein functions as a phosphomannos isomerase. Sodium dodecyl sulfa analysis of lipopolysaccharide (LPS) extracted from the mutant was smaller than that from wild-type. All these data suggest that the cupin protein is a phosphomannos isomerase involved in LPS synthesis, and LPS is an important determinant of pathogenicity and phage susceptibility of A. citrulli.
Bacteriophages, the most abundant entity in all microbial environments, were discovered by dŌĆÖHerelle (1926), and were proposed for use as antibacterial agents. The use of phages to infect Vibrio cholera and other human pathogens were among the early studies of phages (Marks and Sharp, 2000). In addition to human pathogens, phages were investigated for the control of plant diseases in the pre-antibiotic era. For example, Coons and Kotila (1925) showed that phage application could prevent Pectobacterium atrosepticum from causing soft rot in potato tubers. However, most commercial use of phages was replaced with the introduction of antibiotics in the 1920s. Since then, awareness and consideration of phages has increased for three main reasons: widespread application of antibiotics, development of resistant bacteria, and the need for new antibacterial agents against antibiotic-resistant. Therefore, numerous studies have focused on the use of phages against pathogenic bacteria, and phages were recently reconsidered as an alternative solution for control (Park et al., 2018; Vu and Oh, 2020). However, similar to other biocontrol strategies, several factors limit bacteriophage biocontrol. Most of these factors are related to the nature of bacteriophages. For example, to successfully apply bacteriophages under phyllosphere conditions, factors such as temperature, ultraviolet radiation, and pH must be optimized.
In addition to these limitations, which are generally related to bacteriophage structure, the occurrence of bacteriophage-resistant bacteria is a major issue in the development of bacteriophages as a control agent (Hassan et al., 2021). Coevolution of bacteriophages and bacteria occurs constantly through spontaneous mutations (Brockhurst et al., 2021) that allow bacteria to structurally change the composition of their cell wall, capsule, lipopolysaccharide (LPS), and flagella, which act as receptors for phages. For example, LPS alteration and OmpC surface protein silencing were found to be the two main factors associated with phage resistance in Escherichia coli (Pirhonen et al., 1988). Therefore, studies on phage resistance mechanisms and detailed analysis of the interactions between phages and bacteria are necessary to increase the chance of successful long-term application of phages.
The potential mechanisms through which bacteria become resistant to phages were previously found to be dependent on genotype (Barbu et al., 2016). Immunity of bacteria against bacteriophages may involve various mechanisms, such as mutations, restriction modification systems, and adaptive immunity via the CRISPR/Cas system. For example, the type I CRISPR/Cas system, which is abundant in bacteria, provides protection against the virulent phages ╬”TE and ╬”M1 in P. atrosepticum (Watson et al., 2019). Bacteriophage-resistant bacteria increase rapidly, as phage-susceptible genotypes are killed by phages in the same environment. A population of bacteria that has rapidly acquired resistance may have either positive or negative implications for the host. For example, diarrhea-causing E. coli showed decreased virulence after becoming resistant to bacteriophages. By contrast, Enterococcus faecalis resistant to bacteriophage ╬”VPE25 was able to colonize the intestine, in addition to growing faster than the wild-type in the intestine (Chatterjee et al., 2020). Therefore, understanding bacteriophage-host interaction is essential prior to application of phages.
Acidovorax citrulli (previously known as Acidovorax avenae subsp. citrulli) is a gram-negative bacterium that causes bacterial fruit blotch (BFB) in Cucurbitaceae. A. citrulli strains have been divided into groups, with strains in group I showing virulence against plants in the Cucurbitaceae family other than watermelon, and group II strains primarily affecting watermelon. Like other plant pathogenic bacteria, A. citrulli disease relies on the type III secretion system that is responsible for secretion of effector proteins and mutation in the genes encoding type III secretion system and type IV secretion system affected virulence, motility, and biofilm formation of A. citrulli (Eckshtain-Levi et al., 2014; Tian et al., 2015). In addition to these secretion systems, several genes involved in pathogenicity of A. citrulli have been identified in the analysis of Tn5 mutants with significantly decreased the virulence but the detailed role of each gene remains unclear (Luo et al., 2015).
Despite the economic importance of A. citrulli, effective chemical control agents are limited and resistant cultivars are not available yet. Use of bacteriophages against A. citrulli has been proposed and studied since 2016, after the initial isolation of phage ACP17 (Rahimi-Midani et al., 2018), the issue of phage resistance has not been investigated yet. As phage-host interactions can provide insights into the bacterial genome and possibly pathogenic behavior, understanding such interactions is essential. Therefore, in this study, we aimed to identify the nature of bacteriophage-resistant in A. citrulli and any gene involved in this characteristic. Also, the relationship between phage resistance and pathogenicity was analyzed.
The A. citrulli mutant library of 3,264 strains was kindly provided by Professor Sang-Wook Han of Chung-Ang University, Republic of Korea. These mutants were constructed from the wild-type A. citrulli strain KACC17005 using the transposon mutation system EZ-Tn5 Transposome Kit (Lucigen, Madison, WI, USA) according to the manufacturerŌĆÖs instructions.
Bacteriophage ACPWH, isolated previously, was used to screen phageresistant mutants (Rahimi-Midani et al., 2020). Bacteria from the mutant library were grown in KingŌĆÖs B (KB) medium containing 100 ╬╝g/ml each of kanamycin and rifampicin overnight at 37┬░C. Then, 300 ╬╝l of bacterial culture was mixed with 100 ╬╝l 0.7% agar in KB medium and poured into each well of a 48-well culture plate for spot assay. After hardening, 3 ╬╝l of bacteriophage ACPWH (titer, 106 pfu/ml) was spotted onto each well and the plates were incubated at 28┬░C for 24 h. Mutants resistant to bacteriophage ACPWH, which showed no clearing in the spot assay, were selected; the plaque assay was repeated three times to confirm resistance in the selected mutant.
In total, 39 strains of A. citrulli mutant resistant to phage ACPWH were used for pathogenicity test by seed coating of watermelon seeds as described previously (Rahimi-Midani et al., 2020). Briefly, commercial seeds were washed with 1% sodium hypochlorite for 20 min. After washing, the seeds were mixed with a suspension of bacterial mutants (105 cfu/ml, in phosphate-buffered saline). Nine seeds coated with each bcterium were sown in three plastic pots of 12cm diameter (3 seeds per pot) and maintained for 2 weeks, during which germination data were recorded. Screening was conducted in triplicate.
Gene identification was carried out using the rescue cloning method, as described in the manual of the EZ-Tn5 Tnp Transposome Kit. Briefly, bacteria were grown in 100 ml KB broth medium containing kanamycin (100 ╬╝g/ml). Bacterial DNA was extracted using the AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). One microgram of total extracted DNA was treated with EcoRI enzyme and incubated for 1 h at 37┬░C. Digested DNA was ligated with the EcoRI adaptor (GGATCCGCGGCC) and PCR was conducted using the EcoRI adaptor primer (GGATCCGCGGCCNN) under the following conditions: 95┬░C for 5 min; 32 cycles of 94┬░C for 20 s, annealing for 20 s at 52┬░C, and extension at 72┬░C for 20 s; followed by an extension phase at 72┬░C for 3 min, as described previously (Doherty et al., 1999). The PCR product was used for direct sequencing without cloning.
The gene encoding a cupin protein was amplified from wild-type A. citrulli using cupin protein gene promoter forward and reverse (F and R, respectively) primers, which are listed in Table 1. PCR was performed using rTaq plus (Elpisi-Biotech, Daejeon, Korea), as follows: 95┬░C for 5 min; 32 ycles of 94┬░C for 20 s, annealing for 20 s at 60┬░C, and extension at 72┬░C for 20 s; followed by an extension phase at 72┬░C for 3 min. DNA was purified using the Expin PCR SV kit (GeneAll Biotechnology, Seoul, Korea). The 1.3-kb PCR product was cloned into Takara T-Vector pMD9 (Takara Korea Biomedical Inc., Seoul, Korea) and used for complementation.
A phage-resistant and avirulent mutant strain was used for complementation with the cloned cupin protein gene. One hundred milliliters of exponential-phase bacterial culture (OD600 of ~0.6) was centrifuged at 2,800 ├Śg for 30 min. The pellet was resuspended in chilled 10% glycerol and centrifuged at 2,800 ├Śg for 30 min. Glycerol washing and centrifugation were repeated three times. The electrocompetent cells were resuspended in 10% glycerol and stored at ŌłÆ80┬░C. Plasmids were electroporated into the electrocompetent cells under the conditions of 2.0 kV, 200 ohms, and 25 ╬╝F using Gene Pulser II electroporator (Bio-Rad, Hercules, CA, USA), and recovered in super optimal broth with catabolite repression medium at 37┬░C with shaking for 1 h. Electroporated cells were plated onto KB agar plates containing 100 ╬╝g/ml of ampicillin. Transformation was confirmed through PCR with the M13 forward and reverse primers. Positive strains were used for the plaque assay and seed coating experiment. Thirty surface-sterilized watermelon seeds were coated with the avirulent mutant or the complemented strain. The seeds were sown in six pots measuring 15 cm ├Ś 8 cm ├Ś 12 cm per treatment, which were maintained for 20 days to record the germination rate.
In order to confirm the function of the cupin protein gene in pathogenicity and phage susceptibility, this gene was knocked from the wild-type A. citrulli by the CRISPR/Cas9 system. A single guide RNA (sgRNA) for the gene was designed using online tools (Broad Institute), which are listed in Table 2. This sgRNA was then ligated into the all-in-one vector (Addgene plasmid no. 78098, http://n2t.net/addgene:78098) using the site-directed mutagenesis (SDM) method, as described by Liu and Naismith (2008) (Supplementary Fig. 1) with the SDM primers listed in Table 1. The vector was then electrically transformed into wild-type A. citrulli as described above. The transformed bacteria were grown on KB medium containing ampicillin (100 ╬╝g) and selected colonies were grown on KB ampicillin medium, and used for spot assays, as described previously (Khan Mirzaei and Nilsson, 2015).
As one major group of bacterial cupin protein is the phosphomannose isomerase (PMI) that catalyzes the interconversion of D-fructose-6-phosphate (F6P) and D-mannose-6-phosphate (M6P), the growth of wild-type A. citrulli and the phage-resistant and avirulent mutant AC-17-G1 were compared in KB medium containing 5 mM each of M6P and F6P. The wild-type and mutant strains were cultured overnight in KB medium and KB medium containing kanamycin (40 ╬╝g/ml), respectively and were then inoculated into KB medium, KB medium containing kanamycin, and KB medium containing 5 mM each of M6P or F6P and incubated at 26┬░C. The optical density was recorded hourly. Plaque assay was conducted with phage ACPWH and mutant AC-17-G1 cultured in these mediums.
LPS was extracted from the wild-type and the mutant AC-17-G1 as described previously (Szermer-Olearnik and Boraty┼äski, 2015). Briefly, bacteria were grown overnight in KB medium and centrifuged at 2,800 ├Śg for 30 min. The pellet was resuspended in 55% hot water-phenol solution. The resulting mixture was incubated at 60┬░C for 1 h and centrifuged at 21,206 ├Śg for 10 min. The upper phase was removed and treated with DNase (100 ╬╝g/ml), RNase (50 ╬╝g/ml, Takara Korea), and proteinase K (100 ╬╝g/ml, Bioneer). The treated sample was subjected to precipitation in 100% ethanol, as described previously (Darveau and Hancock, 1983), and the extracted LPS was analyzed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
A total of 3,264 transposon mutant strains were used for the selection of mutants resistant to phage ACPWH, which can lyse the wild-type host A. citrulli KACC17005. Thirty-nine mutants showing no lysis in the spot assay were obtained and further screened for pathogenicity. Seeds coated with 37 mutants had no germination or germinated with severe symptoms resulting in death within a few days as the seeds coated with wild-type A. citrulli. Seeds coated with one mutant designated as AC-5-E1 showed a germination rate of 55% and less severe symptoms of BFB. By contrast, seeds coated with the mutant AC-17-G1 showed 88.8% germination with reduced or no BFB symptoms (Fig. 1A). Therefore, this mutant was used for the identification of genes involved in both phage susceptibility and pathogenicity (Fig. 1B).
DNA extracted from the phage-resistant less-virulent mutant AC-17-G1 was digested with EcoRI, which cut one site in the transposon gene and an unknown site near the integration site. An EcoRI linker was ligated to the digested DNA and amplified with a primer specific to the adaptor. A PCR product of almost 1 kb was obtained, and sequence analysis of this product showed that Tn5 was integrated into the cupin protein gene of A. citrulli KACC17005, as annotated to the genome sequence (Gen-Bank accession no. NZ_CP023687.1). This finding was further confirmed through PCR amplification with primers specific to Tn5 and the cupin protein gene, as shown in Fig. 2. Transposon is inserted between position 672 and 673 of the cupin protein open reading frame. The adenine residue of the AUG start codon and the last guanine residue of the TAG stop codon correspond to the nucleotide number 3094447 and 3095583 of NZ_CP023687.1, respectively. PCR with the cupin protein gene promoter F primer and transposon left-end R primer resulted in a DNA fragment of 1,155 bp (Fig. 2, lane F), while PCR with the transposon right-end F primer and cupin protein gene downstream R primer resulted in a DNA fragment of 874 bp (Fig. 2B, lane R). Sequence analysis of these PCR products confirmed that Tn5 is integrated 672 bp from the AUG codon of the cupin protein gene.
The phage-resistant and less-virulent mutant AC-17-G1 was complemented with the cupin protein gene from the wild-type. Transformed cells were selected on KB medium containing ampicillin, to which the wild-type is sensitive. The presence of introduced DNA was confirmed through PCR with the primers M13 F and m13 R, and a DNA fragment of the expected size was detected. The presence of the wild-type cupin protein gene was further confirmed through PCR with cupin-specific primers.
The selected complemented line was then tested for sensitivity through a plaque assay. As shown in Fig. 3, the complemented mutant showed sensitivity to phage ACP-WH, in contrast to mutant ACP-17-G1, which was resistant to this phage.
The mutant AC-17-G1 that had been complemented with the cupin protein gene from wild-type A. citrulli was further investigated for the recovery of pathogenicity. As shown in Fig. 4, watermelon seeds coated with the mutant AC-17-G1 showed 90% germination with no BFB symptoms. However, the mutant complemented with the wild-type cupin protein gene led to virulence as severe as in wild-type A. citrulli, with less than 10% germination and severe BFB symptoms that resulted in the death of all germinated plants.
SDM was performed on the cupin protein gene of wild-type A. citrulli using the CRISPR/Cas 9 system. We designed four sgRNA for the all-in-one vector and introduced them into wild-type A. citrulli. Among colonies selected on ampicillin plates, those recovered from the transformant with sgRNA4 showed resistance against ACPWH (Fig. 5A). The cupin protein gene from this mutant was amplified through PCR with the cupin protein gene promoter F and R primers, and its sequence was analyzed. As shown in Fig. 5B, the mutant with sgRNA showed the deletion of four nucleotides in the middle of the cupin protein gene, which resulted in a frame shift of the open reading frame (ORF) and premature termination of the protein.
To determine whether the cupin protein is a PMI, we compared the growth of mutant AC-17-G1 in the presence of M6P and F6P. As shown in Fig. 6A, mutant AP-17-G1 exhibited slow growth in KB medium. However, when M6P or F6P was added to the medium, the mutant grew at the same rate as the wild-type in KB medium. In addition, plaque assay showed that mutant AC-17-G1 grown in media containing M6P or F6P was sensitive to phage ACPWH contrast to the bacterium grown in KB medium was resistant to this phage (Fig. 6B).
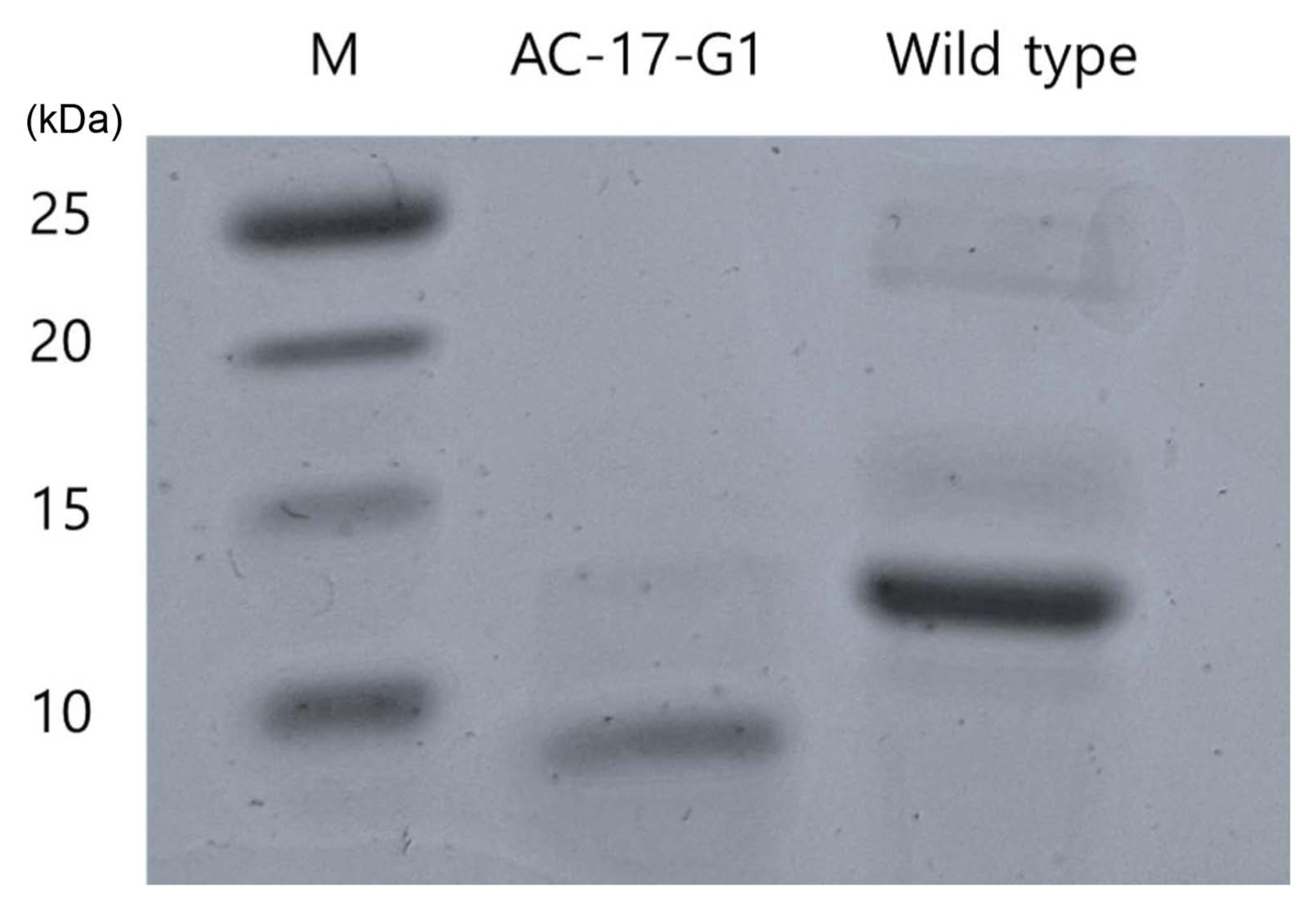
LPS was extracted from wild-type and phage-resistant avirulent mutant AC-17-G1 and analyzed using SDS-PAGE. As shown in Fig. 7, LPS extracted from the mutant was approximately 10 kDa in size, whereas that from the wild-type was approximately 12 kDa.
Recently, the use of bacteriophages for biocontrol has been revisited as an alternative to antibiotics. To develop this novel alternative strategy, detailed characterization of bacteriophages and their host interactions are essential. Bacteria can resist attachment of bacteriophages through various mechanisms to avoid infection in nature. Bacteria can also stop phage adsorption, degrade phage DNA, and initiate cell death upon infection. However, bacteria-phage coevolution not only increases the level of population diversity, but has also been shown to affect bacterial ecological behavior (Koskella and Meaden, 2013). For example, reports have suggested that bacterial strains resistant to lytic phages were more susceptible to other phages (Koskella and Brockhurst, 2014), had reduced carbon metabolism (Xu et al., 2019), and even showed structural changes (Oechslin, 2018). The fitness cost for bacteria has been shown to depend on the timing of infection and exposure time to phages.
In this study, we used the T5 transposon mutation method for mutation of A. citrulli strains, resulting in 39 of 3,624 total mutants showing resistant to phage infection in spot assay. Our study focuses on phage-resistant bacteria with reduced virulence. Through seed coating assay for pathogenicity with these phage-resistant mutants, a mutant AC-17-G1 with severely reduced pathogenicity was selected (Fig. 1). Sequence analysis of the selected mutant AC-17-G1 with phage resistance and reduced virulence showed that Tn5 is inserted in a gene that encodes a protein with the cupin domain, which has two histidine motifs with variable length regions (Fig. 2).
Cupin superfamily proteins exhibit extremely diverse functions and contain a considerable number of proteins with unknown functions. Khuri et al. (2001) showed that cupin proteins are a diverse group of proteins present in all three kingdoms of life. Moreover, cupin protein are found in most of microorganism including bacteria with various functions.
The function of the cupin protein in phage resistance and virulence was confirmed by complementation of the mutated gene with the corresponding gene cloned from wild-type. The complemented bacteria recovered phage sensitivity as shown in Fig. 3 and showed the same pathogenicity as the wild-type (Fig. 4). These results indicated that the cupin protein gene of A. citrulli encodes a protein involved in phage sensitivity. These was further confirmed by generation of a site-directed mutant by using CRIPR/Cas9 system. A mutant showing no lysis in spot assay with phage ACPWH indicating the loss of phage sensitivity (Fig. 5) was selected and sequence analysis of the targeted area showed deletion of four nucleotides in the cupin protein gene. Pathogenicity test of this mutant showed similar virulence in seed coating assay (data not shown). Therefore, it was concluded that the cupin protein gene of A. citrulli encodes a protein that affect both of phage sensitivity and pathogenicity.
Although we confirmed that the cupin protein of A. citrulli could be one of, if there any, complete determinant for phage resistance, this protein is not the only gene involved in pathogenicity. Mutant AC-17-G1 showed complete resistance to phage ACPWH but showed reduced virulence and was not non-pathogenic. In the analysis of A. citrulli Tn5 mutants, Luo et al. (2015) identified insertion of Tn5 at eight genes including cation diffusion facilitator family transporter, alpha/beta hydrolase fold, and inosine monophosphate dehydrogenase which resulted in reduced pathogenicity. Zhang et al. (2018) showed that deletion mutation of the genes hrpG or hrpX that involved in the regulation of type 3 secretion system reduced virulence on watermelon seedlings as well as hypersensitive responses. In addition, mutation of the acrR gene of A. citrulli that involved in the motility and flagella formation resulted in virulence with disease index of 31.7 compare to 48.88 of the wild-type (Guan et al., 2020). Compare to the virulence of these mutants, mutant AC-17-G1 severe decrease in virulence, 0% vs. 88.8% germination in seeds coated with wild-type and mutant AC-17-G1, respectively. This suggested that the cupin protein is a major factor for virulence.
The pathogenicity of A. citrulli strains is related to multiple factors, including the type III and type IV secretion systems and flagella. However, genes in this bacterium are spread through the genome in structures called pathogenicity islands (PAIs). PAIs are responsible for various virulence factors, and the ORF of the cupin protein gene is located within a PAI in the genome of A. citrulli. Homology of cupin protein-coding sequences was identified in three A. citrulli strains (AA78-5, AA99-2, and ATCC19860), which all showed similar locations in PAIs and similar roles in pathogenicity. Due to differing annotation methods, homologues of cupin protein have generally been reported as hypothetical proteins, but this gene has been confirmed to play a role in pathogenicity in A. citrulli. Notably, the genes responsible for virulence in the type III secretion system have not been fully clarified; therefore, this result will help elucidate PAIs in A. citrulli and further characterization of gene involved in pathogenicity is required.
The importance of phage-resistant bacteria to the development of antibacterial agents has attracted researchers, who have identified genes related to phage resistance. Studies have divided the mechanisms of phage resistance in bacteria into two groups, LPS-related and non-LPS-related genes. For example, six receptors for bacteriophages were found in the LPSs of Yersinia pestis, the loss of which led to resistant bacteria with reduced virulence (Filippov et al., 2011). One of the major bacterial cupin proteins is the PMI which facilitates the interconversion F6P and M6P that subsequently utilized in several metabolic pathways including glycolysis and capsular polysaccharide biosynthesis, such as LPS of gram-negative bacteria (Khuri et al., 2001). In order to determine whether the cupin protein of A. citrulli is PMI we tested the growth of mutant AC-17-G1 that showed retarded growth in KB medium in the same medium supplemented with F6P or M6P. As shown in Fig. 6, the growth was recovered in medium containing either of them, probably because the products are functions in many biochemical pathways in bacterial cells. Furthermore, the mutants grown in the KB media supplemented with F6P or M6P showed sensitivity to phage ACPWH. Weisser et al. (1996) showed that PMI can increase growth of the bacterium Zymomonas mobilis, and mutation of genes in mannose synthesis pathways has been reported to not only enhance growth and cell wall formation of the bacterium, but also to play a major role in phage ╬”C31 susceptibility.
LPS has been reported as receptor for phages such as two closely related myoviridae, phages ╬”R1-RT and TG1, that infect Yersinia enterocolitica (Leon-Velarde et al., 2016). Since the products of PMI are used as substrates for LPS synthesis, mutants grown media supplemented with F6P or M6P could synthesize normal LPS, which served as the receptor phage. This was further confirmed by SDS-PAGE analysis of LPS extracted from wild-type and mutant. LPS from mutant AC-17-G1 was smaller than that from wild-type (Fig. 7). Studies with mutant strains defective in LPS biosynthesis-related genes have indicated that LPS is required for bacterial virulence. For example, mutations in LPS biosynthesis-related genes of Salmonella enterica resulted in the loss of its virulence (Kong et al., 2011). Similarly, mutation of genes involved in the synthesis of the core oligosaccharide (OS) of LPS, resulted in clear reduction in the virulence of Burkholderia glumae that causes bacterial panicle blight and seedling rot in rice (Lee et al., 2019). Since PMI is involved in the production of intermediates used for the synthesis of the OS of LPS, our data coincide with these previous data.
It has been suggested that selection of host populations for the resistance to parasites can results in the alteration of other traits as trade-off. For example, mutants of Pseudomonas syringae that have evolved resistance via a single mutation showed significant reduction of the growth within plants tissue and reduction in pathogenicity (Meaden et al., 2015). Since the mutants were screened for phage resistance first and then tested for pathogenicity, mutant AC-17-G1 is not the case of natural selection for resistance against phage and loss of its pathogenicity as the cost. However, similar evolutional changes could happen especially considering the prevalence of phages infection A. citrulli as previously reported (Rahimi-Midani et al., 2020) and this could be an obstacle for the application of phages as the biocontrol agents of BFB. However, these results also support the hypothesis that phage resistance not only reduces pathogenicity, but can also increase the number ŌĆ£good strainsŌĆØ of A. citrulli in the presence of phages.
Acknowledgments
The authors appreciate Professor Sang-Wook Han of Chung-Ang University, Republic of Korea for proving the Tn5 mutants.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig.┬Ā1
Pathogenicity of selected phage-resistant Acidovorax citrulli Tn5 mutants. (A) Germination of watermelon seeds coated with designated bacterial. (B) Representative pots sown with watermelon seeds without coating (N), seeds coated with mutant strain AC-5-E1 (E1), seeds coated with mutant AC-17-G1 G1) and wild-type (W).
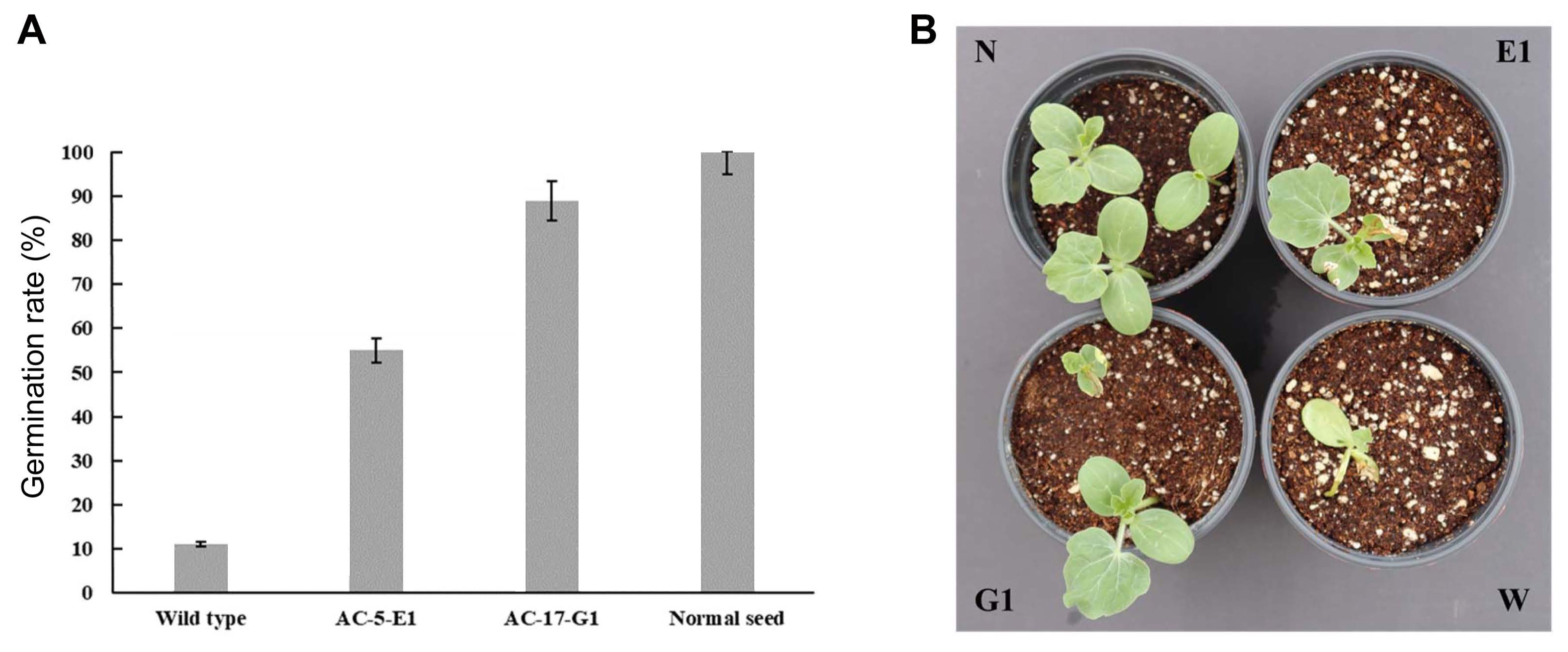
Fig.┬Ā2
Schematic description of Tn5 integration in the genome of Acidovorax citrulli mutant AC-17-G1 and confirmation by PCR. (A) The nucleotide sequences of integration junctions are presented with vertical lines at the junctions. The locations of PCR primers for the analysis and expected size pf PCR products are presented. (B) PCR confirmation of the integration site. M, 1-kb plus base pair ladder; N, PCR of wild-type A. citrulli with transposon right-end R and transposon left-end F primers; F, PCR with the CPF and TLR primers; R, PCR with the TRF and primers.

Fig.┬Ā3
Recovery of mutant AC-17-G1 for phage sensitivity by complementation with wild-type gene. Plaque assay with phage ACPWH was conducted with mutant AC-17-G1 (A) and the same mutant complemented with wild-type cupin protein gene (B).
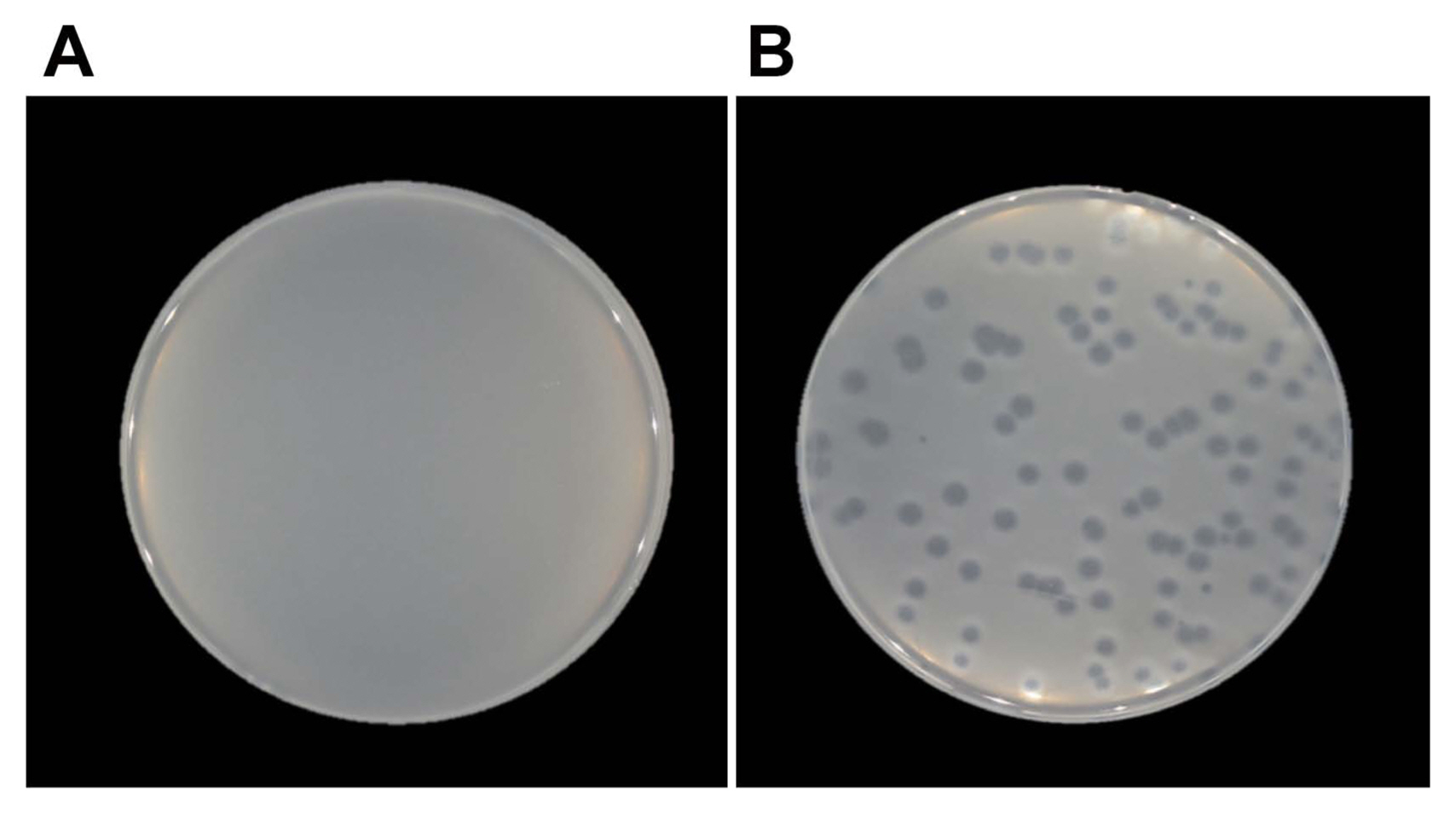
Fig.┬Ā4
Pathogenicity test of the complemented mutant. (A) Germination of watermelon seeds coated with wild-type, mutant AC-17-G1 and mutant AC-17-G1 complemented with wild-type gene. (B) Representative pots sown with watermelon seeds coated with designated bacterial strains.
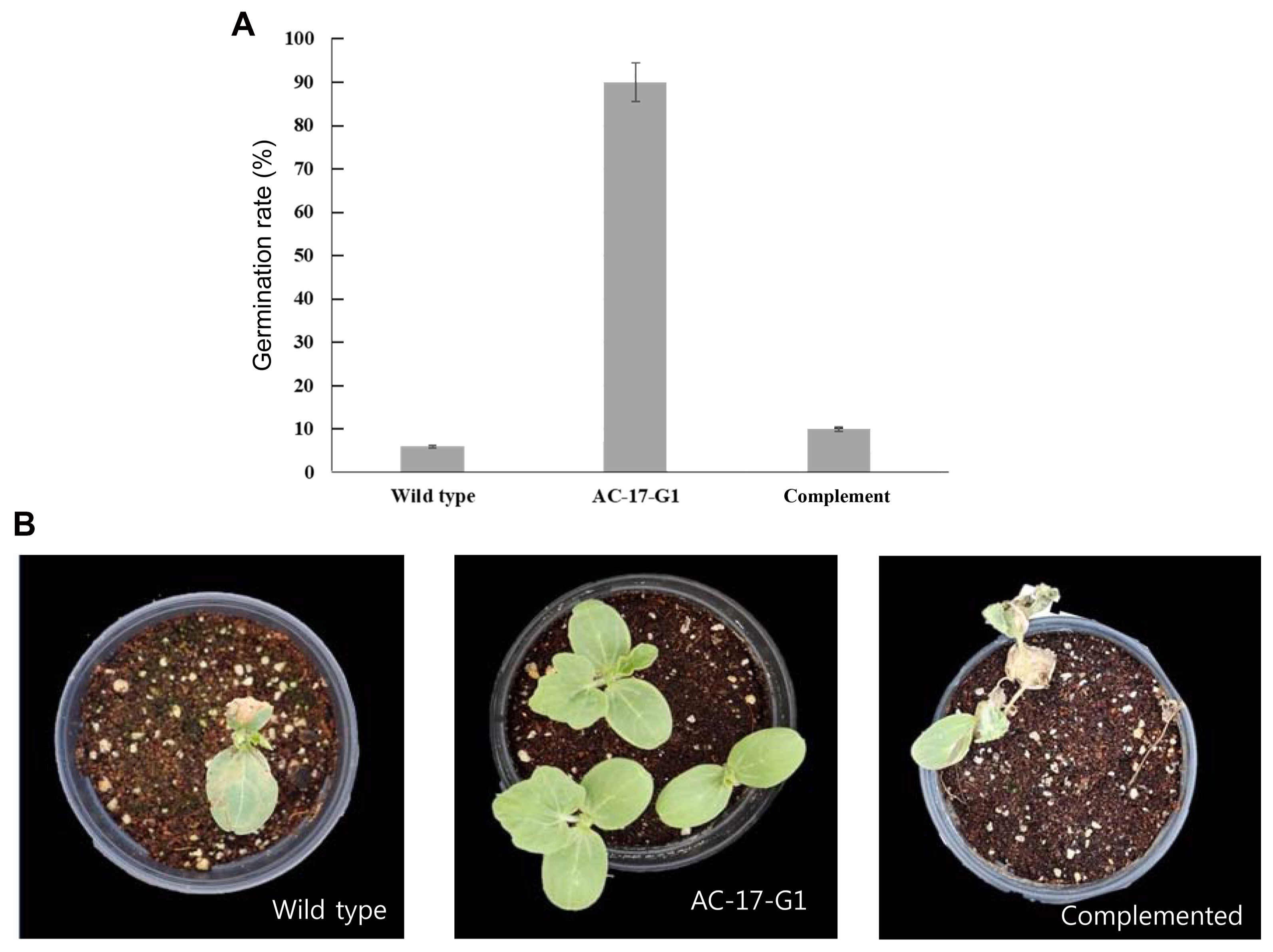
Fig.┬Ā5
Confirmation of site-directed mutagenesis of the cupin protein gene using the CRISPR/Cas 9 system. (A) Spot assay with phage ACPWH against wild-type Acidovorax citrulli (W), mutant AC-17-G1 (G), and a cupin protein gene deleted mutant generated by CRISPR/Cas9 (C). (B) Sequence analysis of the mutant produced by CRISPR/Cas9 system. Four nucleotides were deleted in the middle of the cupin protein gene in the mutant created using sgRNA4. The deleted four nucleotides are shown in bold. The single guide RNA sequence is underlined and the PAM sequence is indicated with dashes.
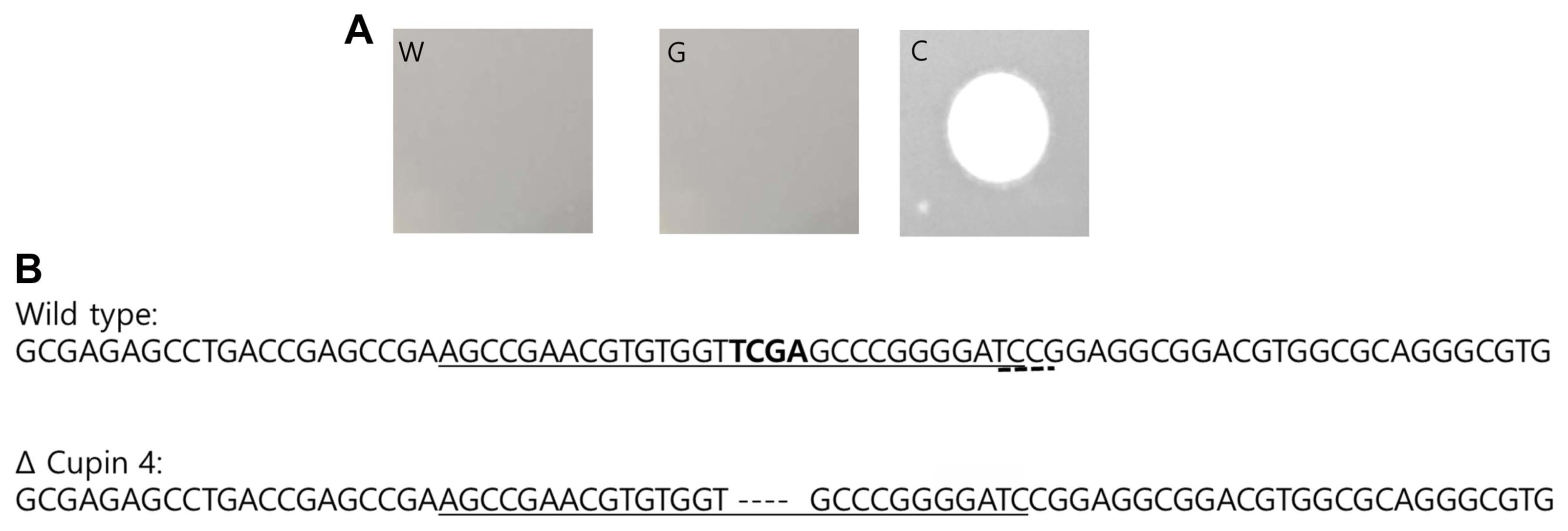
Fig.┬Ā6
Growth of Acidovorax citrulli mutant AC-17-G1 in KingŌĆÖs B (KB) medium supplemented with D-mannose-6-phosphate (M6P) and D-fructose-6-phosphate (F6P) and recovery of phage sensitivity. (A) The growth of wild-type and mutant strains in KB medium and KB medium supplemented with M6P and F6P. (B) Plaque assay with phage ACPWH and mutant AC-17-G1 grown in KB medium (K), KB medium supplemented with F6P (F), and M6P (M).

Fig.┬Ā7
Sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of lipopolysaccharide extracted from mutant strain AC-17-G1 and wild-type Acidovorax citrulli. Molecular weights of standard protein are presented.
Table┬Ā1
Primers used for PCR in this study
References
Barbu, EM, Cady, KC and Hubby, B 2016. Phage therapy in the era of synthetic biology. Cold Spring Harb Perspect Biol. 8:a023879.



Brockhurst, MA, Koskella, B and Zhang, Q-G 2021. Bacteriaphage antagonistic coevolution and the implications for phage therapy. In: Bacteriophages: biology, technology, therapy, eds. by DR Harper, ST Abedon, BH Burrowes and ML McConville, 1-21. Springer, Cham, Denmark.

Chatterjee, A, Willett, JLE, Nguyen, UT, Monogue, B, Palmer, KL, Dunny, GM and Duerkop, BA 2020. Parallel Genomics uncover novel enterococcal-bacteriophage interactions. mBio. 11:e03120-19.



Coons, GH and Kotila, JE 1925. The transmissible lytic principle (bacteriophage) in relation to plant pathogens. Phytopathology. 15:357-370.
Darveau, RP and Hancock, REW 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J Bacteriol. 155:831-838.



dŌĆÖHerelle, F 1926. The bacteriophage and its behavior. Williams and Wilkins, Baltimore, MD, USA. 629.
Doherty, M, Todd, D, McFerran, N and Hoey, EM 1999. Sequence analysis of a porcine enterovirus serotype 1 isolate: relationships with other picornaviruses. J Gen Virol. 80:1929-1941.


Eckshtain-Levi, N, Munitz, T, ┼Įivanovi─ć, M, Traore, SM, Spr├Čer, C, Zhao, B, Welbaum, G, Walcott, R, Sikorski, J and Burdman, S 2014. Comparative analysis of type III secreted effector genes reflects divergence of Acidovorax citrulli Strains into three distinct lineages. Phytopatholgy. 104:1152-1162.

Filippov, AA, Sergueev, KV, He, Y, Huang, X-Z, Gnade, BT, Mueller, AJ, Fernandez-Prada, CM and Nikolich, MP 2011. Bacteriophage-resistant mutants in Yersinia pestis: identification of phage receptors and attenuation for mice. PLoS ONE. 6:e25486.



Guan, W, Wang, T, Huang, Q, Tian, E, Liu, B, Yang, Y and Zhao, T 2020. A LuxR-type regulator, AcrR, regulates flagellar assembly and contributes to virulence, motility, biofilm formation, and growth ability of Acidovorax citrulli
. Mol Plant Pathol. 21:489-501.



Hassan, AY, Lin, JT, Ricker, N and Anany, H 2021. The age of phage: friend or foe in the new dawn of therapeutic and biocontrol applications? Pharmaceuticals. 14:199.



Khan Mirzaei, M and Nilsson, AS 2015. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE. 10:e0118557.



Khuri, S, Bakker, FT and Dunwell, JM 2001. Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol Biol Evol. 18:593-605.


Kong, Q, Yang, J, Liu, Q, Alamuri, P, Roland, KL and Curtiss, R 3rd 2011. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect Immun. 79:4227-4239.



Koskella, B and Brockhurst, MA 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 38:916-931.


Koskella, B and Meaden, S 2013. Understanding bacteriophage specificity in natural microbial communities. Viruses. 5:806-823.



Lee, C, Mannaa, M, Kim, N, Kim, J, Choi, Y, Kim, SH, Jung, B, Lee, H-H, Lee, J and Seo, Y-S 2019. Stress tolerance and virulence-related roles of lipopolysaccharide in Burkholderia glumae
. Plant Pathol J. 35:445-458.



Leon-Velarde, CG, Happonen, L, Pajunen, M, Leskinen, K, Kropinski, AM, Mattinen, L, Rajtor, M, Zur, J, Smith, D, Chen, S, Nawaz, A, Johnson, RP, Odumeru, JA, Griffiths, MW and Skurnik, M 2016.
Yersinia enterocolitica-specific infection by bacteriophages TG1 and ╬”R1-RT is dependent on temperature-regulated expression of the phage host receptor OmpF. Appl Environ Microbiol. 82:5340-5353.



Liu, H and Naismith, JH 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8:91.



Luo, J, Qiu, W, Chen, L, Anjum, SI, Yu, M, Shan, C, Ilyas, M, Li, B, Wang, Y and Sun, G 2015. Identification of pathogenicity-related genes in biofilm-defective Acidovorax citrulli by transposon Tn5 mutagenesis. Int J Mol Sci. 16:28050-28062.



Marks, T and Sharp, R 2000. Bacteriophages and biotechnology: a review. J Chem Technol Biotechnol. 75:6-17.

Meaden, S, Paszkiewicz, K and Koskella, B 2015. The cost of phage resistance in a plant pathogenic bacterium is context-dependent. Evolution. 69:1321-1328.



Oechslin, F 2018. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses. 10:351.



Park, J, Lee, GM, Kim, D, Park, DH and Oh, CS 2018. Characterization of the lytic bacteriophage phiEaP-8 effective against both Erwinia amylovora and Erwinia pyrifoliae causing severe diseases in apple and pear. Plant Pathol J. 34:445-450.



Pirhonen, M, Heino, P, Helander, I, Harju, P and Palva, ET 1988. Bacteriophage T4 resistant mutants of the plant pathogen Erwinia carotovora
. Microb Pathog. 4:359-367.


Rahimi-Midani, A, Kim, J-O, Kim, JH, Lim, J, Ryu, J-G, Kim, M-K and Choi, T-J 2020. Potential use of newly isolated bacteriophage as a biocontrol against Acidovorax citrulli
. Arch Microbiol. 202:377-389.


Rahimi-Midani, A, Lee, YS, Kang, S-W, Kim, M-K and Choi, T-J 2018. First isolation and molecular characterization of bacteriophages infecting Acidovorax citrulli, the causal agent of bacterial fruit blotch. Plant Pathol J. 34:59-64.



Szermer-Olearnik, B and Boratyński, J 2015. Removal of endotoxins from bacteriophage preparations by extraction with organic solvents. PLoS ONE. 10:e0122672.



Tian, Y, Zhao, Y, Wu, X, Liu, F, Hu, B and Walcott, RR 2015. The type VI protein secretion system contributes to biofilm formation and seed-to-seedling transmission of Acidovorax citrulli on melon. Mol Plant Pathol. 16:38-47.


Vu, NT and Oh, C-S 2020. Bacteriophage usage for bacterial disease management and diagnosis in plants. Plant Pathol J. 36:204-217.



Watson, BNJ, Vercoe, RB, Salmond, GPC, Westra, ER, Staals, RHJ and Fineran, PC 2019. Type I-F CRISPR-Cas resistance against virulent phages results in abortive infection and provides population-level immunity. Nat Commun. 10:5526.



Weisser, P, Kr├żmer, R and Sprenger, GA 1996. Expression of the Escherichia coli pmi gene, encoding phosphomannose-isomerase in Zymomonas mobilis, leads to utilization of mannose as a novel growth substrate, which can be used as a selective marker. Appl Environ Microbiol. 62:4155-4161.



- TOOLS
-
METRICS

- ORCID iDs
-
Tae-Jin Choi

https://orcid.org/0000-0002-4923-5121 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print



