Identification of Leonurus sibiricus as a Weed Reservoir for Three Pepper-Infecting Viruses
Article information
Abstract
In plant virus ecology, weeds are regarded as wild reservoirs of viruses and as potential sources for insect-mediated transmission of viruses. During field surveys in 2013–2014, three Leonurus sibiricus plants showing virus-like symptoms were collected from pepper fields in Daegu, Seosan, and Danyang in Korea. Molecular diagnosis assays showed that the collected L. sibiricus samples were infected with either Tomato spotted wilt virus (TSWV), Pepper mild mottle virus (PMMoV), or Beet western yellow virus (BWYV), respectively. Since this is the first identification of TSWV, PMMoV, and BWYV from L. sibiricus, complete genome sequences of three virus isolates were determined to examine their phylogenetic relationships with the previously reported strains and isolates. Phylogenetic analyses performed using full genome sequences of the viruses showed the isolates of TSWV and PMMoV obtained from L. sibiricus are closely related to the pepper isolates of the corresponding viruses. Our results suggest that L. sibiricus could act an alternative host and reservoir of viruses that cause damages in pepper fields.
Pepper (Capsicum annuum L.) is an economically important vegetable crop worldwide. While consumption of pepper is increasing, viral diseases cause a significant loss of the yield and commercial value of pepper every year in Korea. While various viruses are known to infect pepper worldwide (Choi et al., 2005; Green and Kim, 1991), some of the viruses including Cucumber mosaic virus (CMV; Cucumovirus), Broad bean wilt virus 2 (BBWV2; Fabavirus), Pepper mottle virus (PepMoV; Potyvirus), Pepper mild mottle virus (PMMoV; Tobamovirus), and Tomato spotted wilt virus (TSWV; Tospovirus) often cause epidemics in pepper fields in Korea (Choi et al., 2005; Kim et al., 2012; Kim et al., 2011). Continuous and localized occurrence of viral diseases is often closely associated with the ecological factors such as the presence of alternative hosts and insect vectors (Gilbert, 2002). While various weeds of family Solanaceae, which are widely dispersed near cultivated crop fields, were found to be the major reservoir hosts of CMV (Hobbs et al., 2000), the alternative hosts for the other pepper-infecting viruses are still largely unknown.
Leonurus sibiricus L. (family Lamiaceae), commonly called “motherwort”, is a native weed plant in Asia, thus widely distributed and commonly found near crop fields in Korea. Currently, a few economically important viruses including BBWV2 and Tomato yellow spot virus (TYSV) have been identified from L. sibiricus (Barbosa et al., 2013; Seo et al., 2014). During the surveys performed in 2013–2014, we found some L. sibiricus plants showing typical virus symptoms near pepper fields. We examined three of the collected L. sibiricus samples to identify casual viruses from the samples. The first L. sibiricus sample collected in Danyang in 2013 showed symptoms of severe mosaic and yellowing (Fig. 1A). The second sample collected in Daegu in 2013 showed symptoms of mild mottle, yellowing, and stunting (Fig. 1B). The third sample showing mild mottle, mosaic, and chlorosis symptoms was collected in Seosan in 2014 (Fig. 1C). Since the L. sibiricus samples were collected near pepper fields, we sought to examine whether L. sibiricus acts as a weed reservoir for pepper-infecting viruses in Korea. Thus, the collected samples were subjected to reverse transcription (RT)-polymerase chain reaction (PCR) assay using the specific primer pairs designed to detect 6 representative pepper-infecting viruses, including CMV, BBWV2, PepMoV, PMMoV, TSWV, and Potato virus Y (PVY; Potyvirus) (Table 1). Total RNA was extracted from the symptomatic leaves of the collected samples using PureLink™ RNA Mini Kit (Ambion, Austin, USA) and RT-PCR reactions were performed using AccessQuick™ RT-PCR system (Promega, Madison, USA) and the virus-specific primer pairs.

Symptoms appeared on the leaves of Leonurus sibiricus plants. The symptomatic leaf samples were collected from Danyang in 2013 (A), Daegu in 2013 (B), and Seosan in 2014 (C).
The RT-PCR detection results showed that the second sample was infected with TSWV and the third sample was co-infected with two pepper-infecting viruses, BBWV2 and PMMoV. However, no amplification was detected for the first sample. Thus, to identify causal virus from the first sample, total RNA extracted from the first sample was analyzed by Next-generation sequencing (NGS) as described previously (Lee et al., 2015; Seo et al., 2015). Briefly, a transcriptome library was constructed using the TruSeq Stranded Total RNA with Ribo-Zero Plant kit (Illumina, San Diego, CA) according to the manufacturer’s instruction. NGS was carried out using an Illumina HiSeq2000 sequencer and the obtained data was analyzed by BLASTn and BLASTx searches against the viral reference genome database in Genbank (Schelhorn et al., 2013). The entire NGS procedure was carried out by Macrogen Inc. (Seoul, South Korea). Among the analyzed contigs, one large contig (5727 bp) was of viral origin among the analyzed contigs. Nucleotide blast searches showed that the contig has a maximum identity of 92% (with 97% coverage) to the USA strain (Genbank Accession No. AF473561) of Beet western yellows virus (BWYV, Polerovirus). BWYV infection of the first sample was confirmed by RT-PCR with the specific primers for the 5′ and 3′ ends of the BWYV contig (5′-GCGAGAAACCAGCGAATGCT-3′ and 5′-ACACCGAAGTGCCGTAAGGATT-3′, respectively).
As a result, in this study, we identified four pepper-infecting viruses including TSWV, BBWV2, PMMoV, and BWYV from three L. sibiricus samples. Although BBWV2 infection of L. sibiricus and the complete genome sequence of BBWV2 isolate obtained from L. sibiricus were already reported (Seo et al., 2014), this study is the first report of TSWV, PMMoV, and BWYV infection of L. sibiricus to our knowledge. Therefore, we sought to analyze complete genome sequences of isolates of the viruses from L. sibiricus and compare the sequences with those of the previously reported strains and isolates. To this end, complete genome sequences of the isolates of TSWV (LS-DG), PMMoV (LS-SS), and BWYV (LS-DY), which were identified from L. sibiricus were determined. In brief, first strand cDNAs were synthesized using Superscript® III RTase (Invitrogen, Carlsbad, USA) with 3′ end specific reverse primer for PMMoV (5′-GAAAAATAATTACGTCGTTCGC-3′) and specific primers for TSWV as described previously (Lee et al., 2011; Yoon et al., 2005). PCR amplification of individual cDNAs was carried out using Platinum® Taq DNA polymerase (Invitrogen, Carlsbad, USA) with 5′ end primer for PMMoV (5′GGCTTACACACAACAAGCTACC-3′) and appropriate primers for TSWV described in the previous studies (Lee et al., 2011; Yoon et al., 2005). The resulting PCR products were purified using QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and cloned into pGEM-T easy Vector (Promega, Madison, USA) and sequenced using specific primers (Lee et al., 2011; Yoon et al., 2005). End sequences of each virus were obtained by the 5′ and 3′ rapid amplification of cDNA ends-PCR (RACE-PCR) as described previously (Kwon et al., 2014). The obtained full-length sequences of the segments L, M, and S of TSWV isolate LS-DG were 8913, 4787, and 2974 nucleotides in length, respectively, and were deposited in Genbank under the accession numbers, KM076651, KM076652, and KM076653, respectively. The assembled complete genome sequences of PMMoV isolate LS-SS and BWYV isolate LS-DY were 6357 and 5740 nucleotides in length, respectively, and were deposited in Genbank under the accession numbers, KR108207 and KM076647, respectively.
Using the determined full-length nucleotide sequences, phylogenetic analyses were performed to examine epidemiological relationships of the isolates of TSWV, PMMoV, and BWYV from L. sibiricus with the corresponding virus strains and isolates that were reported previously. The isolates of Impatiens necrotic spot virus (INSV; GenBank accession number GU112505 for L RNA; GenBank accession number GU112503 for M RNA; GenBank accession number GU112504 for S RNA), Tobacco mosaic virus (TMV; GenBank accession number X68110), and Pepper yellow leaf curl virus (PYCLV; GenBank accession number HM439608) were included as outgroups for TSWV, PMMoV, and BWYV, respectively, in the phylogenetic analyses. The phylogenetic trees were reconstructed by the neighbor joining (NJ) method and Kimura 2-parameter methods with bootstrap (1000 replicates) using MEGA program (version 6). Since the TSWV genome is distributed on three RNA molecules designated as L RNA, M RNA, and S RNA, phylogenetic analyses of TSWV were performed for each RNA segment (Fig. 2). When the tree was reconstructed using L RNA sequences, TSWV isolate LS-DG was grouped with 6 other Korean isolates from peppers (K4 and K7) and weeds (K5 and K10, from Stellaria aquatica; K6 and K17 from Stellaria media) (Fig. 2A). The phylogenetic tree for M RNA indicated that the isolate LS-DG was most closely related to two Korean isolates obtained from weeds (K5 and K10, both from Stellaria aquatica) (Fig. 2B). For S RNA, the isolate LS-DG was closely related to other Korean isolates (K4, K5, K6, K7, and K10) (Fig. 2C). For PMMoV, full genome sequences of twelve PMMoV pepper isolates collected from Korea, Japan, India, Brazil, and Spain were available in Genbank and retrieved to examine phylogenetic position of PMMoV isolate LS-SS (Fig. 3). The comparative analysis of full-length nucleotide sequences of PMMoV strains and isolates revealed that the isolate LS-SS has more than 94% identity to other pepper isolates (data not shown). In addition, phylogenetic analysis showed that the isolate LS-SS is most close to the isolate J collected from Japan (Fig. 3). For BWYV, full genome sequences of twelve BWYV isolates retrieve from GenBank were included in phylogenetic analysis. Among them, two isolates (Shimizu and Kiyosato) were isolated from spinach and the other isolates were collected from sugar beet. Unfortunately, no full genome sequence of BWYV isolated from pepper was yet reported. As shown in Fig. 4, phylogenetic analysis showed that the isolate LS-DY is most closely related to the USA isolate collected from sugar beet rather than other isolates from Japan and China despite of geographical distance (Fig. 4). In all, the results from phylogenetic analyses showed that TSWV isolate LS-DG and PMMoV isolate LS-SS are closely related to the pepper isolates of the viruses, suggesting that L. sibiricus could act an alternative host and reservoir of viruses that cause damages in pepper fields.

Neighbor joining tree inferred from genomic nucleotide sequences of 13 TSWV isolates and Impatiens necrotic spot virus (INSV) as an outgroup. The numbers on the nodes represent the percentage of 1,000 boostrap replications. GenBank accession numbers are given in parentheses. Phylogenetic trees of TSWV RNA L segments (A), TSWV RNA M segments (B), and TSWV RNA S segments (C) were reconstructed by Neighbor joining method. Full genome sequence of the isolate LS-DG was newly determined in this study. K2, K3, K4, and K7 isolated from pepper in Korea; K5, K6, K10 and K17 from weeds (Stellaria aquatic and S. media) in Korea; YNrP from pepper in China; PVR from pepper in Spain; P105 and P202WT from pepper in Italy.
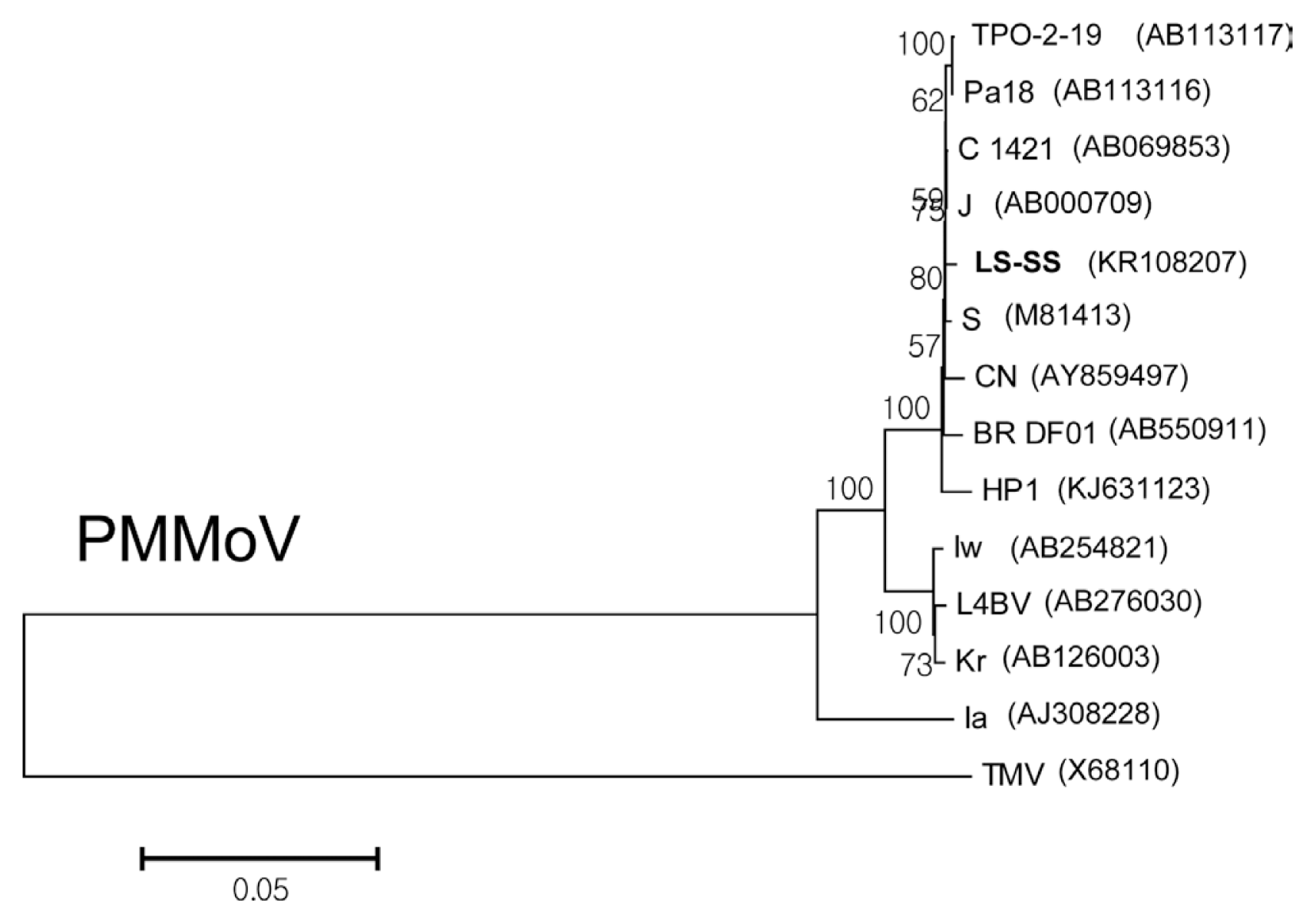
Neighbor joining tree inferred from full genome sequences of 13 PMMoV isolates and tobacco mosaic virus (TMV) as an outgroup. The numbers on the nodes represent the percentage of 1,000 boostrap replications. GenBank accession numbers are given in parentheses. Full genome sequence of the isolate LS-SS was newly determined in this study. TPO-2-19, Pa18, C1421, J, Iw, and L4BV isolated from pepper in Japan; S and Ia from pepper in Spain; Kr from pepper in Korea; CN from pepper in China; BR-DF01 from pepper in Brazil; HP1 from pepper in India.
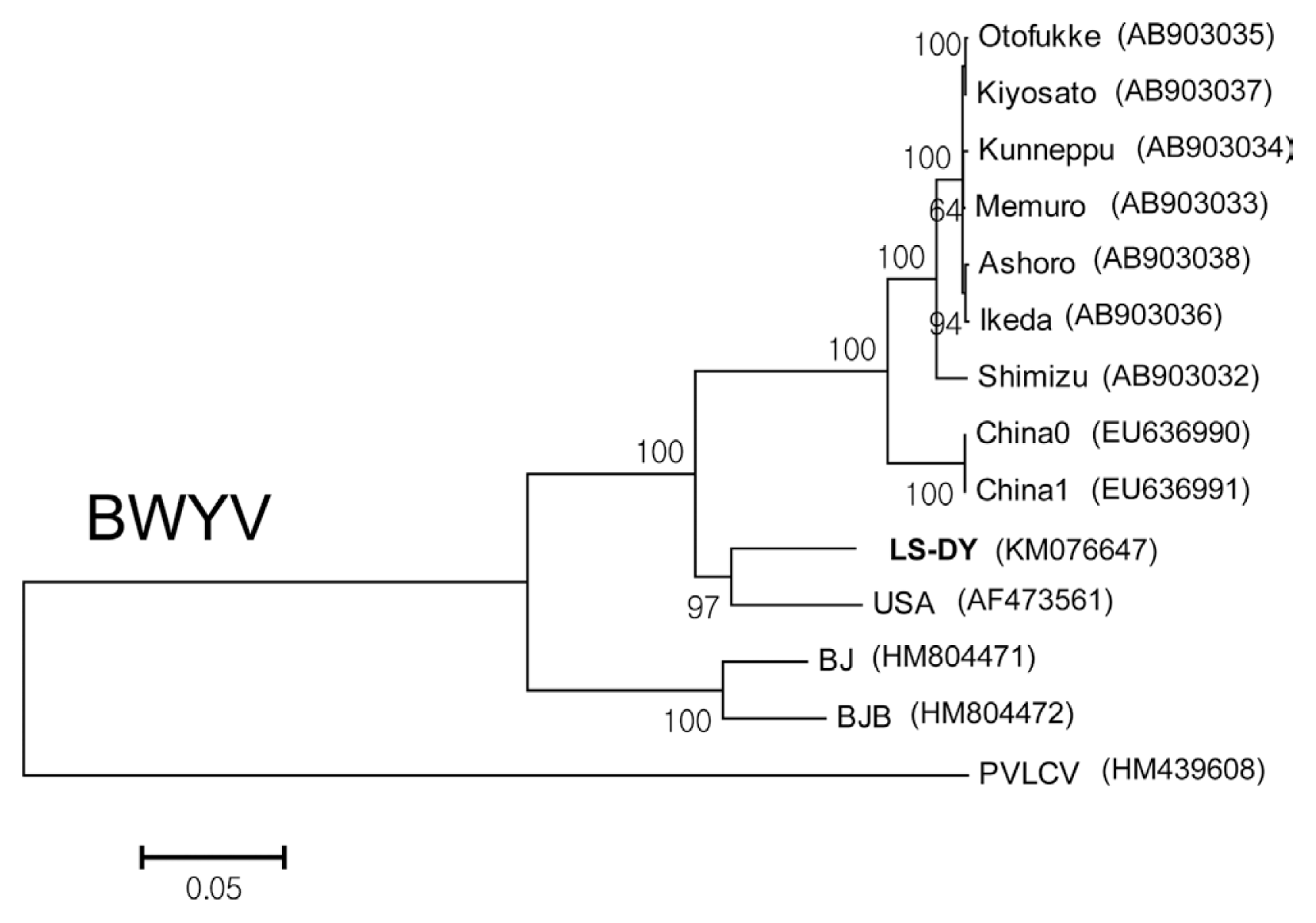
Neighbor joining tree inferred from full genome sequences of 13 BWYV isolates and Pepper yellow leaf curl virus (PYLCV) as an outgroup. The numbers on the nodes represent the percentage of 1,000 boostrap replications. GenBank accession numbers are given in parentheses. Full genome sequence of the isolate LS-DY was newly determined in this study. Otofukke, Kunneppu, Ashoro, Memuro, and Ikeda isolated from sugar beet in Japan; Kiyosato and Shimizu from spinach in Japan; China 0, China 1, BJ, and BJB from sugar beet in China; USA from sugar beet in USA.
In Korea, TSWV and BWYV were firstly reported in paprika in 2004 and 2010, respectively (Kim et al., 2004; Kim et al., 2012). Since then, the incidence of TSWV and BWYV has been increasing in pepper fields in Korea. Our study supports the possibility that alternative weed reservoirs such as L. sibiricus might act important roles in rapid spread and continuous occurrence of TSWV and BWYV in Korea. PMMoV is a seed-borne virus. Therefore, seeds of weed plants infected with PMMoV can be a primary source of virus infection in pepper fields. This makes PMMoV difficult to control in open crop fields, because various weeds are abundant in areas where crop plants are grown. Understanding ecology of plant viruses in relation to their natural hosts and insect vectors is critical to design appropriate virus control measures. In this study, L. sibiricus was newly identified as a weed host of TSWV, PMMoV, and BWYV. Phylogenetic analyses suggested that L. sibiricus could act as an important reservoir of these viruses for infection of pepper fields. Our results further suggest the requirement of developing management practices for weeds that are potential reservoirs of plant viruses and important sources for feeding or reproduction of insect vectors to control virus diseases.
Acknowledgments
This work was supported by a grant from the Basic Research Program (PJ010878012015) of National Institute of Horticultural and Herbal Science, Rural Development Administration, Republic of Korea.