Identification and Characterization of Fungal Pathogens Associated with Boxwood Diseases in the Republic of Korea
Article information
Abstract
Boxwood is a representative ornamental shrub that is widely used in landscaping horticulture. After pruning, damaged leaves or stems of boxwoods are unavoidably vulnerable to infection by various plant pathogens. Several boxwood diseases caused by fungi, such as Volutella blight and Macrophoma leaf spot, have been reported worldwide including Republic of Korea. In this study, we isolated and identified fungal pathogens of boxwood diseases that occurred in Korea and characterized their morphological and taxonomic characteristics. Boxwood samples showing blight symptoms were collected in Seoul, Republic of Korea, and the putative fungal pathogens Pseudonectria buxi, P. foliicola, and Neofusicoccum buxi were successfully identified. Investigation of the morphological features of the field isolates, including mycelial growth and conidial morphology, and phylogenetic analysis of multiple DNA barcode loci revealed that there were some morphological and genetic variations among isolates, but all of the analyzed isolates were closely related to the corresponding reference strains. We also found that P. foliicola strains were more virulent than P. buxi, and the N. buxi strains isolated in this study were weak pathogens or saprophytes. The results of our study will contribute to the development of control strategies for boxwood diseases caused by fungi and accelerate research on the complex ecology of boxwood diseases.
Boxwood (Buxus spp.) are small evergreen shrub widely used in ornamental landscaping as individual specimens or in hedges, parterres, and groups worldwide (Batdorf, 2004). In Korea, boxwood has become a popular landscaping plant, with the production of boxwood accounting for 10.4% of the total ornamental tree market (Korea Forest Service, 2015). Boxwood plants subjected to environmental stress conditions such as winter injury, exposure to bright sunlight, scale insects, and stem wounds are at a greater risk of subsequent fungal infection. Several boxwood diseases caused by fungi have been reported, including leaf and stem blight, root rot, cankers on leaves and stems, and leaf spot (Castroagudín et al., 2020; Jacobi et al., 2003; LeBlanc et al., 2018; Şimşek et al., 2019; Yang et al., 2021). Major boxwood diseases include boxwood blight, Volutella blight, Macrophoma leaf spot, and Phytophthora root rot (Bezerra, 1963; LeBlanc et al., 2019; Lombard et al., 2015; Spetik et al., 2020).
Volutella blight mainly caused by Pseudonectria buxi and P. foliicola results in dieback of leaves and twigs, with pink sporodochia, which is an obvious sign of a fungal pathogen developing on the underside of leaves (Shi and Hsiang, 2014b; Yang et al., 2021). This characteristic distinguishes Volutella blight from winter injury and other diseases with similar symptoms. Although the entire plant does not die, plants with symptoms of Volutella blight are not commercially acceptable; therefore, its widespread occurrence and disfiguring symptoms have caused substantial economic losses to the ornamental industry. Volutella blight has been found worldwide, including in Europe and Asia (Garibaldi et al., 2016; Shi and Hsiang, 2014a; Spetik et al., 2020). A few P. buxi strains causing Volutella blight were recently identified in Korea (Shin et al., 2022).
The other well-known fungal disease of boxwood is Macrophoma leaf spot, which is caused by Neofusicoccum buxi, formerly known as Macrophoma candollei and Dothiorella candollei (Yang et al., 2017). It is a weak and parasitic pathogen and thus acts as a secondary pathogen infecting boxwood when boxwood plants are damaged by primary infection with fungal pathogens or physiological disorders (Hansen, 2009; Jacobi et al., 2003). The most obvious sign of Macrophoma leaf spot is visible fruiting bodies produced on dying or dead straw-colored boxwood foliage. It is considered as an indication of the disease that can be confused with signs and symptoms of Volutella blight (Jacobi et al., 2003; Yang et al., 2021). Despite being the most commonly observed diseases affecting boxwood, little is known about the genetics, biology, and etiology of the causal agents of Volutella blight and Macrophoma leaf spot thus far. Moreover, few taxonomic and morphological characteristics of these pathogens are available.
In this study, we aimed to identify and characterize the morphological and taxonomic features of fungal pathogens associated with boxwood diseases in Korea. We collected boxwood samples showing blight symptoms and recovered a total of 142 fungal isolates, including putative boxwood pathogens, P. buxi, P. foliicola, and N. buxi. The collection of pathogens showed varied morphological characteristics, as well as genetic variations. The results of this study will be helpful to develop efficient disease management strategies for horticulture practices in Republic of Korea.
Materials and Methods
Fungal isolation
Collected symptomatic leaves and stem cuttings (10 mm long) were surface sterilized by soaking in 1% sodium hypochlorite for 1 min, 70% ethanol for 1 min, and were then rinsed with sterile water three times (Park et al., 2020). Eight or nine sterilized samples were placed on 90-mm-diameter Petri plates containing potato dextrose agar (PDA; Difco, Sparks, MD, USA) and incubated at 25°C for 3–5 days. Then, fungal colonies were transferred to complete media (CM) (Leslie and Summerell, 2006). All isolates were pure-cultured using the single-spore isolation method (Ho and Ko, 1997) and stored in 20% glycerol at −80°C until use. Representative isolates of P. buxi and N. buxi (NL1-1-1, B-2, B3L6, and B2L2-1) were deposited to the Korean Collection for Type Cultures (KCTC) under deposit numbers KCTC56889-56892, respectively.
Morphological identification
Each fungal isolate was grown on PDA and CM agar at 25°C for 10 days. Spores were collected from cultures grown on oatmeal agar (OA) or pine needle agar (PNA) at 25°C under near-UV light for 21 days. The structure of spores was observed with light microscopy at 400× (Leica DM500, Jena, Germany). The average dimensions of spores were recorded from 100 spores per isolate. Pictures of fungal colonies were taken 10 days after inoculation. ImageJ was used to measure the length and width of spores (Schneider et al., 2012).
Molecular identification and phylogenetic analyses
Fungal genomic DNA was extracted following the standard protocol (Leslie and Summerell, 2006). Initially, the internal transcribed spacer (ITS) region of 18S–28S nuclear DNA (rDNA) was amplified using the primers ITS4 and ITS5 (White et al., 1990). Polymerase chain reaction (PCR) was performed in 20 μl reactions of AccuPower ProFi Taq PCR PreMix (Bioneer, Daejeon, Korea), 20 pmol of each primer, and approximately 20 ng of template DNA followed by previously described parameters (White et al., 1990). Amplified PCR products were purified using the MEGAquick-spin Plus Total Fragment DNA Purification Kit (Intron Biotechnology, Seongnam, Korea) and were then sequenced (Macrogen, Seoul, Korea). The consensus sequences of each isolate were used as queries to conduct a BLASTn search against the NCBI GenBank database to obtain preliminary identification.
Additional loci, including β-tubulin and the D1/D2 region of the large subunit of the rDNA gene cluster (LSU), were sequenced for phylogenetic analyses using the primer pairs Btub4Rd/Btub2Fd (Woudenberg et al., 2009) and LR0R/LR5 (Rehner and Samuels, 1994; Vilgalys and Hester, 1990), respectively. The sequences of the representative isolates were deposited in GenBank (GenBank accession nos. M169273-OM169276, OM176571-OM176574, ON340774-ON340791). The reference strains of P. buxi (CBS 114049 and CBS 324.53), P. foliicola (CBS 122566 and CBS 123190), and N. buxi (CBS 113714, CBS 116.75, and CBS 214.31) were obtained from the culture collection of the Westerdijk Fungal Biodiversity Institute (Utrecht, The Netherlands). A consensus sequence was manually generated for each set of forward and reverse sequences in MEGA-X (Kumar et al., 2018). Additional sequences of N. parvum strains (CBS 123652, CBS 115186, and CBS 121486) were obtained from the GenBank database (Yang et al., 2017). Sequences of Sarocladium kiliense and Pseudofusicoccum ardesiacum were used as the outgroups in the analysis of Pseudonectria species and N. buxi, respectively.
Pathogenicity tests on boxwood leaves
Pathogenicity tests using boxwood leaves were performed as previously described (Shi and Hsiang, 2014b). Detached leaves of 1-year-old boxwood plants were surface-sterilized by soaking in 1% sodium hypochlorite for 1 min, placing in 70% ethanol for 1 min, rinsing with sterilized distilled water three times, and drying. For each fungal isolate, 5-mm agar plugs containing fungal culture were placed on a wounded spot on the adaxial side of ten detached leaves. Then, inoculated leaves were placed on moist filter papers in transparent plastic boxes incubated at 25°C with 12 h light/12 h dark for 7 to 21 days. Fungal pathogens were reisolated from symptomatic leaves to fulfill Koch’s postulates.
Results
Survey of fungal distribution on boxwood
Diseased tissues of boxwood that had symptoms ranging from yellow leaves to entirely dead shoots were found in some regions of Seoul, Republic of Korea. Leaves and branches were dry and dead, but the whole plants were still alive (Fig. 1A). On some symptomatic leaves, black fruiting bodies appeared as small black dots on the lower leaf surface (Fig. 1B). To isolate fungal strains associated with these disease symptoms, dozens of samples were collected from symptomatic boxwood hedges during July and August 2020. A total of 142 fungal isolates including previously reported four P. buxi strains (B2S72-1, B2S72-3, B3B2, and B4L3-3) (Shin et al., 2022) were successfully obtained and identified as 34 species using ITS sequence analysis (Table 1). The most frequently isolated species was Diaporthe eres (29 isolates), followed by Alternaria alternata (24 isolates) and various Fusarium species (13 isolates). Thirteen isolates were identified as putative pathogens of boxwood; P. buxi was found the most often, with eight isolates, along with two isolates of P. foliicola and three isolates of N. buxi.
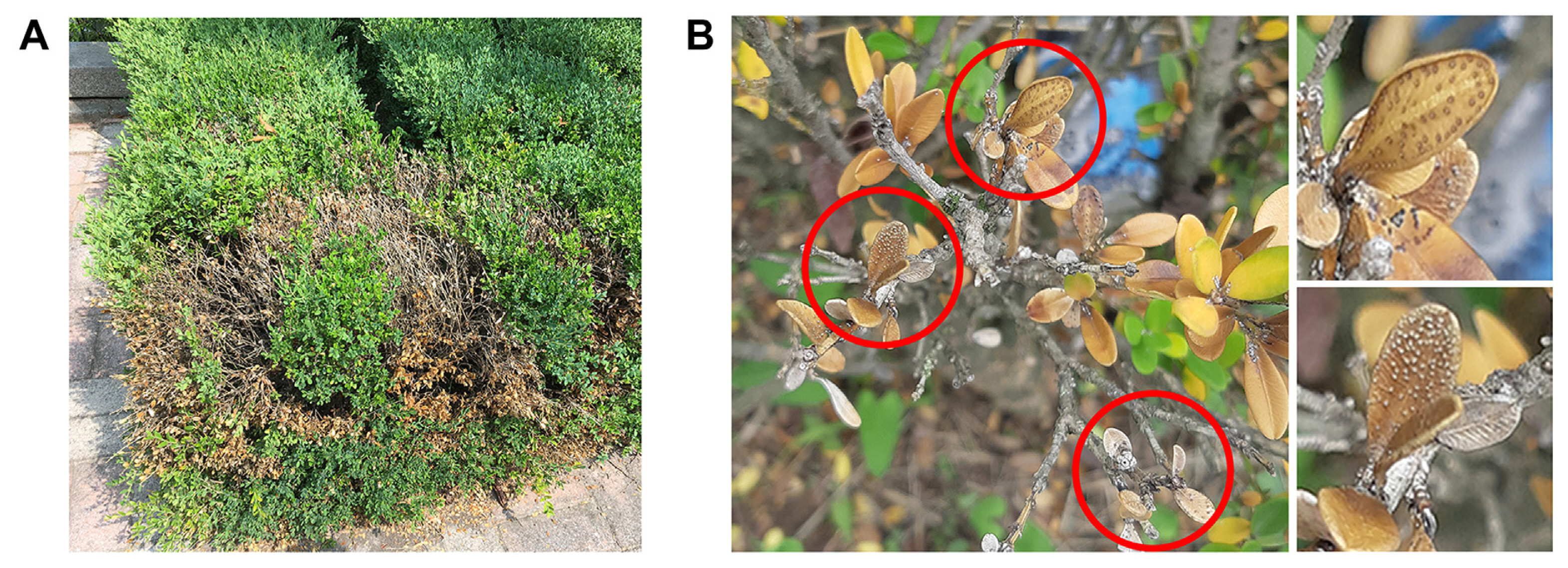
Blight disease symptoms of boxwood. (A) Blight symptoms on boxwood shrub. Diseased leaves were dried and yellowed. (B) Black sporodochia were observed on some boxwood leaves associated with blight symptoms in the growing season.
Morphological characterization
Fungal isolates of P. buxi, P. foliicola and N. buxi with corresponding reference strains (CBS 114049 and CBS 324.53 for P. buxi, CBS 122566 and CBS 123190 for P. foliicola, CBS 113714, CBS 116.75, and CBS 214.31 for N. buxi) (Lombard et al., 2015; Yang et al., 2017) were inoculated on PDA, OA, CM, and PNA to compare colony and spore morphologies (Fig. 2, Supplementary Fig. 1). Both P. buxi and P. foliicola isolates grown on CM showed pink and white fluffy mycelia with pink conidia (Fig. 2A and B). However, the conidia of P. foliicola were markedly smaller than those of P. buxi, as previously reported (Figs. 2 and 3) (Bezerra, 1963). P. buxi field isolates identified from this study produced distinctly shorter conidia compared to those of CBS reference strains CBS 114049 and CBS 324.53 (Fig. 3A), and the conidia of all tested P. buxi strains showed slightly variable widths (Fig. 3B). The average spore length of P. foliicola isolates (B4L1f and B4L2f) was longer than that of the CBS 123190 strain and shorter than that of the CBS 122566 strain (Fig. 3C). Both P. foliicola CBS reference strains produced slightly wider conidia than P. foliicola strains isolated from this study (Fig. 3D).
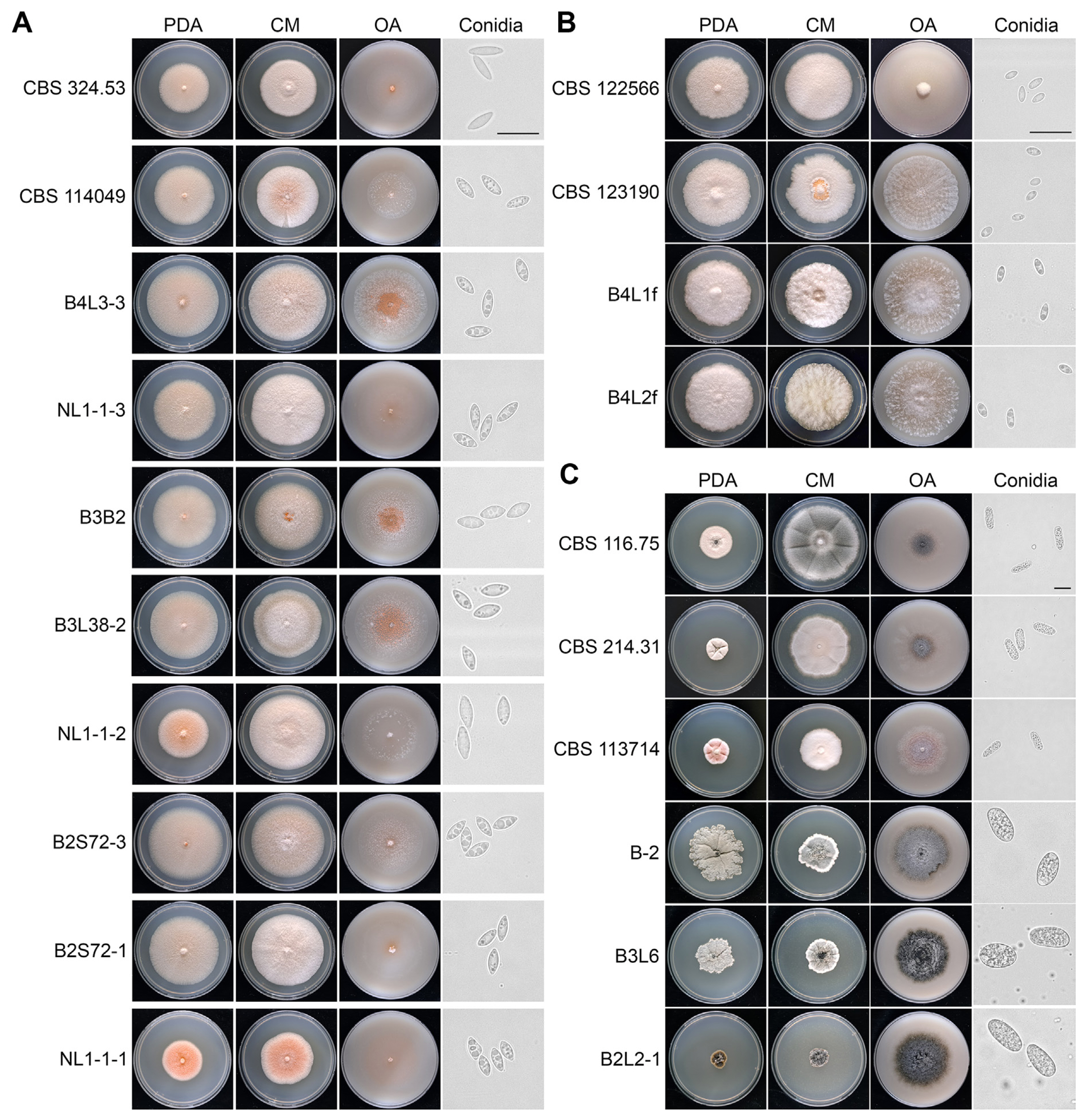
Morphological characterization of Pseudonectria buxi, P. foliicola, and Neofusicoccum buxi. Morphological characteristics of P. buxi (A), P. foliicola (B), and N. buxi (C) strains isolated from symptomatic boxwood leaves and reference strains (CBS strains). Mycelial growth is shown on potato dextrose agar (PDA), complete media (CM) and oatmeal agar (OA) at 18 days post inoculation (dpi). Microscopic pictures of conidia are on the right side. Scale bars = 20 μm.
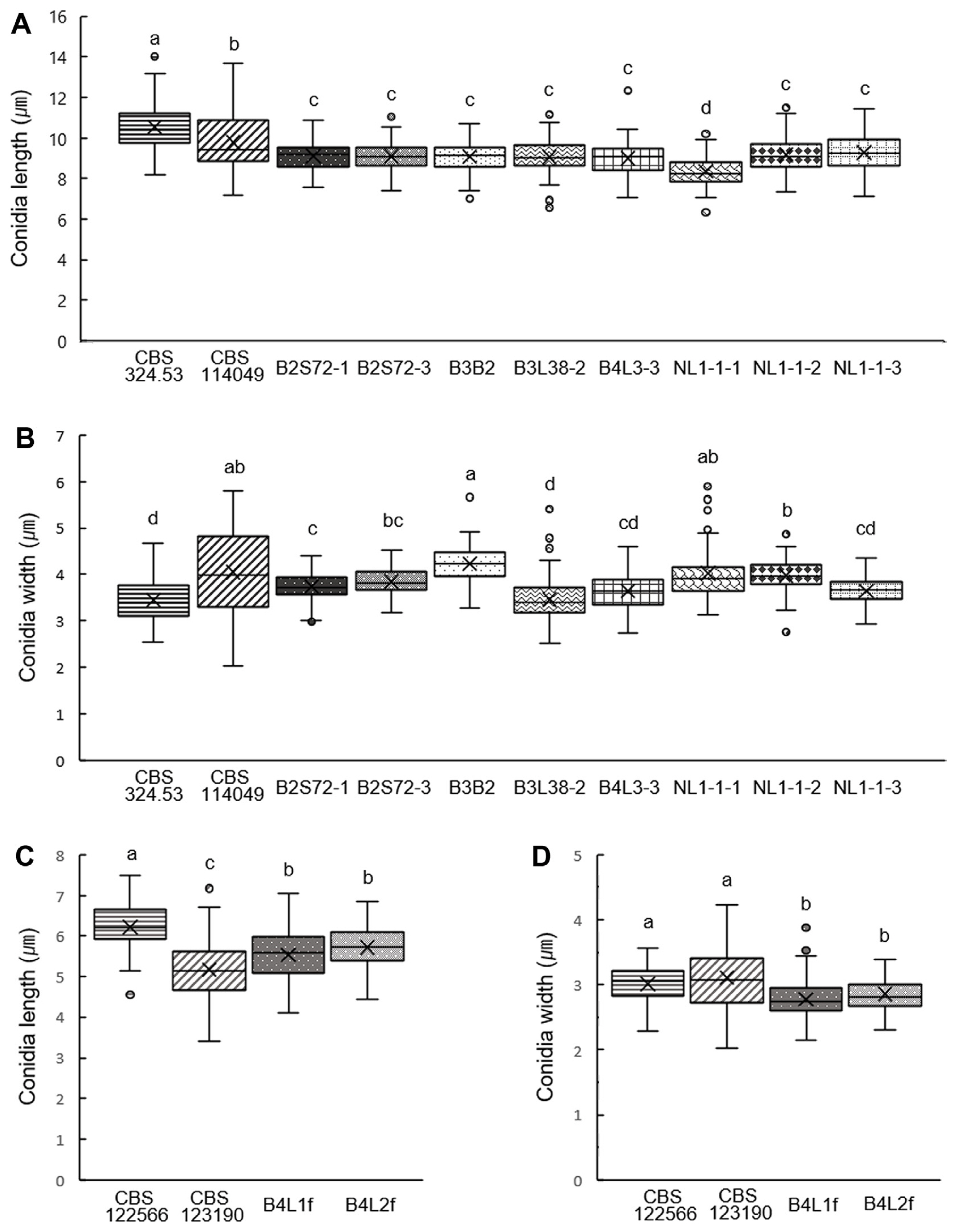
Conidia size measurement of Pseudonectria buxi and P. foliicola. Measurement of conidia length (A, C) and width (B, D) from 100 conidia of each isolate of P. buxi (A, B) and P. foliicola (C, D). Conidia were collected at 18–21 dpi on oatmeal agar. The ImageJ program was used for conidia measurement. Different letters above values indicate significant differences (P < 0.05, Tukey’s test in one-way ANOVA).
Three CBS N. buxi strains (CBS 116.75, CBS 214.31, and CBS 113714) have been reported until now (Yang et al., 2017), and they showed various morphological characteristics on CM, PDA, and OA (Fig. 2C). Furthermore, field isolates (B-2, B3L6, and B2L2-1) identified as putative N. buxi showed notably different phenotypes compared to CBS strains. Colonies of N. buxi isolates showed more wrinkled mycelia and blackish gray color. Furthermore, all N. buxi isolates produced significantly larger spores than the CBS N. buxi strains (Figs. 2C and 4). Conidia of CBS strains were 18–20 × 5–7 μm in size, while conidia of isolates annotated as N. buxi were 25–27 × 10–12 μm in size (Fig. 4). The results suggest that fungal strains predicted as N. buxi in this study have different morphological and physiological characteristics compared to previously reported CBS strains (Yang et al., 2017).
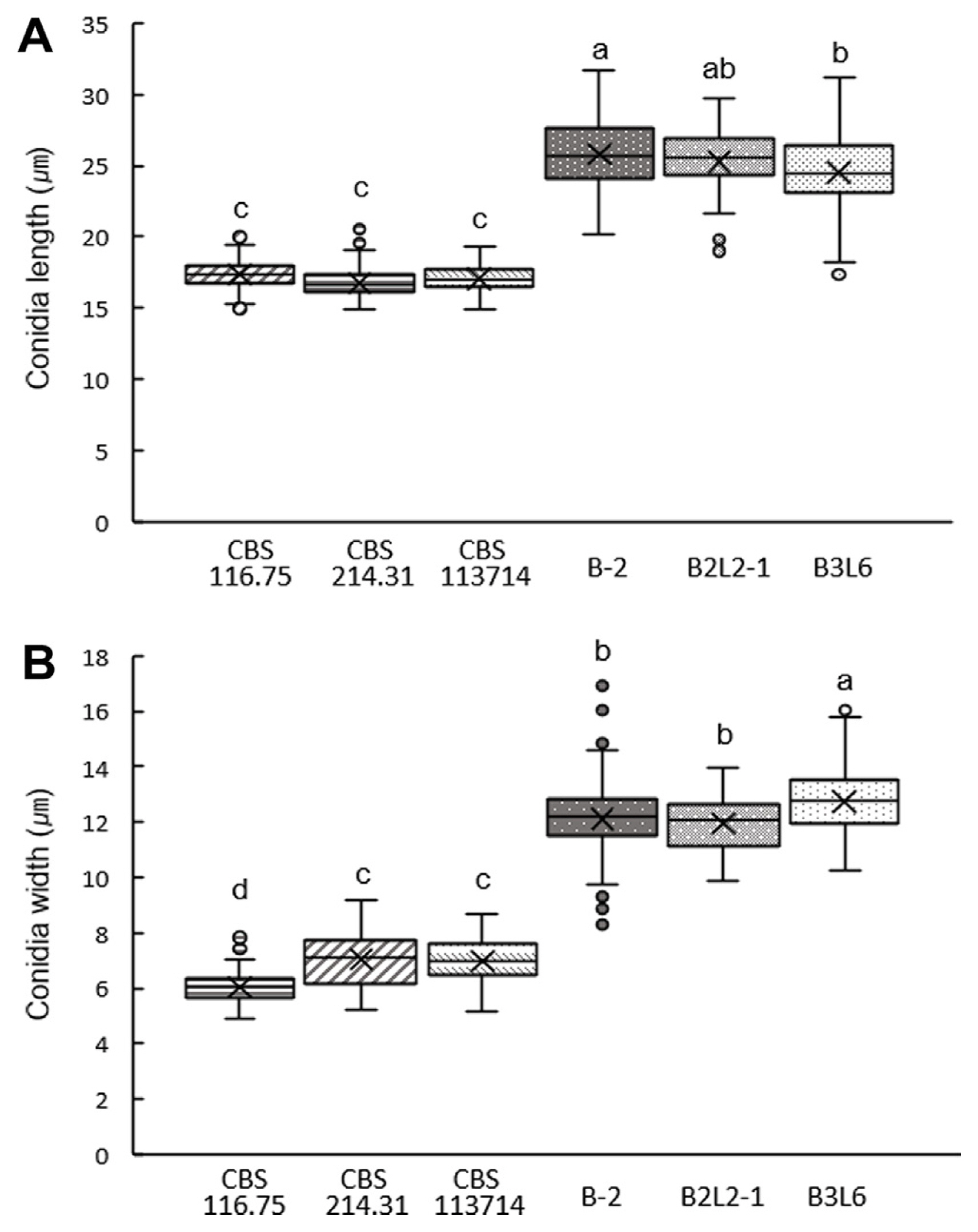
Conidia size measurement of Neofusicoccum buxi. Measurement of conidia length (A) and width (B) from 100 conidia of each isolate of N. buxi. Conidia were collected at 18–21 dpi on pine needle agar. Different letters above values indicate significant differences (P < 0.05, Tukey’s test in one-way ANOVA).
Phylogenetic analyses
Multigene alignment was performed using DNA barcode genes of ITS, LSU, and β-tubulin. We used the neighbor-joining and maximum likelihood methods to analyze the alignment. Sequence analysis of P. buxi and P. foliicola strains produced a total length of 1,576 bp (ITS + LSU + β-tubulin). Fungal isolates identified as P. buxi were segregated into three subclades, and two CBS P. buxi strains were separated (Fig. 5A). However, we could not distinguish distinct morphological characteristics of mycelial colonies and/or spores among the three subclades (Figs. 2 and 5A).

Phylogenetic analyses of Pseudonectria spp. and Neofusicoccum buxi. Neighbor-joining phylogenetic trees based on the concatenated ITS, β-tubulin, and LSU sequences of Pseudonectria spp. (A) and N. buxi (B). Sarocladium kiliense and Pseudofusicoccum ardesiacum were used as outgroups.
We confirmed that isolates obtained in this study were closely related to N. buxi strains separated from N. parvum (Fig. 5B). Moreover, they formed a separate branch in the phylogenetic tree, distinguishable from CBS N. buxi strains (Fig. 5B). Each isolate had 30 variable sites compared to CBS N. buxi strains, showing 98.05% identity. All three field isolates of N. buxi showed identical sequences, although there were morphological variances (Figs. 2C and 5B).
Pathogenicity tests
Virulence tests were conducted by inoculation of mycelial plugs of fungal strains on healthy boxwood leaves and incubated for 7 days in the case of P. foliicola and 21 days in the case of P. buxi and N. buxi (Fig. 6). Fungal strains were reisolated from inoculated leaves to fulfill Koch’s postulates. It was previously reported that P. buxi can invade through wound sites in boxwood leaves (Shi and Hsiang, 2014b). For this reason, we predicted that P. foliicola, would have an invading mechanism similar to that of P. buxi. Detached leaves with isolates of P. buxi and P. foliicola turned dark brown, and pink sporodochia were produced on the back sides of leaves. Leaves that were inoculated with CBS strains and field isolates of P. foliicola showed faster symptom progression than those inoculated with P. buxi strains (Fig. 6A and B). All isolates of N. buxi, as well as CBS strains, produced black perithecia on the back sides of detached leaves (Fig. 6C). Moreover, all isolates of N. buxi caused straw-colored leaves.

Pathogenicity tests of Pseudonectria buxi, P. foliicola, and Neofusicoccum buxi. Pathogenicity tests were conducted, and symptoms were evaluated on detached leaves using isolates of P. buxi (A), P. foliicola (B), and N. buxi (C). Detached boxwood leaves were cut and inoculated with hyphal plugs. After 7–21 days of incubation, brownish or yellowish colored leaves were detected, whereas no obvious symptoms were observed on the mock-inoculated leaves.
Discussion
Boxwoods are unavoidably vulnerable to infection by various plant pathogens through wound sites since pruning of ornamental trees should be performed periodically. In particular, occurrences of fungal diseases on boxwoods such as Volutella blight and Macrophoma leaf spot lead to decreases in economic value and aesthetic quality (Daughtrey, 2019; Jacobi et al., 2003). Because common diseases of boxwood cause similar symptoms on infected plants, characterization and understanding of the pathogens is important for accurate diagnosis and management of the diseases. In this study, we isolated the pathogens causing Macrophoma leaf spot and Volutella blight on boxwood in Korea. Based on the morphological and taxonomic features, we successfully identified 13 fungal isolates (P. buxi, P. foliicola, and N. buxi) responsible for blight symptoms of boxwood.
In Botryosphaeriales, to which N. buxi belongs, species identification based on morphological characteristics or single DNA barcode sequence analysis is not accurate (Slippers et al., 2017). Because of these taxonomic difficulties, the classification of N. buxi has continued to change. The first report of the Macrophoma leaf spot described the causative pathogen as Hyponectria buxi (Dodge, 1944). Then, fungal isolates included in H. buxi were changed into N. buxi through multiple DNA barcode-based species delimitation (Yang et al., 2017). Meanwhile, Dothiorella candollei, formerly known as Macrophoma candollei, was reported as the pathogen of leaf spot of boxwood (Batdorf, 1995). Recently, D. candollei was correctly identified as N. buxi, included in the order Botryosphaeriales (Yang et al., 2017). In this study, three fungal strains (B-2, B3L6, and B2L2-1) of N. buxi were predicted based on the BLASTn results and phylogenetic analysis of multiple DNA barcode loci. However, all isolates annotated as N. buxi showed markedly different morphologies of mycelial colonies and conidia compared to those of the CBS reference strains, suggesting that they have the potential to be novel species. Further isolation and in-depth fungal taxonomical studies are required for these isolates annotated as N. buxi.
All 13 isolates of P. buxi, P. foliicola, and N. buxi we collected in this study fulfilled Koch’s postulates. Boxwood leaves inoculated with CBS strains and isolates of P. buxi and P. foliicola showed blight symptoms, such as brownish colored leaves. Interestingly, P. foliicola caused dieback of leaves faster than P. buxi, suggesting that P. foliicola strains have stronger virulence than P. buxi. Leaves inoculated with isolates of N. buxi also showed straw-colored leaves and produced black perithecia at 21 dpi. Taken together, the N. buxi strains isolated in this study seemed to be weak pathogens or saprophytes, similar to previous reports (Jacobi et al., 2003).
This study included investigations of an outbreak of boxwood diseases and the determination of the morphological and taxonomical characteristics of associated fungal pathogens in Republic of Korea. Symptomatic boxwood samples were collected in a limited area for this study. If boxwood samples are collected in a wide range of areas in further research, more diverse and indigenous fungal pathogens of Republic of Korea can be analyzed. Our study will contribute to the development of control strategies for boxwood diseases caused by fungi and accelerate research on the complex ecology of boxwood diseases.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This work was supported by Crop Viruses and Pests Response Industry Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (321101-03 and 321107-03) and the National Research Foundation of Korea (2021R1A2C1004673).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).

