A Rapid and Simple Method for DNA Preparation of Magnaporthe oryzae from Single Rice Blast Lesions for PCR-Based Molecular Analysis
Article information
Abstract
Rice blast is one of the most destructive diseases of rice worldwide, and the causative agent is the filamentous ascomycete Magnaporthe oryzae. With the successful cloning of more and more avirulence genes from M. oryzae, the direct extraction of M. oryzae genomic DNA from infected rice tissue would be useful alternative for rapid monitoring of changes of avirulence genes without isolation and cultivation of the pathogen. In this study, a fast, low-cost and reliable method for DNA preparation of M. oryzae from a small piece of infected single rice leaf or neck lesion was established. This single step method only required 10 min for DNA preparation and conventional chemical reagents commonly found in the laboratory. The AvrPik and AvrPi9 genes were successfully amplified with the prepared DNA. The expected DNA fragments from 570 bp to 1,139 bp could be amplified even three months after DNA preparation. This method was also suitable for DNA preparation from M. oryzae strains stored on the filter paper. All together these results indicate that the DNA preparation method established in this study is reliable, and could meet the basic needs for polymerase chain reaction-based analysis of M. oryzae.
The filamentous ascomycete fungus Magnaporthe oryzae (synonym Pyricularia oryzae) (Couch and Kohn, 2002; Zhang et al., 2016), is the causal agent of rice blast disease in rice-growing areas worldwide (Ou, 1985). Typical symptoms of the disease are diamond shape lesions on leaves and necrotic spots on the panicle stem, also called neck blast. Genetic resistance is the most economic, effective and environmentally responsible method for its control. However, resistance genes typically lose their effectiveness after a short period in commercial production, due to the emergence of M. oryzae pathotypes (also called races) overcoming resistance (Ou, 1985). Monitoring of pathogenic changes of M. oryzae populations was carried out in various parts of the world in the past decades in order to identify resistance genes useful to breed for efficient and durable resistance (Kawasaki-Tanaka et al., 2016; Lara-Álvarez et al., 2010; Nguyet et al., 2020; Odjo et al., 2014; Pagliaccia et al., 2018; Park et al., 2008; Peng et al., 2021; Wang et al., 2017). Several sets of differential cultivars carrying single or few resistance gene(s) have been extensively used for monitoring pathotypes (Kiyosawa 1984; Kobayashi et al., 2007; Tsunematsu et al., 2000). However, the conventional pathotyping analysis of M. oryzae isolates with differential cultivars takes at least 4-weeks from sowing seeds to disease scoring. It also requires greenhouse space to grow plants and important manpower for isolating, growing, inoculating and scoring pathogen strains. These constraints limit the application of pathotyping to small samples that are not sufficient for monitoring pathotype changes at large scales and over long periods of time.
On the pathogen side avirulence/virulence to specific resistance genes is controlled by so called avirulence (Avr) genes in a gene-for-gene manner (Silué et al. 1992). Populations of the blast fungus adapt to rice resistance genes by the selection of individuals carrying mutated avirulence genes. Monitoring avirulence genes allows to detect potential resistance breakdown and to propose appropriate resistance gene deployment strategies. Cloning of avirulence genes in M. oryzae was initiated by genetic studies in the 1990 and boosted more recently by the development of rapid and low-cost high throughput sequencing methods. To date, nine avirulence (Avr) genes, including ACE1, AvrPi9, AvrPia, AvrPib, AvrPii, AvrPik/km/kp, AvrPita, AvrPiz-t, and AVR1-CO39 have been successfully cloned from rice-infecting strains (Böhnert et al., 2004; Li et al., 2009; Orbach et al., 2000; Ribot et al., 2013; Wu et al., 2015; Yoshida et al., 2009; Zhang et al., 2015). Transposable element insertion in regulatory or coding sequences, complete or partial deletion, and point mutation in Avr genes of M. oryzae can abolish the direct or indirect recognition by the plant of the cognate effector, resulting in the susceptibility of rice cultivars carrying the cognate resistance genes (Chuma et al., 2011; Li et al., 2009; Zhang et al., 2015). Availability of Avr gene sequences opens the way to determine pathotypes by a direct molecular characterization of Avr genes, instead of pathotyping by inoculation on differential cultivars (Chen et al., 2014; Chuma et al., 2011).
To perform molecular characterization of M. oryzae Avr genes, DNA extraction of the pathogen is needed. The conventional method for preparation of fungal DNA consists of several steps, including isolation of single spore from diseased rice samples, cultivation of mycelium on solid medium, cultivation of mycelial plug on liquid or solid medium, disrupting cell wall by enzyme digestion or mechanic actions, removal of proteins with solvent, and precipitation of DNA (Huang et al., 2014; Saleh et al., 2012b; Singh et al., 2014; Sweigard et al., 1990). Although these methods are suitable to isolate large amount of DNA with high quality, they are time consuming, labor intensive and even expensive (when using a kit). To timely carry out the analysis and monitoring of avirulence genes and genetic diversity of M. oryzae populations in rice-growing areas, it is necessary to develop a rapid and simple method for direct amplification from infected rice samples. Harmon et al. (2003) have extracted DNA of M. oryzae from infected leaf tissues of perennial ryegrass with a kit. To date, a rapid and simple method is not available for rice.In this study, we developed a one-step method for DNA extraction and polymerase chain reaction (PCR) amplification of M. oryzae directly from infected leaf and neck of rice.
Twenty-two typical leaf blast and neck blast lesions were collected from blast nursery located at Yuxi of Yunnan province of China, respectively. The samples were allowed to dry and stored in the laboratory at room temperature in paper bags for one week. The healthy leaf and neck without blast symptoms of rice collected from greenhouse were used as negative control, and the M. oryzae strain CH0997 (Saleh et al., 2012a) was used as positive control. Additionally, 96 M. oryzae single spore strains isolated previously in laboratory from different rice-growing areas of Yunnan Province of China and stored on filter papers, were also used for DNA preparation and PCR amplification.
DNA extraction buffer consisting of 25 mM Tris-HCl (pH 8.7), 2.5 mM EDTA (pH 8.0), 250 mM KCl, 0.02% Tween 20 was prepared and sterilized before use. A small piece of leaf of about 2 mm × 2 mm from single leaf blast lesion, or neck of about 2 mm in length from single neck blast lesion were cut and then put into a well of 96-well PCR plate. For amplification from −20°C stock, a piece of filter paper about 2 mm × 2 mm was cut and deposited into 96-well PCR plate. For both leaf lesion and filter paper samples, 100 μl of DNA extraction buffer were added to each well. The PCR plate was then caped and heated at 95°C for 10 min in a thermocycler (C1000, Bio-Rad, Hercules, CA, USA). DNA quality was checked by migration of 1 μl of DNA solution on 1.0% agarose gel; DNA concentration was determined by Qubit 3 Fluorometer (Invitrogen, Carlsbad, CA, USA) with Qubit dsDNA HS (high sensitivity, 0.2–100 ng) Assay Kit, and then the DNA solution was stored at −20°C until use for PCR amplification. The genomic DNA of M. oryzae strain CH0997 extracted from mycelium using the HP Fungal DNA Mini Kit (Omega BIO-TEK, Norcross, GA, USA) was used as positive control.
Two sets of PCR primers were used for amplification to determine quality and quantity of extracted DNA. The first set was designed with Primer3 software for amplification of a 1,139-bp fragment containing the full AvrPi9 gene of M. oryzae, based on the gene sequence of AvrPi9 gene (Wu et al., 2015). The primers were AvrPi9Fw (5′-GGTCCACTGCTCCATCTTGTTTG-3′) and AvrPi9Rv (5′-CCCTTCTGCGCATTACTGATACC-3′). The second set was designed with Primer3 software for amplification of a 570 bp fragment containing the full AvrPik gene of M. oryzae, based on the gene sequence of AvrPik locus (Yoshida et al., 2009). The primers were AvrPikFw (5′-CCCTCGTAAAATCCAATTCC-3′) and AvrPikRv (5′-CTTATGAGCCGTCAACCAAG-3′).
All PCR reactions were carried out using 2× Es Taq MasterMix (Beijing CoWin Biotech Co., Ltd., Beijing, China). Each PCR consisted of the following components: 10 μl of 2× Es Taq PCR Master Mix (containing 0.5 U of Es Taq DNA polymerase, 2× Es Taq PCR Buffer containing loading buffer, 1.5 mM MgCl2, 0.2 mM of each dNTP), 1 μl of 10 μM for each primer, 1 μl of DNA solution prepared with the method described above, or 1 μl (10 ng/μl) DNA of CH0997 (positive control), and added with distilled H2O to final reaction volume of 20 μl. PCR amplification conditions consisted of a denaturing step of 95°C/3 min, followed by 35 cycles of 95°C/30 s, 55°C/30 s, and 72°C/90 s, ending with an extension step of 72°C/7 min. Amplicons were separated by 1.2% agarose gel electrophoresis and visualized by straining with Dured Nucleic Acid Gel Stain (Beijing Fanbo Biochemicals Co., Ltd., Beijing, China). Three PCR amplicons for each set of primers were randomly selected for sequencing.
To determine the quantity and quality of DNA extracted from leaf blast lesions with the method described in this study, one microliter of DNA extracts was checked by electrophoresis on 1% agarose gel. Since the extraction was done on a small fraction of infected tissues, low DNA quantity was extracted and no fragments could be seen (data not shown). The DNA concentrations evaluated with a more sensitive method (Qubit 3 Fluorometer, Life Technologies, Penang, Malaysia) showed that DNA concentrations of extracts ranged from 0.33 ng/μl to 2.04 ng/μl, and the average was about 0.81 ng/μl. Subsequently, the DNA extracted from 22 leaf blast samples and one healthy leaf sample were used as PCR template for amplification with primers for AvrPik and AvrPi9. Fragment of the expected sizes were observed for both genes and for all infected leaves and the positive control (Fig. 1A and C), demonstrating successful and specific direct amplification. To validate whether the amplified fragments correspond to the targeted AvrPik and AvrPi9 genes, three randomly selected amplicons from each primer set were sequenced. The sequenced fragments showed identical sequences to AvrPik and AvrPi9, respectively (data not shown), indicating that the amplicons are the expected target genes. In order to evaluate the stability of extracted DNA, PCR reactions were carried out with extraction products stored at −20°C for over 3 months after preparation. Again, the expected PCR products were obtained (Fig. 1B and D). Similarly to DNA extracted from leaf blast lesions, AvrPik and AvrPi9 genes were successfully amplified with DNA solution of the 22 neck blast samples and positive control CH0997 (Fig. 1E and G). The concentration of DNA extracts from neck blast samples were also determined with the same method as mentioned above. The DNA concentrations ranged from 1.29 ng/μl to 8.87 ng/μl, and the average was about 4.42 ng/μl. When using the extracted DNA from infected necks stored at −20°C for over 3 months as template, the expected PCR products were also amplified clearly (Fig. 1F and H). These results showed that both infected leaves and neck blast lesions are suitable for pathogen DNA extraction with this described method.
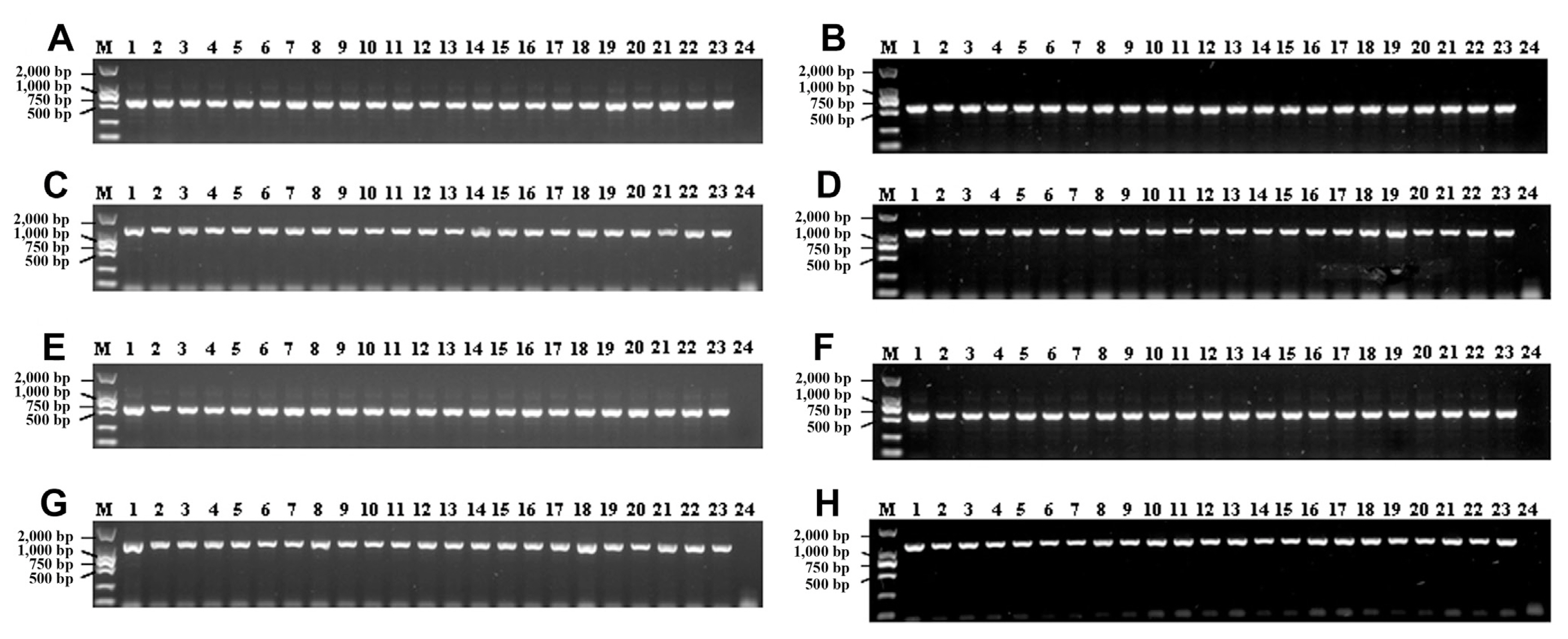
Polymerase chain reaction products amplified by using primers specific to AvrPik and AvrPi9 genes. Comparisons of amplifications with DNA template prepared the same day (A, C, E, G) and after three months of storage at −20°C (B, D, F, H). (A, B) Amplicons of 570-bp amplified with AvrPik gene-specific primers from leaf blast lesions. (C, D) Amplicons of 1,139-bp amplified with AvrPi9 gene-specific primers from leaf blast lesions. (E, F) Amplicons of 570-bp amplified with AvrPik gene-specific primers from neck blast lesions. (G, H) Amplicons of 1,139-bp amplified with AvrPi9 gene-specific primers from neck blast lesions. M, DNA marker DL2000; lane 1, CH0997 (positive control); lane 2 to 23, DNA templates of M. oryzae prepared directly from single blast lesions; lane 24, negative control (healthy leaf [A, C] or healthy neck [E, G] of rice).
M. oryzae strains isolated from blast lesions are usually stored on dried filter paper for long term storage at −20°C (Valent et al., 1986). In order to test our DNA extraction method on these samples, the DNA extracted from the paper stock of 96 M. oryzae strains were used for PCR amplification with AvrPik and AvrPi9 specific primers. As a result, the PCR fragments of the expected size were also obtained from each sample (Fig. 2A and B), indicating that this method is also suitable for M. oryzae DNA extraction from filter paper stock.

Polymerase chain reaction products of DNA extracted from filter paper stock of 96 Magnaporthe oryzae strains amplified with specific primers of AvrPik and AvrPi9 gene. (A) Amplicons of 570-bp amplified with AvrPik specific primers. (B) Amplicons of 1,139-bp amplified with AvrPi9 specific primers. M, DNA marker DL2000.
DNA extraction is the first step in many molecular assays including molecular diagnostic, monitoring, population genetics and genomics. Some simplified methods were used for DNA extraction from spores or mycelium of M. oryzae cultures (Jia et al., 2014; Saitoh et al., 2006; Xu and Hamer, 1995). In this study, we developed a fast, low-cost and reliable method for DNA preparation of M. oryzae directly from infected rice tissues with common chemical reagents in a regular laboratory. This one-step protocol requires a small piece of infected rice tissue or filter paper containing M. oryzae, and takes only 10 min to prepare pathogen DNA. Although DNA extracts could not be seen on agarose gel by electrophoresis method, DNA concentration could be determined with a more sensitive method, such as Qubit 3 Fluorometer in this study. The concentration of DNA extracts from neck blast samples were higher than that of leaf blast lesions; this result might be partly attributed to the fact that the cylindrical neck blast samples could contain more pathogen mycelium, compared to that in thin leaf blast samples of the same length. The extracted DNA could be used directly as PCR template to detect the target DNA fragments of M. oryzae from 570 bp to 1,139 bp in length, indicating that prepared DNA with the method established in this study can meet the basic needs of PCR-based analysis, such as population genetics of M. oryzae with simple sequence repeat markers, diagnostic and Avr genes detection with gene-specific molecular markers, etc. Usually, single typical blast lesions are larger than 3 mm in length on infected rice in the field (Bonman et al., 1987). Then, a single lesion is sufficient for both direct DNA preparation and, at the same time, isolation of M. oryzae single spore for further studies. With more and more Avr genes being cloned from M. oryzae, the available Avr gene sequences would facilitate the development of PCR-based Avr gene-specific primers to determine the Avr genes in M. oryzae population. The rapidly prepared DNA samples from single blast lesion will meet the needs of timely monitoring of dynamic changes of Avr genes in large M. oryzae populations in rice-growing areas.
Acknowledgments
This work was supported by the Applied Basic Research Key project of Yunnan (202001AS070006), the National Natural Science Foundation of China (31560493, 31860524), and the Major Special Project of Yunnan Province (202102AE090003).
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
