 |
 |
| Plant Pathol J > Volume 32(2); 2016 > Article |
Abstract
Fusarium graminearum Schwabe causes Fusarium head blight (FHB), a devastating disease that leads to extensive yield and quality loss of wheat and other cereal crops. Twelve isolates of F. graminearum were collected from naturally infected spikes of wheat from Assiut Egypt. These isolates were compared using SRAP. The results indicated distinct genetic groups exist within F. graminearum, and demonstrated that these groups have different biological properties, especially with respect to their pathogenicity on wheat. There were biologically significant differences between the groups; with group (B) isolates being more aggressive towards wheat than groups (A) and (C). Furthermore, Trichoderma harzianum (Rifai) and Bacillus subtilis (Ehrenberg) which isolated from wheat kernels were screened for antagonistic activity against F. graminearum. They significantly reduced the growth of F. graminearum colonies in culture. In order to gain insight into biological control effect in situ, highly antagonistic isolates of T. harzianum and B. subtilis were selected, based on their in vitro effectiveness, for greenhouse test. It was revealed that T. harzianum and B. subtilis significantly reduced FHB severity. The obtained results indicated that T. harzianum and B. subtilis are very effective biocontrol agents that offer potential benefit in FHB and should be harnessed for further biocontrol applications. The accurate analysis of genetic variation and studies of population structures have significant implications for understanding the genetic traits and disease control programs in wheat. This is the first known report of the distribution and genetic variation of F. graminearum on wheat spikes in Assiut Egypt.
Fusarium head blight (FHB), caused by the fungal plant pathogen Fusarium graminearum, is an important disease in cereal crops causing major economic losses of 20-100% (McMullen et al., 1997 and Manning et al., 2000). Diseased spikelets exhibit symptoms of premature bleaching shortly after infection. The fungus produces a mycotoxin that poses a significant threat to the health of domestic animals and humans. The major toxin produced by F. graminearum in association with FHB in wheat and barley is deoxynivalenol (Pestka and Smolinski, 2005 and McMullen et al., 1997). Mycotoxins produced by Fusarium species result in a loss of yield and reduced quality of grains. Fusarium toxins including the trichothecenes nivalenol (NIV), deoxynivalenol (DON) and its derivatives 3- and 15-acetyldeoxynivalenol (3-ADON, 15-ADON) contaminate cereal products and have been shown to be harmful to humans, animals, and plants (Desjardins and Hohn, 1997; Desjardins et al., 1993 and Goswami and Kistler, 2004). Sequence related amplified polymorphism (SRAP) technology has been recognized as one of the most variable types of DNA sequences found in plants. This SRAP system has been employed for mapping and gene tagging in Brassica (Li and Quiros, 2001). SRAP marker is homogenously distributed in the genome and could produce higher polymorphism than those from AFLP, RAPD, and SSR markers. It has been employed to evaluate genetic diversity and phonetic relationships among Turfgrass species (Budak et al., 2004a). The polymorphism produced by SRAP (95%) marker technique was higher than those produced by ISSR (81%), RAPD (79%), and SSR (87%) (Budak et al., 2004b). The SRAP marker technique was used as a new technique to assess genetic relationships and diversity among genotypes of Saccharum. The level of observed polymorphism proved that the SRAP system was robust at amplifying markers across species and genera and did so according to the evolutionary history interconnecting members of the Saccharum complex (Suman et al., 2008). Cloning and sequencing of a set of cDNA to visualize transcript polymorphism are reported using SRAP technology in three Bentgrass species. The ESTs identified in this study could potentially be used in Turfgrass breeding and genetics programs as functional markers. Integration of these ESTs to the existing linkage map of Turfgrass species provides high-density coverage in selected genomic regions. Minimum evolutionary tree clustering indicated that ESTs obtained using SRAP could be used for comparative genomics analysis of transcribed genes among the grass species (Dinler and Budak, 2008). Furthermore, Baysal et al., 2009 use SRAP primers to study the population and genetic relationships within and among Fusarium oxysporum f. sp. lycopersici races. Mutlu et al., 2008 reported the tagging of the gene for resistance to Fusarium wilt (FOM) in eggplant using SRAP, RGA, SRAP-RGA and RAPD markers. Molecular markers are useful tools in the analysis of genetic variation in populations of plant-pathogenic fungi. A number of molecular techniques are available for studying the genetic relationships within and among fungal populations within a species. Sixty isolates of F. graminearum, the causal pathogen of Fusarium head blight, were compared using vegetative compatibility analysis and Polymerase Chain Reaction (PCR)-based Sequence Related Amplified Polymorphisms (SRAP) (Fernando et al., 2006). SRAP is based on two-primer amplification to amplify the ORFs. In a gene, ORFs are located between the start-code sequence (initiation codon) and the stop-code sequence (termination codon) (Li and Quiros, 2001). The analysis of sequenced SRAP fragments targets into hypothetical proteins from different Fusarium species showing that the SRAP technique not only allows studying F. poae genetic variability, but also targets coding regions into the F. poae genome. Genetic variability of F. poae using SRAP technique also demonstrates the efficacy of this molecular marker to amplify open reading frames in fungus (Dinolfo, et al., 2015). In this study SRAP analysis was used to determine the genetic variation of F. graminearum isolates.
Biological control of F. graminearum has shown promise in previous studies due to their low environmental impact, and their ability to help reduce growersã dependency on chemicals, thereby slowing the development of fungicide resistance in pathogen populations (Crane et al., 2013; Jochum et al., 2006). Several bacteria or fungal strains have been reported to have antagonistic effects against F. graminearum (Hue et al., 2009). Trichoderma species are biological control agents that control ascomycetous and basidiomycetous fungi, which are mainly soil-borne but also airborne pathogens. Antagonists of phytopathogenic fungi have been used to control plant diseases, and 90% of such applications have been carried out with different strains of the fungus Trichoderma (Monte, 2001). The genus Trichoderma comprises a great number of fungal strains that act as biological control agents, the antagonistic properties of which are based on the activation of multiple mechanisms. Trichoderma can indirectly biocontrol phytopathogens by competing for nutrients and space nutrients, through the secretion of antibiotic volatiles and/or diffusible metabolites, which modify soil conditions promoting growth and plant defense mechanisms. Moreover, mycoparasitism is considered a direct biocontrol mechanism (BenûÙtez et al., 2004; Howell, 2003). The addition of Trichoderma metabolites that may act as elicitors of plant resistance, or the expression in transgenic plants of genes whose products act as elicitors, also results in the synthesis of phytoalexins, PR proteins and other compounds, and in an increase in resistance against several plant pathogens, including fungi and bacteria (Dana et al., 2001; Elad et al., 2000). Bacterial isolates obtained from rhizosphere and kernel of wheat was reported for control Fusarium head blight (Stockwell et al., 1999). Among them, Bacillus strains are well-known antibiotic producers, which have advantage over other biocontrol microorganisms due to their inherent property to form endospores and resistance to extreme conditions. The antagonistic effects of Bacillus strains have been shown by in vitro antibiosis (Chan et al., 2003) and in situ disruption of spikelet infection leading to reduced disease severities (Khan et al., 2001). B. subtilis was demonstrated to be the most effective in reduction, affecting fungal growth parameters and toxin production in vitro at all the tested incubation periods (Cavaglieri et al., 2005). Bacillus species, as a group, offer several advantages over other bacteria for protection against root pathogens because of their ability to form endospores and the broad-spectrum activity of their antibiotics. There are numerous reports of Bacillus species which repress pathogens (Bacon et al., 2001; Estûˋvez de Jensen et al., 2002). Therefore, this study was carried out with the objective of evaluating the efficacy of Trichoderma harzianum and Bacillus subtilis in management of FHB caused by F. graminearum.
A total of fifteen wheat fields in three zones of Assiut governorate were sampled during March and April 2011. Ear bleaching and spikelet bleaching with FHB symptoms were collected. The infected wheat heads were cut into 0.5 cm long pieces. These were surface sterilized in 3% sodium hypochlorite solution, rinsed twice in sterile distilled water and blot dried between sterile filter paper. The surface sterilized pieces were placed onto potato dextrose agar (PDA) amended with streptomycin sulphate (120 mg/l) and incubated for five days at 25ô¯C. Fungal colonies were identified based on cultural and morphological characteristics like mycelial colour, pigmentation, spore shape, septation and sporophores. All colonies with characteristic growth patterns of FHB pathogens were transferred on to fresh PDA; those with growth patterns typical of Fusarium species were also plated onto Sucrose Nutrient Agar (SNA) (Nirenberg, 1981). Plates were incubated at 25ô¯C under 12 hrs day light and 12 hrs darkness cycles for 10 days and cultures identified to species level based on colony characteristics on PDA and spore morphology on SNA according to Nelson et al., 1983. Hyphae were stained with 0.05% trypan blue in lacto-phenol (Meyer et al., 1998) and examined under a compound microscope to determine hyphal morphology. All isolates of F. graminearum were purified by single spore isolation.
Twelve isolates of F. graminearum were tested for the ability to infect wheat plants in greenhouse of Plant Pathology Department, Faculty of Agriculture, Assiut University in October 2011. A medium susceptible Egyptian wheat cultivar (Sakha-69) was used for the investigation. Wheat seeds were planted in sterilized pots (25 û 25 cm2) containing a peat/sand mixture (ten seeds per pot) with four pots per isolate (replicates). The pots were kept moist and moved to random positions on the greenhouse bench following a complete randomized design, and grown at 25 ôÝ 2ô¯C during day and 17 ôÝ 2ô¯C during night. F. graminearum conidial inoculums was prepared with Mung Bean Agar Medium (MBA) (Bai and Shaner, 1996). Each isolate was cultured separately at 25 ôÝ 1ô¯C for14 days and used to inoculate wheat plants.
Conidia suspension of each isolate was harvested and adjusted to 5 û 105 conidia/ml. Three drops (0.01%) of Tween 20 was added to ensure uniform conidia dispersion. Wheat spikes were inoculated at 50% flowering (GS65, Zadoks et al., 1974) by spraying with hand sprayer, exposing all spikelets to the inoculum. Controls were treated similarly with distilled water only. After inoculation, the spikes were incubated under polythene bags for 48 hrs to ensure high relative humidity for optimal infection. Each isolate was inoculated separately and replicated three times during 10 days.
Fusarium head blight was assessed as a percentage of heads showing disease symptoms, on ten average sized spikes per replicate. The number of infected spikelets/head was recorded at two dates (14 and 28 days after inoculation) and adjusted to the total number of spikelets/heads. The relative number of infected spikelets of the two assessment dates was averaged (Cumagun and Miedaner, 2003; Mesterhazy, 2002; Snijders and Perkowski, 1990).
Isolates were cultured on potato dextrose agar (PDA) for 7 days at 25ô¯C. Mycelia of each isolate were prepared in a flask (250 ml) with 100 ml of Potato Dextrose Broth (PDB). To obtain mycelia, PDB flasks were inoculated with a 0.5 ml suspension of approximately105 conidial spores per milliliter of an isolate. The flasks were incubated at 25ô¯C for 7 days without agitation. The mycelia were harvested by filtration through two layers of sterilized miracloth, frozen with liquid nitrogen and stored at ã80ô¯C until lyophilized. Before proceeding with nucleic acid extraction, the mycelium was ground in liquid nitrogen in a sterile mortar to obtain a mycelium powder.
Total genomic DNA was extracted from lyophilized mycelium according to the CTAB (hexacetyltrimethylammonium bromide; Sigma-Aldrich) a modification of miniprep protocol described by OãDonnell et al., 1997 and Voigt et al., 1999 was used. Approximately 50 mg of pulverized mycelium was re-suspended in 700 ö¥l of CTAB extraction buffer (100 mM Tris-HCl [pH 8.4], 1.4 M NaCl, 25 mM EDTA, 2% CTAB) and vortexed for 10 seconds. Following extraction, an equal volume of chloroform was added to each tube, vortexed for 5 seconds, and then spun for 10 minutes at 12,300 û g in a microcentrifuge (Eppendorf AG-Centrifuges 5415D). A 500 ö¥l portion of the upper phase was removed to a new 1.5 ml tube, and DNA was precipitated by the addition of an equal volume of ã20ô¯C isopropanol. After the DNA was pelleted at 12,300 û g in a microcentrifuge for 1 min, the supernatant was discarded and resulting pellets were washed twice with 70% ethanol. Pellets were each air-dried and re-suspended in 100 ö¥l of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]). RNA contamination was removed by incubating each preparation with 40 ö¥g/ml RNase A (Sigma) at 37ô¯C for 30 min. The concentration of DNA was determined by spectrophotometry with a nano-drop spectrophotometer ND-1000 (NanoDrop Technologies) at A260.
The SRAP analysis was carried out using 16 primer combinations (Table 1). The PCR reaction was set up in a final volume of 20 ö¥l containing 50 ng of template DNA, 1û PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs mix, 0.1 mM forward primer, 0.1 mM reverse primer and 1 unit of Taq polymerase; final volume completed to 20 ö¥l with sterile ddH2O (Li and Quiros, 2001 and Budak et al., 2004a with minor modifications). Amplifications were performed in mastercycler gradient-thermal cycler (Eppendorf, Germany) programmed for 5 min at 95ô¯C for initial denaturation (one cycle); followed by 35 cycles of 1 min at 94ô¯C for denaturation, 1 min at 47ô¯C for annealing, 1 min at 72ô¯C for extension, and ending with 5 min at 72ô¯C for a final extension (one cycle) (Buddak et al., 2004c). Amplified DNA was analyzed by electrophoresis in 1.0% agarose gel, then stained with ethidium bromide (0.5 ö¥g/ml) and observed under UV light in gel documentation system (Bio-Rad).
Digital images were scored as ã1ã for presence and ã0ã for absence of clear and unambiguous DNA fragments. Similarity matrix was constructed from the binary data with Jaccardãs coefficients (Jaccard, 1908). The genetic distance matrix was subjected to cluster analysis using the Unweighted Pair-Group Method with Arithmetic mean (UPGMA) in NTSYS-pc version 2.1 program (Rohlf, 2000).
Trichoderma harzianum and Bacillus subtilis were isolated from wheat kernels. T. harzianum was isolated on PDA medium at 26ô¯C and identified based on morphological characteristics of mycelia and conidiophores as described by Domsch et al., 1980 and Dhingra and Sinclair, 1995. Whereas, B. subtilis was isolated on Nutrient Agar (NA) at 28ô¯C, and identified based on morphological, culture and biochemical activities according to Skinner and Lovelock, 1979 and Sneath et al., 1986.
Antagonistic capability of seven isolates of T. harzianum and five isolates of B. subtilis were tested against the highly pathogenic isolate (F.g.8) of F. graminearum in vitro. Dual culture technique was followed; mycelial disks 5 mm in diameter were cut from the edges of actively growing colonies of F. graminearum and Trichoderma isolates, and were placed opposite each other, 1.5 cm from the edge of 9 cm Petri dishes containing PDA. Petri dishes inoculated with F. graminearum alone served as controls. Each pair was replicated four times and incubated for four days at 25ô¯C in darkness, then scored for degree of antagonism using the 1-5 scale of Bell et al. (1982) and Mahmoud and Abo-Elyousr (2014): 1, Trichoderma completely overgrew the pathogen and covered the entire Petri dish; 2, Trichoderma overgrew at least two thirds of the Petri dish; 3, Trichoderma and F. graminearum each colonized 50% of the medium surface and neither organism appeared to dominate the other; 4, F. graminearum colonized at least two-thirds of the medium surface and appeared to withstand the encroachment of Trichoderma; 5, F. graminearum completely overgrew the entire Petri dish. An isolate of Trichoderma was considered to be antagonistic to the pathogen if the mean score for a given comparison was ãÊ2, but not highly antagonistic if the score was ãË3. For test the efficacy of B. subtilis, the pathogen agar disc was inoculated at the middle of plate and the antagonist at two equidistant points located 1.5 cm from plate edge. Degree of antagonism was determined by measuring the pathogen colony diameters and percentage inhibition calculated:
Where: (A) is the colony diameter of pathogen alone (control); (B) is the colony diameter of pathogen after antagonist effect.
Ten seeds of susceptible wheat cultivar (Sakha-69) were sown, and the inoculum of the highly pathogenic isolate (F.g.8) of F. graminearum was produced in MBA (Bai and Shaner, 1996) as described previously in pathogenicity tests. Conidia suspension of F. graminearum was harvested and adjusted to 5 û 105 conidia/ml. Three drops (0.01%) of tween 20 was added to ensure uniform conidia dispersion. T. harzianum isolate (T. h.3) and B. subtilis isolate (B. s.2) with great inhibition zone in vitro against F. graminearum were investigated for their ability to reduce the incidence of head blight in wheat. Inoculum of T. harzianum was prepared using PDB for 14 days at 26ô¯C. Inoculums was harvested by passing the liquid culture through double layer cheesecloth, and adjusted to 5 û 105 spore/ml. While B. subtilis was grown on Nutrient Broth medium for 48 hrs at 28ô¯C and inoculums was adjusted to 2 û 105 cfu/ml with fresh medium. Wheat spikes were inoculated at 50% flowering (GS65, Zadoks et al., 1974) by spraying with hand sprayer, exposing all spikelets to the inoculum. Inoculation with F. graminearum began at 5 hr after the inoculation with T. harzianum and B. subtilis. Positive controls were inoculated similarly with F. graminearum only, while negative controls were treated with distilled water. There were four replicate pots per treatment. Pots were arranged in complete randomized design. After inoculation, the spikes were incubated under polythene bags for 48 hrs to ensure high relative humidity for optimal infection. Each isolate was inoculated separately and replicated three times during 10 days. Head blight was evaluated as described previously in pathogenicity tests.
The results were analyzed using ANOVA test and the means differences were regarded as significant using LSD test at 5% level of probability according to Gomez and Gomez, 1984.
Wheat fields were surveyed to access the incidence of FHB during wheat growing season, from March to the beginning of May, in 2011. Fifteen fields representing three districts of Assiut governorate namely Dirout, Manfalout and Abuteeg were surveyed. The geographic origins of the isolates collected are given in Table 2. The results indicated that, the morphological characteristics of twelve isolates were found to be identical to those of F. graminearum. Isolates were identified as F. graminearum on the basis of growth rate, pigmentation of colonies on PDA, spore morphology on SNA as well as morphology and size of microconidia and macroconidia according to Nelson et al. (1983) and Summerell et al. (2003). Hyphae were stained with 0.05% trypan blue in lacto-phenol (Meyer et al., 1998) and examined under a compound microscope to determine hyphal morphology.
FHB was assessed at two dates (14 and 28 days) after inoculation. All F. graminearum isolates caused visible head blight symptoms under greenhouse conditions. No symptoms of disease occurred in uninfected control. The wheat cultivar Sakha-69 was extremely susceptible to all of the isolates tested under greenhouse conditions. Means of FHB severity ranged from 54.25 to 92.50 %, averaging 68% in total. There were significant differences in disease severity among the twelve F. graminearum isolates. These differences were observed on the two dates on which the percentage of diseased spikelets was calculated. The percentage of diseased spikelets increased with time for the twelve F. graminearum isolates (Table 2). Based on the obtained results, the most aggressive isolates were F.g.8 followed by F.g.6 and F.g.9 which isolated from Manfalout. Whereas, the least aggressive isolates were F.g.11 and F.g.12, which isolated from Abuteeg. On the other hand, results show that, F. graminearum isolates obtained from Dirout were varied significantly, as some of these isolates associated with the low level of disease severity F.g.2 (62.75%) and some have a high level of aggressiveness such as F.g.5 (79.50%). The results also, revealed that, the percentage of diseased spikelets differed significantly among isolates of Manfalout. The most aggressive isolate showing the highest disease severity was the isolate F.g.8 (92.50%), while the least aggressive one was the isolate F.g.7 (78.25%). Therefore, isolate F.g.8 was selected for further work in biological control. On the other hand, the results showed that, F. graminearum group (B) is more pathogenic to wheat than groups (A) and (C), although the pathogenicity of individual isolates within each group was varied. The virulent of group (B) is potentially due to the differences in Fusarium mycotoxins production. Mycotoxins, predominantly are trichotecenes i.e nivalenol (NIV) and deoxynivalenol (DON). Previous publications confirmed that producing mycotoxins; such as trichothecenes, zearalenone and fumonisins, are associated with the most aggressive isolates. DON is necessary to suppress plant defense enabling the pathogen to break through the rachis node. DON production is strongly induced, most likely by the host, at this point of infection (Carter et al., 2002; Ilgen et al., 2009; Jansen et al., 2005). Moreover, in wheat and maize, trichothecene biosynthesis alters strain aggressiveness (Proctor et al., 1995), with DON-producing strains being perceived as more virulent than NIV-producing strains (Desjardins et al., 2008). Also, this observation was supported by results obtained by Kimura et al. (1998) and Alexander et al. (1998).
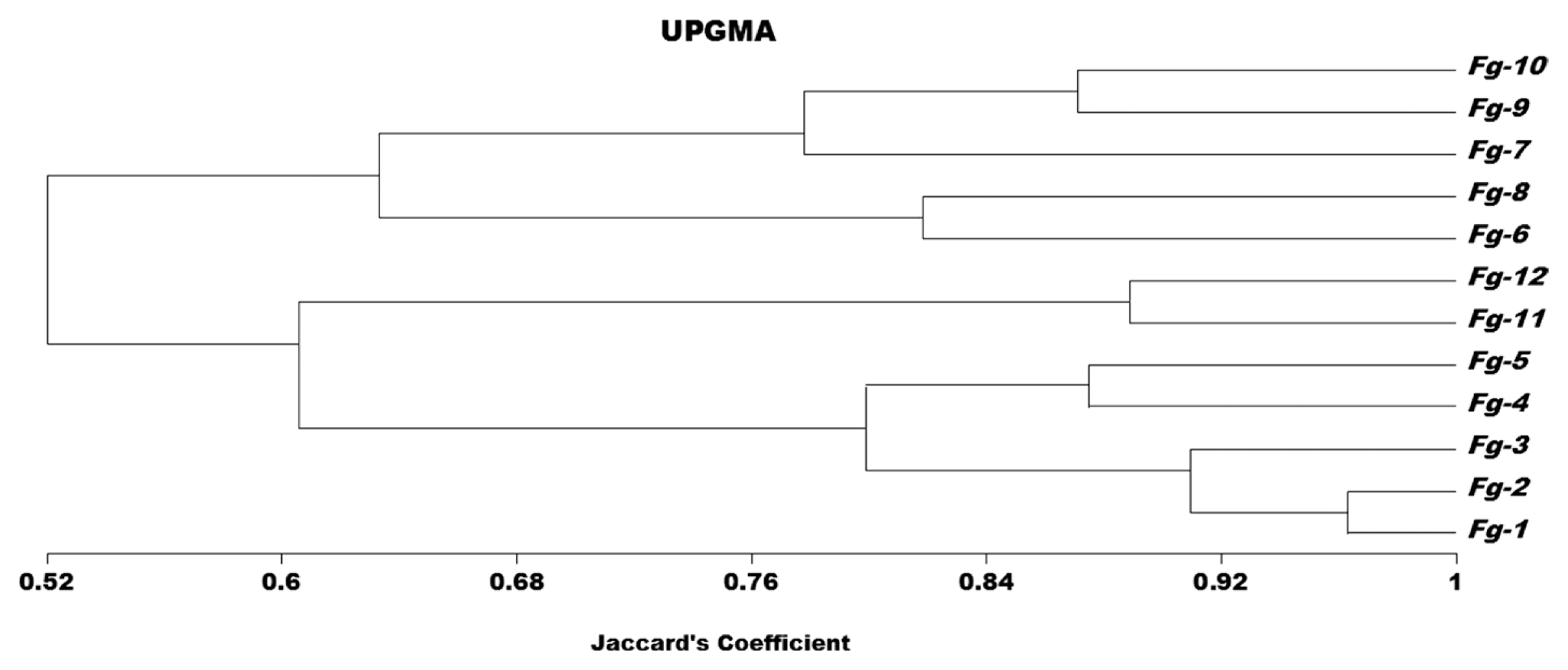
Genetic variation was detected among twelve isolates of F. graminearum using Sequence Related Amplified Polymorphism (SRAP) and it has been indicated a wide variation among all isolates of F. graminearum. Among the sixteen SRAP primer combinations, seven were amplified the genomic DNA of F. graminearum, and produced 2-8 bands ranging from 75-5,000 bp. The number of amplified DNA fragments varied, depending upon the primers and isolates used. The primers Em2- Me6, Em3- Me4, Em3- Me10, Em6- Me4, Em11- Me4, Em11- Me6 and Em14- Me3 amplified the genomic DNA of all isolates of F. graminearum and producing fingerprint profiles, which were clearly distinguished among the different isolates of F. graminearum. A dendrogram constructed using the SRAP data (Fig. 1) shows the isolates to be divided into three groups. The tested isolates were clustered together according to the geographic areas. The isolates of F. graminearum obtained from Dirout, Manfalout and Abuteeg were clustered together with a genetic similarity of 52%. Isolates obtained from Dirout (group A), were clustered together with a genetic similarity of 79%. While, isolates obtained from Manfalout (group B), were clustered together with a genetic similarity of 63%. Isolates F.g.11 and F.g.12 obtained from Abuteeg (group C) were clustered together and displayed high genetic similarity of 88%. The analysis also indicated that isolates: F.g.4, F.g.5, F.g.9 and F.g.10 (33%) of the F. graminearum isolates were highly similar to one another; they exhibited similarity coefficients of (87%). The genetic relationships among twelve isolates of F. graminearum were determined by Jaccardãs coefficient, Table 3. The matrix of similarity values of SRAP profiles ranged from 42 to 96% among all the isolates. The highest genetic similarity (96%) was recorded between F.g.1 and F.g.2 which obtained from Dirout followed by (88%) between F.g.11 and F.g.12. Whereas, the lowest genetic similarity (42%) was recorded between F.g.5 and F.g.9. The three F. graminearum groups identified by SRAP analysis did not share the same pathogenicity to wheat, as well as isolates from within region have been shown to vary in pathogenicity. There were biologically significant differences between the groups, with group B isolates being more aggressive towards wheat than groups A and C.
T. harzianum and B. subtilis were isolated from wheat kernels. T. harzianum was isolated on PDA medium at 26ô¯C and identified based on morphological characteristics of mycelia and conidiophores as described by Domsch et al., 1980 and Dhingra and Sinclair, 1995. Whereas, B. subtilis was isolated on nutrient agar (NA) at 28ô¯C, and identified based on morphological, culture and biochemical activities that summarized in Table 4 (Skinner and Lovelock, 1979; Sneath et al., 1986; Bergeyãs Manual of systematic bacteriology, 2001).
Seven isolates of T. harzianum and five isolates of B. subtilis were obtained from wheat kernels. In vitro and greenhouse studies were conducted to evaluate the efficacy of Trichoderma harzianum and Bacillus subtilis in control of F. graminearum. In vitro assay was carried out by dual culture technique and the antagonism was measured as reduction in pathogen colony diameter. T. harzianum isolates significantly reduced the growth of F. graminearum colonies in culture with the inhibition rate of 51%, 49%, 65%, 63%, 60%, 46% and 48% respectively. Whereas, B. subtilis isolates exhibited a medium antifungal effect on the mycelium growth of F. graminearum with the inhibition rate of 45%, 55%, 46%, 43% and 42% respectively (Table 5).
In order to gain insight into biological control effect in situ, T. harzianum and B. subtilis were applied in greenhouse. Based on their in vitro effectiveness isolate (T.h.3) of T. harzianum and isolate (B.s.2) of B. subtilis were selected for greenhouse test. Based on the data under greenhouse conditions, T. harzianum was more effective than B. subtilis in reducing severity of FHB. T. harzianum showed highly reduction in head blight severity (55%), while B. subtilis reduced head blight severity by (36.7%) compared with the untreated controls, Table 6. The obtained results revealed that T. harzianum and B. subtilis significantly reduced the percentage of diseased spikelets under greenhouse conditions. Obtained results indicate that the use of microorganisms that antagonize plant pathogens is risk-free when it results in enhancement of resident antagonists.
In the current study, wheat fields in three districts of Assiut governorate were surveyed for the occurrence of FHB. The results of pathogenicity tests indicated significant differences in aggressiveness among isolates of F. graminearum; some isolates produced average of 92.50% disease severity, whereas others were much less virulent with average 54.25%. Isolates from Manfalout appeared to be more variable than the isolates from Dirout and Abuteeg. This difference in pathogenicity between F. graminearum groups confirms the need for the vigilant monitoring of potentially infected material and selection of suitable plant breeding strategies.
The SRAP-PCR results of this study give a clear evidence for existence of variations within a small geographical area for F. graminearum. The present investigation will help in formulating control measures against the pathogen showing high variability. The SRAP marker allowed the identification of genetic variability of twelve isolates of F. graminearum where all the twelve isolates from Assiut have a genetic similarity of 52%. The results of SRAP analysis are generally in compatible with the pathogenicity tests of the isolates of F. graminearum which indicate significant level of variation among the twelve isolates of F. graminearum. Differences in aggressiveness among isolates may due to genetic recombination, mutation, or selection. In F. graminearum, there is large genetic variation within a given population, even in samples collected from a small area within a field (McMullen et al., 1997; Mert-Tû¥rk et al., 2014). Several studies reported variation in aggressiveness among F. graminearum isolates sampled from various parts of the world within a country and even within populations from individual fields (Akinsanmi et al., 2006; Bai and Shaner, 1996; Cumagun and Miedaner, 2003; Miedaner et al., 2010). As pointed out by Miedaner et al. (2000), the high level of genetic variation in aggressiveness and other characteristics suggests that F. graminearum isolates possess a high level of genetic plasticity that may threaten resistant host varieties. Continued monitoring of populations is required to detect such events, which might pose a threat to the FHB-resistant varieties being produced in different countries relying on a limited number of resistance genes. The previous Egyptian study was limited in scope of this topic. Therefore, further studies are required to understand the population structure and establish the degree of genetic diversity of F. graminearum from different geographic regions in Egypt. Depending on the results of SRAP profiles, F. graminearum isolates were divided into three groups A, B and C on the basis of the geographical origin of the isolates. Previous publications have confirmed the same finding (Carter et al., 2000; Carter et al., 2002; OãDonnell et al., 2000). Results in the present study indicate a high genotypic diversity among the F. graminearum isolates in Assiut. Although isolates from different regions clustered together, indicating a relatively high level of genetic exchange between different regions, there was also evidence for diversity related to geographic separation. Diversity studies of F. graminearum in other countries (Akinsanmi et al., 2006; Zeller et al., 2004) have revealed a similar population structure (Carter et al., 2000). SRAP markers showed high genetic diversity among G. zeae isolates. The significant proportion of variance accounted by the variety compared with the geographic origin of isolates suggests that seedborne inoculum may be contributed to the genetic diversity within the G. zeae. Because, inoculum migration together with sexual recombination are probably the main factors affecting the genetic diversity of G. zeae populations (Fernando et al., 2006). The current study demonstrated that, SRAP analysis identified considerable diversity within F. graminearum. Moreover, Dinolfo et al., 2015 demonstrated the efficacy of SRAP molecular marker to amplify open reading frames in fungus.
Biological control is an efficient and environmentally friendly way to reduce the disease severity of FHB. The findings of the present study declared that, all the antagonists inhibited the growth of F. graminearum in culture, however T. harzianum were the most effective, inhibiting the growth of the pathogen. In greenhouse, T. harzianum reduced the percentage of diseased spikelets by 55%, while B. subtilis reduced the percentage of diseased spikelets by 36.7%. Trichoderma and Bacillus have the highest inhibitory effect to pathogens in culture (Mû¥llenborn et al., 2007; Perello et al., 2002; Schunmacher et al., 2007). Production of antifungal secondary metabolites by Trichiderma can induce resistance of plants against infection by pathogenic microorganisms. Trichoderma strains exert biocontrol against fungal phytopathogens either indirectly, by competing for nutrients and space, modifying the environmental conditions, or promoting plant growth and plant defensive mechanisms and antibiosis, or directly, by mechanisms such as mycoparasitism (BenûÙtez et al., 2004). Trichoderma strains grow rapidly and they are naturally resistant to many toxic compounds, including herbicides, fungicides and pesticides such as DDT, and phenolic compounds (Chet et al., 1997) and because the strains recover very rapidly after the addition of sublethal doses of some of these compounds. Resistance to toxic compounds may be associated with the presence in Trichoderma strains of ABC transport systems (Harman et al., 2004). Inhibition occurred by B. subtilis against F. graminearum probably due to the late production of antifungal metabolites or competition for nutrients and space rather than inhibition by antimicrobial secretion (Asak and Shoda, 1996; Agarry et al., 2005; Melo, 1998). B. subtilis strains produce a broad spectrum of antimicrobial compounds, including predominantly peptides as well as a couple of non-peptidic compounds such as polyketides, an aminosugar, and a phospholipid (Stein, 2005). The antifungal effects might have been due to one or more antifungal compounds produced by this biocontrol agent. Chitin is a common constituent of fungal cell walls (Cohen-Kupiec and Chet, 1998). B. subtilis could produce chitinase on chitin-amended media. It indicates that B. subtilis could break down cell wall of F. graminearum by producing chitinase. The cell wall of fungi provides both protective and aggressive functions. If removed or weakened, the fungi die unless they are osmotically protected (Latgûˋ, 2007). It may be presumed that growth inhibition of F. graminearum by B. subtilis strains in our study might be due to the production of antimicrobial compounds or competition for nutrients and space.
In conclusion, the results demonstrate that SRAP technique is a useful marker system in determining the genetic characterization of isolates of F. graminearum. This difference in pathogenicity between F. graminearum groups confirms the need for the vigilant monitoring of potentially infected material and selection of suitable plant breeding strategies. Biocontrol agents could play an important role in organic cereal production. In conventional production, such agents may extend protection of spikes past the flowering stage when fungicides can no longer be applied. Certain strains of spore producing bacteria (B. subtilis) and fungi (T. harzianum) have shown promise results for the control of FHB.
Tableô 1
Sequences of the SRAP primers and 16 primer combinations used to amplify F. graminearum genomic DNA
Tableô 2
Pathogenicity tests of F. graminearum isolates on wheat (Sakha-69) under greenhouse conditions
| Isolates No | Geographical origin of isolates | Diseased spikelets (%)* |
|---|---|---|
| F.g.1 |
Group A Dirout- Assiut |
68.00 F |
| F.g.2 | 62.75 G | |
| F.g.3 | 73.25 E | |
| F.g.4 | 75.50 E | |
| F.g.5 | 79.50 CD | |
|
|
||
| F.g.6 |
Group B Manfalout- Assiut |
84.50 B |
| F.g.7 | 78.25 D | |
| F.g.8 | 92.50 A | |
| F.g.9 | 81.00 C | |
| F.g.10 | 78.50 CD | |
|
|
||
| F.g.11 |
Group C Abuteeg- Assiut |
56.50 H |
| F.g.12 | 54.25 H | |
|
|
||
| Uninfected control | 0.0 I | |
Tableô 3
Similarity matrix of F. graminearum isolates (Jaccardãs Coefficient)
Tableô 4
Morphological and physiological characteristics of bacterial isolates
Tableô 5
Effect of T. harzianum and B. subtilis on colony diameter of F. graminearum in dual culture
| Treatments | Antagonism class | Colony diameter of F. graminearum (cm)* | Inhibition of F. graminearum growth (%) |
|---|---|---|---|
| F.g.+T.h.1 | 1.5 | 4.37 EF | 51.38 |
| F.g.+T.h.2 | 1.7 | 4.55 DE | 49.44 |
| F.g.+T.h.3 | 1.0 | 3.12 I | 65.27 |
| F.g.+T.h.4 | 1.5 | 3.30 HI | 63.33 |
| F.g.+T.h.5 | 1.2 | 3.60 GH | 60.00 |
| F.g.+T.h.6 | 1.7 | 4.85 BCD | 46.11 |
| F.g.+T.h.7 | 2.0 | 4.60 CDE | 48.88 |
| F.g.+B.s.1 | - | 4.95 BCD | 45.00 |
| F.g.+B.s.2 | - | 4.05 FG | 55.00 |
| F.g.+B.s.3 | - | 4.80 BCDE | 46.66 |
| F.g.+B.s.4 | - | 5.05 BC | 43.88 |
| F.g.+B.s.5 | - | 5.15 B | 42.77 |
| Control | - | 9.0 A | 0.0 |
Tableô 6
Influence of T. harzianum and B. subtilis on FHB incited by F. graminearum on wheat cultivar Sakha-69 under greenhouse conditions
| Treatments | Diseased spikelets (%)* | Reduction (%) |
|---|---|---|
| F.g.+T.h.3 | 41.00 C | 55.06 |
| F.g.+B.s.2 | 57.75 B | 36.71 |
| F.g. alone (positive control) | 91.25 A | 0.0 |
| Uninfected control | 0.0 D | - |
References
Agarry, OO, Akinyosoye, FA and Adetuyi, FC 2005. Antagonistic property of microorganisms associated with cassava (Manihot esculenta Crantz). Afr J Biotechnol. 4:627-663.

Akinsanmi, OA, Backhouse, D, Simpfendorfer, S and Chakraborty, S 2006. Genetic diversity of Australian Fusarium graminearum and F. pseudograminearum. Plant Pathol. 55:494-504.

Alexander, NJ, Hohn, TM and McCormick, SP 1998. The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P-450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl Environ Microbiol. 64:221-225.




Asak, O and Shoda, M 1996. Biocontrol of Rhizoctonia solani causing damping-off disease of tomato with Bacillus subtilis. Appl Environ Microbiol. 62:4081-4085.




Bacon, CW, Yates, IE, Hinton, DM and Meredith, F 2001. Biological control of Fusarium moniliforme in maize. Environmental Health Perspectives. 109:325-332.

Bai, GH and Shaner, GE 1996. Variation in Fusarium graminearum and cultivar resistance to wheat scab. Plant Dis. 80:975-979.

Baysal, O, Siragusa, M, Ikten, H, Polat, I, Gumrukcu, E, Yigit, F, Carimi, F and Teixeira da Silva, JA 2009. Fusarium oxysporum f. sp. lycopersici races and their genetic discrimination by molecular markers in West Mediterranean region of Turkey. Physiol Mol Plant Pathol. 74:68-75.

Bell, DK, Wells, HD and Markham, CR 1982. In vitro antagonism of Trichoderma species against six fungal plant pathogens. Phytopathology. 72:379-382.

BenûÙtez, T, Rincû°n, AM, Limû°n, MC and Codû°n, AC 2004. Biocontrol mechanisms of Trichoderma strains. Int Microbiol. 7:249-260.

Bergeyãs Manual of Determinative Systematic Bacteriology. 2001. Buchanan, RE and Gibbon, NE2nd. The William and Wilkins Co, Baltimore.
Budak, H, Shearman, RC, Parmaksiz, I and Dweikat, I 2004b. Comparative analysis of seeded and vegetative biotype Buffalograsses based on phylogenetic relationships using ISSRs, SSRs, RAPDs, and SRAPs. Theor Appl Genet. 109:280-288.



Budak, H, Shearman, RC, Parmaksiz, I, Gaussoin, RE, Riordan, TP and Dweikat, I 2004c. Molecular characterization of Buffalograss germplasm using sequence related amplified polymorphism markers. Theor Appl Genet. 108:328-334.



Budak, H, Shearman, RC, Gaussoin, RE and Dweikat, I 2004a. Application of sequence-related amplified polymorphism markers for characterization of Turfgrass species. HortScience. 39:955-958.

Carter, JP, Razanoor, HN, Desjardins, AE and Nicholson, P 2000. Variation in Fusarium graminearum isolates from Nepal associated with their host of origin. Plant Pathol. 49:1-10.

Carter, JP, Rezanoor, HN, Holden, D, Desjardins, AE, Plattner, RD and Nicholson, P 2002. Variation in pathogenicity associated with the genetic diversity of Fusarium graminearum. Eur J Plant Pathol. 108:573-583.
Cavaglieri, L, Orlando, J, RodrûÙguez, MI, Chulze, S and Etcheverry, M 2005. Biocontrol of Bacillus subtilis against Fusarium verticillioides in vitro and at the maize root level. Res Microbiol. 156:748-754.


Chan, YK, McCormick, WA and Seifert, KA 2003. Characterization of an antifungal soil bacterium and its antagonistic activities against Fusarium species. Can J Microbiol. 49:253-262.


Chet, I, Inbar, J and Hadar, I 1997. Fungal antagonists and mycoparasites. In: The Mycota IV: Environmental and microbial relationships, eds. by DT Wicklow and B SûÑderstrûÑm, 165-184. Springer-Verlag, Berlin.
Cohen-Kupiec, R and Chet, I 1998. The molecular biology of chitin digestion. Current Opinion in Biotechnol. 9:270-277.

Crane, J, Gibson, D, Vaughan, R and Bergstrom, G 2013. Iturin levels on wheat spikes linked to biological control of Fusarium head blight by Bacillus amyloliquefaciens. Phytopathology. 103:146-155.


Cumagun, CJR and Miedaner, T 2003. Aggressiveness of 42 isolates of Gibberella zeae (Fusarium graminearum) in wheat under field and greenhouse conditions. J Plant Dis Protec. 110:554-559.
Dana, MM, Limû°n, MC, MejûÙas, R, Mach, RL, BenûÙtez, T, Pintor-Toro, JA and Kubicek, CP 2001. Regulation of chitinase 33 (chit33) gene expression in Trichoderma harzianum. Curr Gene. 38:335-342.


Desjardins, AE and Hohn, TM 1997. Mycotoxins in plant pathogenesis. Mol Plant-Microbe Interact. 10:147-152.

Desjardins, AE, Hohn, TM and McCormick, SP 1993. Trichothecene biosynthesis in Fusarium species: chemistry, genetics, and significance. Microbiol Mol Biol Rev. 157:595-604.


Desjardins, AE, Busman, M, Manandhar, G, Jarosz, AM, Manandhar, HK and Proctor, RH 2008. Gibberella ear rot of maize (Zea mays) in Nepal: distribution of the mycotoxins nivalenol and deoxynivalenol in naturally and experimentally infected maize. J Agr Food Chem. 56:5428-5436.

Dhingra, OD and Sinclair, JB 1995. Basic plant pathology methods. second edition. Lewis Publishers, CRC Press, USA. 400-450.
Dinler, G and Budak, H 2008. Analysis of expressed sequence tags (ESTs) from Agrostis species obtained using sequence related amplified polymorphism. Biochem Genet. 46:663-676.



Dinolfo, MI, CastaûÝares, E and Stenglein, SA 2015. SRAP as an informative molecular marker to study the Fusarium poae genetic variability. J Phytopathol. 163:657-663.


Domsch, KH, Gams, W and Anderson, TH 1980. Compondium of soil fungi. Academic Press. A Subbsidiary of Harcout Brace Jovanovich, Bublishers, London. 1:859.
Elad, Y, Freeman, S and Monte, E 2000. Biocontrol agents. Mode of action and interaction with other means of control. IOBC wprs Bulletin, Vol 24. Sevilla, EspaûÝa.
Estûˋvez de Jensen, C, Percich, JA and Graham, PH 2002. Integrated management strategies of bean root rot with Bacillus subtilis and Rhizobium in Minnesota. Field Crops Research. 74:107-115.

Fernando, WGD, Zhang, JX, Dusabenyagasani, M, Guo, XW, Ahmed, H and McCallum, B 2006. Genetic diversity of Gibberella zeae isolates from Manitoba. Plant Dis. 90:1337-1342.


Gomez, KA and Gomez, AA 1984. Statistical procedures for agricultural research. 2nd Ed. John Willey, New York. 680.
Goswami, RS and Kistler, HC 2004. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol. 5:515-525.


Harman, GE, Howell, CR, Viterbo, A, Chet, I and Lorito, M 2004. Trichoderma species-opportunistic, avirulent plant symbionts. Nat Reviews Microbiol. 2:43-56.


Howell, CR 2003. Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis. 87:4-10.


Ilgen, P, Hadeler, B, Maier, FJ and SchûÊfer, W 2009. Developing kernel and rachis node induce the trichothecene pathway of Fusarium graminearum during wheat head infection. Mol Plant-Microbe Interact. 22:899-908.


Jaccard, P 1908. Nouvelles rescherches sur la distribution florale. Bulletin de la Sociûˋtûˋ vaudoise des sciences naturelles. 44:223-270.
Jansen, C, von Wettstein, D, SchûÊfer, W, Kogel, KH, Felk, A and Maier, FJ 2005. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc Natl Acad Sci USA. 102:16892-16897.



Jochum, C, Osborne, L and Yuen, G 2006. Fusarium head blight biological control with Lysobacter enzymogenes strain C3. Biol Contr. 39:336-344.

Khan, NI, Schisler, DA, Boehm, MJ, Slininger, PJ and Bothast, RJ 2001. Selection and evaluation of microorganisms for biocontrol of Fusarium head blight of wheat incited by Gibberella zeae. Plant Dis. 85:1253-1258.


Kimura, M, Kaneko, I, Komiyama, M, Takatsuki, A, Koshino, H, Yoneyama, K and Yamaguchi, I 1998. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. Cloning and characterization of Tri101. J Biol Chem. 273:1654-1661.

Latgûˋ, JP 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol. 66:279-290.



Li, G and Quiros, CF 2001. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet. 103:455-461.


Mahmoud, AF and Abo-Elyousr, K 2014. Genetic diversity and biological control of Rhizoctonia solani associated with root rot of soybean in Assiut governorate. Egypt J Plant Physiol Pathol. 2:1-5.
Manning, B, Southwell, R, Hayman, P and Moore, K 2000. Fusarium head blight in northern NSW. NSW Agriculture Research Update, AgDex 110/637.
McMullen, MP, Jones, R and Gallenberg, D 1997. Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Dis. 81:1340-1348.


Melo, IS 1998. Agents microbianos de space control de fungus fitopatogenicos. In: Control biologico, eds. by IS Melo and JL Azevedo, Jagua Riuna V.1. Brazil. EMBRAPA, 17-30.
Mert-Tû¥rk, F, Gencer, R and Kahriman, F 2014. Chemotyping of the Fusarium graminearum isolates and variation in aggressiveness against wheat heads. J Animal and Plant Sci. 24:1858-1862.
Mesterhazy, A 2002. Role of deoxynivalenol in aggressiveness of Fusarium graminearum and F. culmorum and in resistance to Fusarium head blight. Eur J Plant Pathol. 108:675-684.
Meyer, L, Wehner, FC, Nel, LH and Carling, DE 1998. Characterization of the crater disease strain of Rhizoctonia solani. Phytopathology. 88:366-371.


Miedaner, T, Bolduan, C and Melchinger, A 2010. Aggressiveness and mycotoxin production of eight isolates each of Fusarium graminearum and Fusarium verticillioides for ear rot on susceptible and resistant early maize inbred lines. Eur J Plant Pathol. 127:113-123.


Miedaner, T, Reinbrecht, C and Schilling, AG 2000. Association among aggressiveness, fungal colonization, and mycotoxin production of 26 isolates of Fusarium graminearum in winter rye head blight. J Plant Dis Protec. 107:124-134.
Monte, E 2001. Understanding Trichoderma: between biotechnology and microbial ecology. Int Microbiol. 4:1-4.

Mû¥llenborn, C, Steiner, U and Oerke, EC 2007. Disease control with Bacillus brevis: update and future prospects. In: Proceeding of the 15th International Symposium on Modern Fungicides and Antifungal Compounds; May 6-10; Ramada Treff Hotel, Friendrichroda, Germany.
Mutlu, N, Boyaci, FH, GûÑûÏmen, M and Abak, K 2008. Development of SRAP, SRAP-RGA, RAPD and SCAR markers linked with a Fusarium wilt resistance gene in eggplant. Theor Appl Genet. 117:1303-1312.



Nelson, PE, Toussoun, TA and Marasas, WFO 1983. Fusarium species: An illustrated manual for identification. Pennsylvania State University Press, 193.
Nirenberg, H 1981. A simplified method for identifying Fusarium spp. occurring on wheat. Can J Botany. 59:1599-1609.

OãDonnell, K, Cigelnik, E, Weber, NS and Trappe, JM 1997. Phylogenetic relationships among ascomycetous truffles and the true and false morels inferred from 18S and 28S ribosomal DNA sequence analysis. Mycologia. 89:48-65.

OãDonnell, K, Kistler, HC, Tacke, BK and Casper, HH 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc Natl Acad Sci USA. 97:7905-7910.



Perello, A, Simon, MR and Arambarri, AM 2002. Interactions between foliar pathogens and the saprophytic microflora of the wheat (Triticum aestivum L.) phylloplane. J Phytopathol. 150:232-243.

Pestka, JJ and Smolinski, AT 2005. Deoxynivalenol: toxicology and potential effects on humans. Journal of Toxicology and Environmental Health Part B: Critical Reviews. 8:39-69.


Proctor, RH, Hohn, TM and McCormick, SP 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant-Microbe Interact. 8:593-601.


Rohlf, FJ 2000. NTSYS-PC: Numerical taxonomy and multivariate analysis system. Version 2.1. New York. Applied Biostatistics.
Schunmacher, R, Stopacher, N, Reithner, B, Omann, M, Zeillinger, S and Krska, R 2007. Peptaibol profiles in cultures of Trichoderma atroviride: Detection and characterization by LC-MS/MS. In: Poceedings of the 15th International Symposium on Modern Fungicides and Antifungal Compounds; May 6-10; Ramada Treff Hotel, Friendrichroda, Germany.
Skinner, FA and Lovelock, DW 1979. Identification methods for microbiologist. 2nd Ed. The Soc For Appl Bacterial Technical Series. Academic Press, London.
Sneath, PHA, Mair, NS, Elisabeth Sharpe, M and Holt, JG 1986. Bergeyãs manual of systematic bacteriology. 2:Section 13, Endospore-forming Gram-positive rods and cocci. The Williams and Wilkings Company, Baltimore Md., USA. 1105-1207.
Snijders, CHA and Perkowski, J 1990. Effects of head blight caused by Fusarium culmorum on toxin content and weight of wheat kernels. Phytopathology. 80:566-570.

Stein, T 2005. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol. 56:845-857.


Stockwell, CA, Bergstrom, GC and da Luz, WC 1999. Selection of microbial antagonists for biological control of Fusarium head blight of wheat. 82-84. Proceedings of the 1999 National Fusarium Head Blight Forum. Michigan State University, University Printing, East Lasting, MI.
Suman, A, Kimbeng, CA, Edme, SJ and Veremis, J 2008. Sequence-related amplified polymorphism (SRAP) markers for assessing genetic relationships and diversity in sugarcane germplasm collections. Plant Genetic Resources: Characterization and Utilization. 6:222-231.

Summerell, BA, Salleh, B and Leslie, JF 2003. Utilitarian approach to Fusarium identification. Plant Dis. 87:117-128.

Voigt, K, Cigelnik, E and Oãdonnell, K 1999. Phylogeny and PCR identification of clinically important Zygomycetes based on nuclear ribosomal-DNA sequence data. J Clin Microbiol. 37:3957-3964.




Hue, AG, Voldeng, HD, Savard, ME, Fedak, G, Tian, X and Hsiang, T 2009. Biological control of Fusarium head blight of wheat with Clonostachys rosea strain ACM941. Can J Plant Pathol. 31:169-179.




 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




