Proteomic and Phenotypic Analyses of a Putative YggS Family Pyridoxal Phosphate-Dependent Enzyme in Acidovorax citrulli
Article information
Abstract
Acidovorax citrulli (Ac) is a phytopathogenic bacterium that causes bacterial fruit blotch (BFB) in cucurbit crops, including watermelon. However, there are no effective methods to control this disease. YggS family pyridoxal phosphate-dependent enzyme acts as a coenzyme in all transamination reactions, but its function in Ac is poorly understood. Therefore, this study uses proteomic and phenotypic analyses to characterize the functions. The Ac strain lacking the YggS family pyridoxal phosphate-dependent enzyme, AcΔyppAc(EV), virulence was wholly eradicated in geminated seed inoculation and leaf infiltration. AcΔyppAc(EV) propagation was inhibited when exposed to L-homoserine but not pyridoxine. Wild-type and mutant growth were comparable in the liquid media but not in the solid media in the minimal condition. The comparative proteomic analysis revealed that YppAc is primarily involved in cell motility and wall/membrane/envelop biogenesis. In addition, AcΔyppAc(EV) reduced biofilm formation and twitching halo production, indicating that YppAc is involved in various cellular mechanisms and possesses pleiotropic effects. Therefore, this identified protein is a potential target for developing an efficient anti-virulence reagent to control BFB.
Acidovorax citrulli (Ac) is a gram-negative, rod-shaped, seed-borne bacterium known worldwide as the causative bacterial fruit blotch (BFB) agent in cucurbits, including watermelon (Schaad et al., 1978; Willems et al., 1992). BFB occurs in various tissues throughout all watermelon growth stages (Rahimi-Midani et al., 2020). After water-soaked lesions form on the cotyledons, the seedlings gradually wither and die (Burdman et al., 2005). Furthermore, when a disease infects the watermelon’s fruit, the surface exhibits water-soaked lesions that eventually turn black, and necrosis transpires inside the fruit (Latin and Hopkins, 1995). BFB causes enormous damage to the cucurbit crop industry and still requires management (Shrestha et al., 2013). Due to difficulties in producing resistant lines or cultivars, it is vital to determine Ac’s virulence factor and related mechanisms to control BFB. Ac strains are categorized into two groups based on genetic information and host range (Zivanovic and Walcott, 2017). Group I strains display high virulence on diverse cucurbits, including melon, whereas group II strains are highly virulent on watermelon but not other plants. In addition, Ac strains in group I display swimming motility but not in group II in in vitro conditions. Thus, Ac strains may possess different abilities due to each group’s genetic background and biological mechanisms.
For effective BFB control, researches regarding diverse Ac virulence factors and mechanisms are still ongoing. Similar to other gram-negative plant pathogenic bacteria, type II, III, and VI secretion systems are indispensable for Ac virulence (Tian et al., 2015; Zhang et al., 2018; Zivanovic and Walcott, 2017). Various type III secretion effectors were recently identified (Jiménez-Guerrero et al., 2020). In addition, Ac mutant strains with impaired twitching motility and biofilm formation were less virulent in melons (Bahar et al., 2009). Another study verified that type IV pili influence twitching motility, surface adhesion, and virulence (Bahar et al., 2009). Quorum sensing genes, ferric uptake regulators, and a putative glycerol-3-phosphate dehydrogenase are required for Ac virulence (Johnson and Walcott, 2013; Lee et al., 2021; Liu et al., 2019). However, most studies only focus on the group I strains.
The YggS family pyridoxal phosphate-dependent enzyme is a pyridoxal 5′-phosphate (PLP)-binding protein (PLBBP) family member, highly conserved in gram-negative bacteria, and acts as a coenzyme in all transamination and certain amino acid decarboxylation, deamination, and racemization reactions (Ito et al., 2019; Vu et al., 2020). These enzymes are also involved in B6 vitamin homeostasis and various amino acid maintenance, including isoleucine, threonine, leucine, and homoserine (Ito et al., 2019; Prunetti et al., 2016). In addition, protein mutation expressed pleiotropic effects in diverse organisms (Côté et al., 2016; Darin et al., 2016; Ito et al., 2013). Recent studies determined that YggS family pyridoxal phosphate-dependent enzyme functions were characterized in Ac’s group I strain (Wang et al., 2022). Ac strains that lacked the enzyme were less virulent on melon, attenuated hypersensitive response on tobacco, and were less tolerant against oxidative stress. However, this enzyme’s roles have not yet been characterized in the group II strain and their watermelon host.
This study reports the YppAc (YggS family pyridoxal phosphate-dependent enzyme in Ac; accession no. ATG95625) function in the group II strain KACC17005, whose genome information was annotated (Park et al., 2017). We investigated Ac’s growth in the presence of L-homoserine and virulence to characterize YppAc’s function in the group II strain. In addition, we used Ac and AcΔyppAc for comparative proteomic analysis to postulate YppAc-related cellular mechanisms. Protein functions were evaluated through diverse phenotypic assays, including biofilm formation, twitching motility, and growth on M9 solid mediums containing different agar percentages.
Materials and Methods
Bacterial strains and growth conditions
Group II’s Ac strain KACC17005 was used as the wild-type strain (Park et al., 2017). TSB (tryptic soy broth, 30 g/l) or TSA (TSB with 1.5% agar) was used for Ac culture. Escherichia coli EC100D and DH5α identified Tn5 transposon insertion site and cloning in LB (Luria Bertani; 1% tryptone, 0.5% yeast extract, and 1% NaCl). In addition, M9 media (47.7 mM Na2HPO4·7H2O, 22 mM KH2PO4, 8.6 mM NaCl, 18.7 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, and 20 ml of 20% glucose in 1 liter) was used as a minimal medium. For selection, appropriate antibiotics were added to each media at the following final concentrations: rifampicin, 50 μg/ml; gentamycin, 10 g/ml; kanamycin, 50 μg/ml; and ampicillin, 100 μg/ml.
AcΔyppAc selection and its complemented strain generation
Supplementary Table 1 organizes all plasmids and bacterial strains used in this study. AcΔyppAc was selected via screening the Tn5 insertional mutant library using a previously reported method (Kim et al., 2020; Lee et al., 2021). A germinated seed inoculation was used to select virulence-deficient mutants. Next, genomic DNA was extracted using DNeasy Blood & Tissue Kits (Qiagen, Hilden, Germany) and digested with the restriction enzyme SmaI. Digested DNA fragments were self-ligated using T4 ligase (Thermo Fisher Scientific, Rockford, IL, USA) and introduced into EC100D. The self-ligated plasmid was extracted, and the Tn5-insertional site was identified through Sanger sequencing. Polymerase chain reaction (PCR) used YppAc-specific primers (F: 5′-CTCGAGATGACGACGATTGGTAACAA-3′ and R: 5′-AAGCTTCAGTGGTGGTGGTGGTGGTGGCGCGCCGGATAGCTGC-3′) to amplify yppAc’s open reading frame and generate the complemented strain. The PCR product was ligated to the pGem-T easy vector (Promega, Madison, WI, USA), generating pGem-YppAc, and Sanger sequencing confirmed the cloned fragment. HindIII and BamHI digested the generated plasmid, and it was cloned into pBBR1-MCS5 (Kovach et al., 1995), creating pMCS5-YppAc. Electroporation introduced this plasmid into AcΔyppAc, generating AcΔyppAc(YppAc). PCR used the YppAc-specific primer to confirm the transformant. An empty pBBR1-MCS5 vector was transformed into the wild-type and mutant strains to eliminate vector side effects, creating Ac(EV) and AcΔyppAc(EV).
Pathogenicity test
As previously published, the patho- genicity test incorporated two methods (germinated seed inoculation and leaf infiltration) (Lee et al., 2022). For germinated seed inoculation, the TSA-grown Ac strain was suspended in 10 mM MgCl2, adjusted to 0.3 at O.D. 600 nm (approximately 108 colony forming units [cfu]/ml), and diluted to a final 106 cfu/ml concentration with 10 mM MgCl2. Ten seeds per strain were soaked in prepared bacterial suspension for 1 h at room temperature. Then, inoculated seeds were sown into 50 pot trays with autoclaved soil and grown for 7 days in a controlled growth room chamber (26 ± 1°C, 16 h day and 18 h night photoperiod, 60 to 70% relative humidity).
As previously reported, disease severity was measured daily with a 0 to 2 scale value (Disease index: *0, no symptom; *1, water-soaked region (spot); *2, seedling wilt).
The infiltration inoculation method was slightly modified from previous studies (Lee et al., 2021). The watermelon seedlings’ four true leaf stages for infiltration were used. The bacterial suspension was adjusted to 0.3 O.D. 600 nm and diluted (10−2) with 10 ml of MgCl2. The prepared inoculum was directly infiltrated onto the second true leaf’s underside with a needleless 3 ml syringe. Thereafter, cfus from two discs (0.4 cm in diameter) were counted at 0, 2, 4, 6, and 8 days after inoculation.
Growth assay
To observe bacterial growth in TSB, the Ac strain suspension was adjusted to 0.3 at O.D. 600 nm and diluted (10−3) in TSB medium with the appropriate antibiotics. The diluted bacterial suspension was then incubated in a shaking incubator at 28°C and measured every 12 h. To check Ac’s growth in M9 minimal media, the bacterial cell concentration was adjusted to 0.5 at O.D. 600 nm and diluted (10−1) in M9. To investigate the roles of L-homoserine and pyridoxine (PN), which have been known to be associated with mechanisms mediated by YggS protein (Ito et al., 2019), 0.05 mM (final concentration) of L-homoserine or PN was added to the M9 medium. The prepared bacterial suspension was incubated in a shaking incubator at 28°C and measured every 24 h. Each strain had three biological replicates for the growth assay, and all experiments were carried out at least four times.
Bacterial growth in M9 containing different agar percentages
We incubated bacteria in an M9 solid medium (minimal media) containing different agar percentages to check bacterial growth relative to hardness. First, Ac suspension was adjusted to 0.5 at O.D. 600 nm bacterial cell concentration. Then, 5 μl of serially diluted bacterial suspension was dropped into M9 solid medium containing 1.5%, 1%, and 0.5% agar. TSA medium containing 1.5% agar was the control. Dropped plates were incubated in a 28°C incubator until colonies formed, which were counted to determine bacterial population. All experiments were repeated at least five times.
Comparative proteomic analysis
Three biological re- plicates from Ac and AcΔyppAc were used for comparative analysis. Sample harvest, protein/peptide preparation, and quantification followed a previously reported protocol (Lee et al., 2022). Briefly, bacterial cells were harvested at 0.6 at O.D. 600 nm and disrupted in a lysis buffer (6 M guanidine HCl, 10 mM dithiothreitol, 50 mM Tris-HCl pH 7.8) using the Ultrasonic Processor (Colo Parmer, Vernon Hills, IL, USA). After sonication, soluble proteins were collected, and protein concentrations were measured using a BCA assay kit (Thermo Fisher Scientific, Rockford, IL, USA). Next, 1 mg of the protein was alkylated by adding 100 mM of iodoacetamide. After the 1 h incubation, dithiothreitol was added to a final 20 mM concentration. Trichloroacetic acid precipitated the proteins, which were then dissolved in 50 mM ammonium bicarbonate (pH 7.8) and digested by trypsin to obtain peptides. Obtained peptides were cleaned with tC18 Cartridges (Waters, Milford, MA, USA) and quantified.
Next, we loaded 1 μg of cleaned samples into a split-free nano-LC (EASY-nLC II, Thermo Fisher Scientific, Bremen, Germany) connected to the LTQ Velos Pro instrument (Thermo Fisher Scientific). Loaded sample elution was performed through a 300 nl/min flow rate with a 420-min gradient by a water per acetonitrile (A: 100% water with 0.1% formic acid; B: 100% acetonitrile with 0.1% formic acid; 7% of solvent B for 5 min, 35% of solvent B for 380 min, 80% of solvent B for 10 min, and 7% of solvent B for 25 min). Then, six data-dependent MS/MS scans obtained the full mass spectrometry spectra. Dynamic exclusion was permitted with one repeat count, 0.5 repeat duration, and 3-min elimination duration. Ion charge state selection was allowed for 2+ and 3+ ions. The maximum six most intense ions from each full mass scan were collected for fragmentation and inspected within the centroid mode’s linear ion trap part.
Protein identification and quantification adhered to a previously established protocol (Lee et al., 2022). Concisely, the SEQUEST search algorithm in Thermo proteome discoverer used NCBI’s Ac strain KACC17005 genome data to identify proteins. The target-decoy method improved this study’s credibility (Elias and Gygi, 2007). The findings were considered meaningful with a 0.01 false discovery rate, despite permitting two missed cleavages. Regarding mass accuracy, the precursor had a 100 ppm tolerance, and the likelihood score exceeded 20.
The Scaffold 4 software (Proteome Software, Portland, OR, USA) conducted comparative analyses after protein identification. Peptide spectrum matches (PSMs) were employed for protein comparisons (Choi et al., 2008), and each protein’s PSMs were normalized to each biological replicate’s total PSMs. The PSMs average value from the three biological replicates was used to identify differentially abundant proteins (more than a 2-fold change) between Ac and AcΔyppAc. Pairwise comparisons were determined through Student’s t-test (P < 0.05). Differentially abundant proteins were classified using clusters of orthologous groups (COG) analysis (Tatusov et al., 2000).
Biofilm formation assay
The biofilm formation assay was performed as previously reported (Lee et al., 2022). Bacterial strains were grown in TSA, adjusted to 0.3 at O.D. 600 nm, and diluted (10−3) in TSB. Then, 190 μl of prepared bacterial suspension was poured into each 96-well polyvinyl chloride plate well and incubated at 28°C for 2 days. The supernatant was carefully removed and washed once with autoclaved water to determine the formed biofilm. Filtered 0.1% crystal violet was added to each well for staining over 30 min. The crystal violet was removed, washed twice, and eluted for 20 min with 95% ethanol. Biofilm formation was evaluated at 590 nm using a Spectramax 190 microplate reader (Molecular Devices, Sunnyvale, CA, USA). In this study, 20 biological replicates were performed for each strain.
Twitching motility assay
Twitching halo production was assessed using a previous investigation method (Lee et al., 2022). Ac strains were grown in TSA, collected, and adjusted to 0.3 at O.D. 600 nm. A 5 μl bacterial suspension was dotted onto the TSA (0.5% agar) and incubated at 28°C. Following a two-day incubation period, colony and twitching halo sizes were measured under a LEICA M205 C microscope (LEICA, Wetzlar, Germany). Five independent experiments were performed, with each strain tested in triplicate.
Statistical analysis
Student’s t-test and one-way analysis of variance with Turkey HSDab assessed statistical significance using SPSS 12.0K software (SPSS Inc., Chicago, IL, USA). A P-value below 0.05 was deemed statistically significant.
Results
YppAc is essential for Ac virulence
Screening a KACC17005-created Tn5-insertional mutant library identified one mutant strain that did not elicit BFB disease symptoms in watermelon. We attributed this phenotype to a gene that encodes a putative YggS family pyridoxal phosphate-dependent enzyme in Ac (YppAc). We generated Ac(EV) and AcΔyppAc(EV), the wild-type and AcΔyppAc strains carrying the empty pBBR1-MCS5 vector, to confirm whether YppAc contributes to virulence. Additionally, we created AcΔyppAc(YppAc), an AcΔyppAc strain possessing the yppAc gene on the vector.
AcΔyppAc(EV) virulence was abolished entirely during germinated-seed inoculation, and the disease index remained at 0 for 7 days (Fig. 1A and B). On the other hand, all watermelon seedlings infected by Ac(EV) and AcΔyppAc(YppAc) exhibited typical disease symptoms, and the disease index reached two 7 days post-inoculation. Leaf infiltration assay results were similar to those from the germinated-seed inoculation (Fig. 1C and D). Based on cfu measurements from the infiltrated leaves, the AcΔyppAc(EV) value was significantly lower than Ac(EV) and AcΔyppAc(YppAc) over the observation period. Furthermore, Ac(EV)- and AcΔyppAc(YppAc)-infected leaves turned brownish-black and expressed wilting symptoms. However, AcΔyppAc(EV)-infiltrated leaves indicated almost no symptoms (Fig. 1D). These results demonstrate that YppAc is indispensable for Ac virulence.
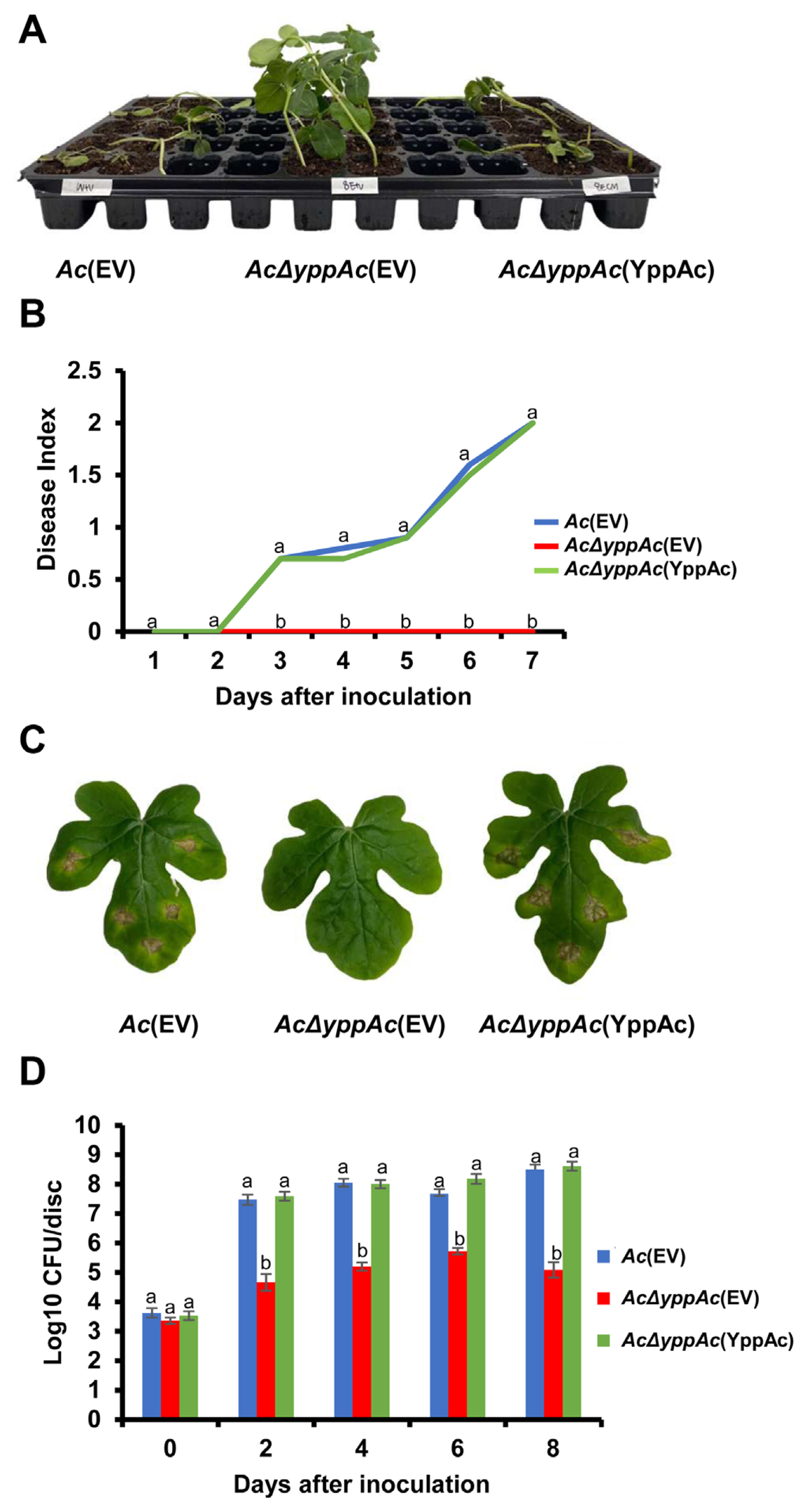
Pathogenicity assay of Ac(EV), AcΔyppAc(EV), and AcΔyppAc(YppAc) using germinated-seed inoculation and leaf infiltration methods on watermelon. (A) The watermelon seedlings infected by three strains in the germinated seed inoculation. The photograph was taken 7 days post-inoculation. (B) Disease index from the germinated seed inoculation. The disease index = [No symptom (numbers of plants) × 0 + spot symptoms (numbers of plants) × 1 + wilt symptoms (numbers of plants) × 2]/Total numbers of seedlings. (C) The infiltrated watermelon true leaves. The photograph was taken 8 days post-inoculation. (D) The colony counting method measured the watermelon true leaves’ viable bacterial population in the infiltration inoculation method. Different letters on error bars in the graph indicate statistically significant differences determined through ANOVA (P < 0.05) combined with Tukey’s HSD.
YppAc does not affect Ac’s growth ability
Leaf infiltration confirmed that viable AcΔyppAc(EV) cell quantities significantly reduced compared to Ac(EV) and AcΔyppAc(YppAc). To determine whether this virulence decrease was due to the AcΔyppAc(EV) multiplication differences, we investigated the three strains’ growth in the TSB and M9 media. All strains in TSB conveyed similar growth patterns and grew exponentially for up to 12 hours post-incubation, reaching the stationary phase (Fig. 2A). Similar to TSB, there was no significant growth difference in the three M9 medium strains for 6 days (Fig. 2B). These data indicate that YppAc is not related to bacterium multiplication.
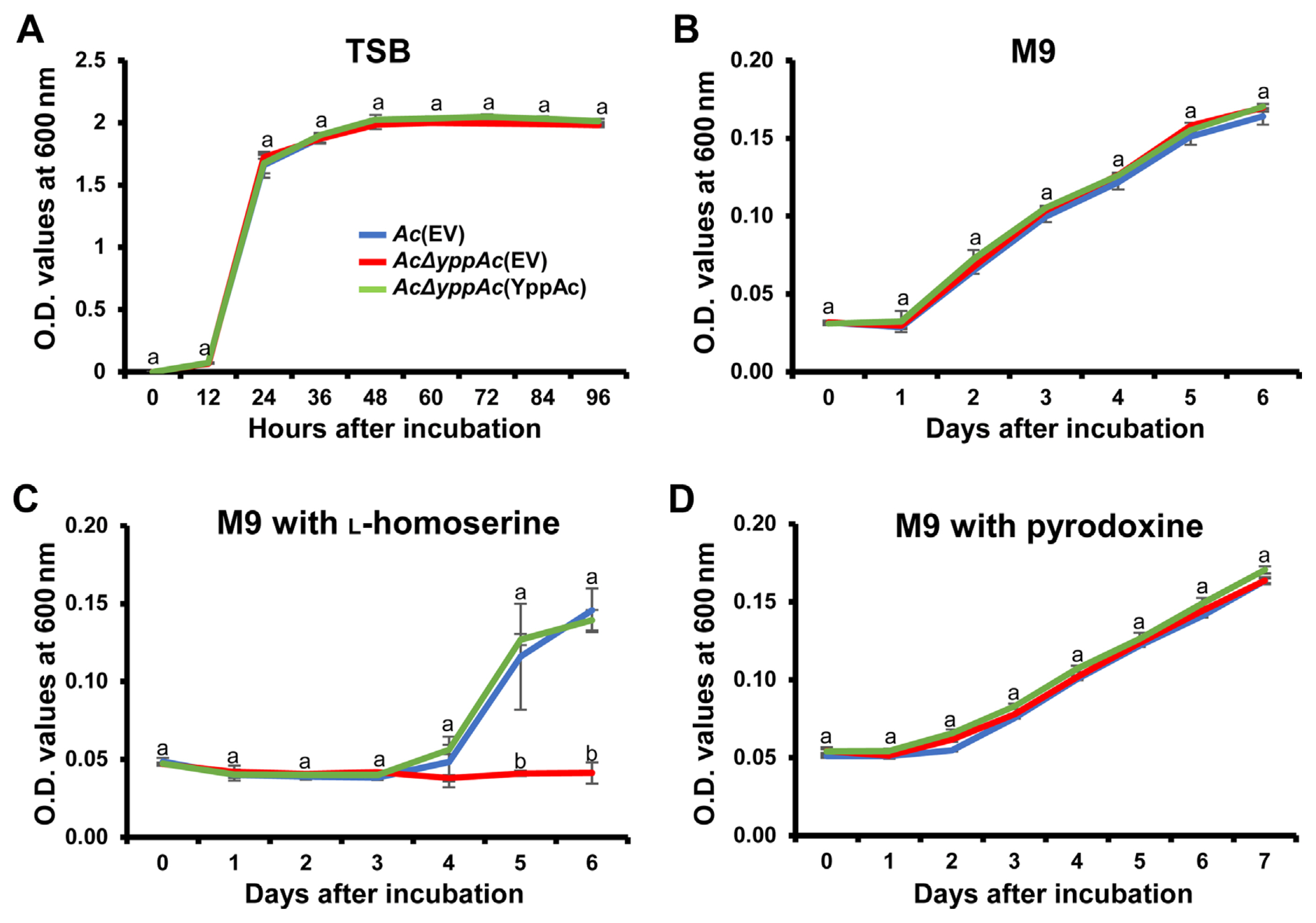
The bacterial growth curve in tryptic soy broth (TSB), M9, and M9 with L-homoserine, or M9 with pyridoxine. Bacterial growth measurement in TSB medium (A), M9 minimal medium (B), and M9 minimal medium exposed to 0.05 mM of L-homoserine (C) or pyridoxine (D). Bacterial suspensions were measured at O.D. 600 nm with a spectrophotometer. Different characters on error bars in the graph indicate statistically significant differences determined through ANOVA (P < 0.05) combined with Tukey’s HSD.
AcΔyppAc(EV) growth is inhibited by homoserine but not PN
A previous study reported that the YggS family protein is involved in the catalytic homoserine (HSE) pathway in E. coli (Ito et al., 2019). Based on this, we evaluated AcΔyppAc(EV) growth when exposed to HSE. As mentioned, the three strains displayed a similar growth curve with no significant difference. Surprisingly, when the three strains were incubated in the M9 medium supplemented with HSE, no strain grew for 3 days (Fig. 2C). In addition, Ac(EV) and AcΔyppAc(YppAc) grew to more than the 0.140 O.D. value, but AcΔyppAc(EV) continued expressing growth defects (Fig. 2C). These data support that the YggS family protein, YppAc, is also associated with HSE-related metabolic processes.
Another previous investigation concluded that E. coli lacking the YggS protein were sensitive to excess PN (Ito et al., 2019). As described above, to confirm PN effects on AcΔyppAc(EV) growth, Ac strains were incubated in an M9 medium supplemented with PN. Contradicting expectations, there was no significant PN sensitivity difference between the tested strains (Fig. 2D). This result suggests that YppAc in Ac has a different metabolism from YggS in E. coli.
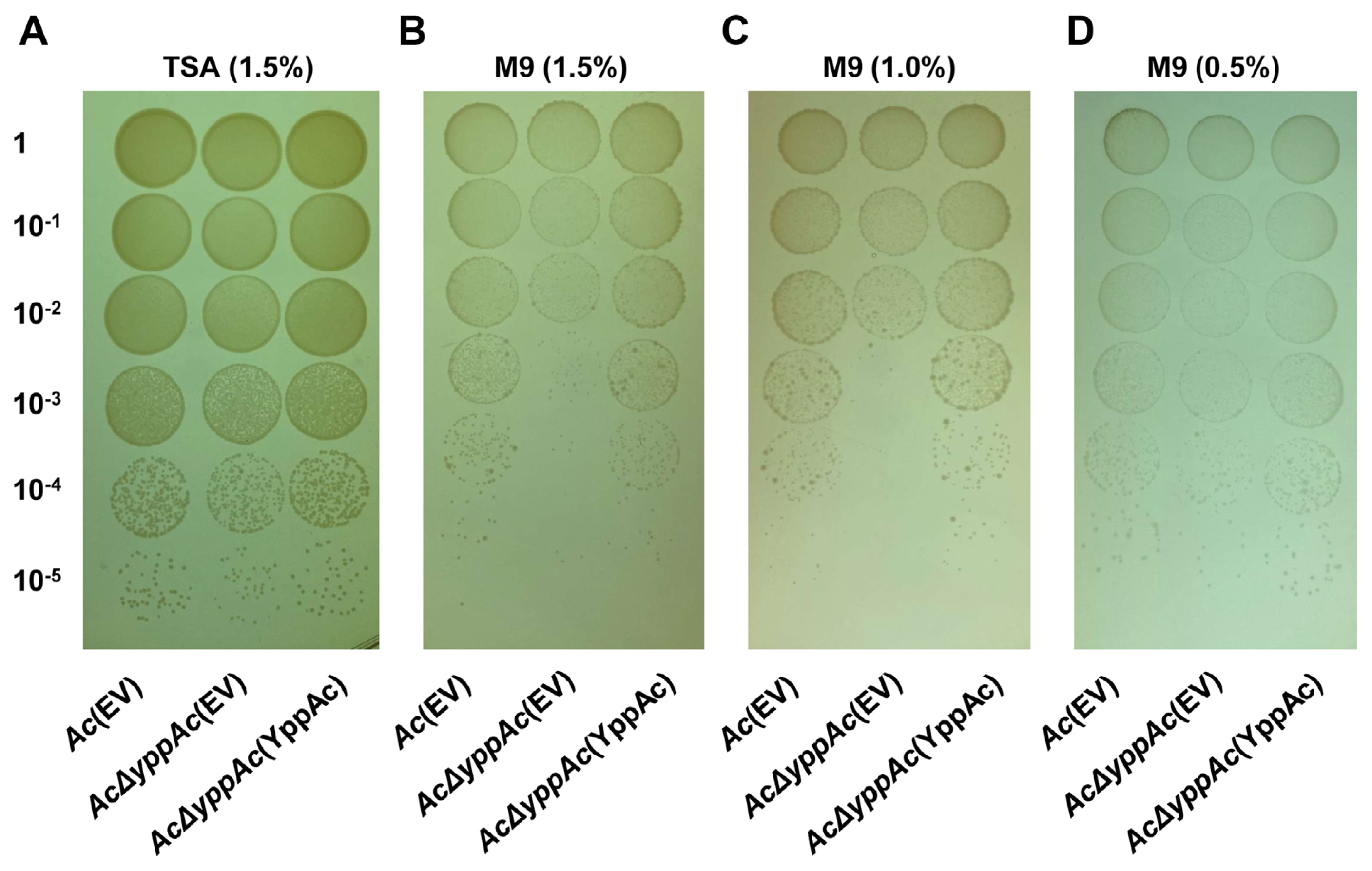
YppAc influences growth ability in M9 solid media
TSB and M9 did not express any growth O.D. value differences. The cell viability was reconfirmed in TSA and M9 solid media to verify whether the O.D. value measurement was accurate. Approximately 108 cfu/ml of Ac suspension was serially diluted and dotted on the solid media. As expected, all three strains exhibited similar population levels on the TSA medium containing 1.5% agar (Fig. 3A). Surprisingly, AcΔyppAc(EV) grew in 10−2-fold diluted suspensions in M9 solid medium with 1.5% agar, whereas Ac(EV) and AcΔyppAc(YppAc) were observed in 10−4-fold diluted suspensions (Fig. 3B). After confirmation, in which AcΔyppAc(EV) displayed a growth defect in the M9 solid medium with 1.5% agar, AcΔyppAc(EV) growth was also examined in the M9 solid medium containing 1% and 0.5% of agar, to determine whether medium hardness affects growth ability. Ac(EV) and AcΔyppAc(YppAc) displayed similar population levels in the M9 solid medium with 1% of agar, but AcΔyppAc(EV) growth was noted in 10−3 suspension (Fig. 3C). In the M9 medium with 0.5% agar, the three strains’ populations were comparable (Fig. 3D). Notably, countable single AcΔyppAc(EV) colonies were rarely observed in M9 solid media. Therefore, it was impossible to calculate the experiment’s cfu value. These results indicate that the wild-type strain could not grow as well in the nutrient-limited M9 solid media compared to the TSA medium. More importantly, YppAc is associated with the growth ability in M9 solid media.
Proteomic analysis
Comparative proteomic analysis aided YppAc in Ac’s altered biological process and property characterization. The wild-type and AcΔyppAc strains were used for the comparative proteomic with a COG analyses. From the three biological replicates, 1,013 and 1,047 proteins were commonly detected in Ac and AcΔyppAc, respectively (Supplementary Table 2). In addition, 49 and 84 proteins were uniquely identified, and 8 and 24 proteins were more (over 2-fold) abundant in Ac and AcΔyppAc, respectively (Fig. 4A). These differentially abundant proteins were categorized through COG analysis (Fig. 4B, Supplementary Tables 3 and 4). Proteins belonging to groups E (amino acid transport and metabolism), K (transcription), and M (cell wall/membrane/envelope biogenesis) were prevalent in Ac. On the other hand, proteins categorized in groups C (energy production and conversion), F (nucleotide transport and metabolism), G (carbohydrate transport and metabolism), H (coenzyme metabolism), I (lipid metabolism), L (replication and repair), N (cell motility), O (post-translation modification), T (signal transduction), and U (intracellular trafficking and secretion) were higher in AcΔyppAc than in Ac.
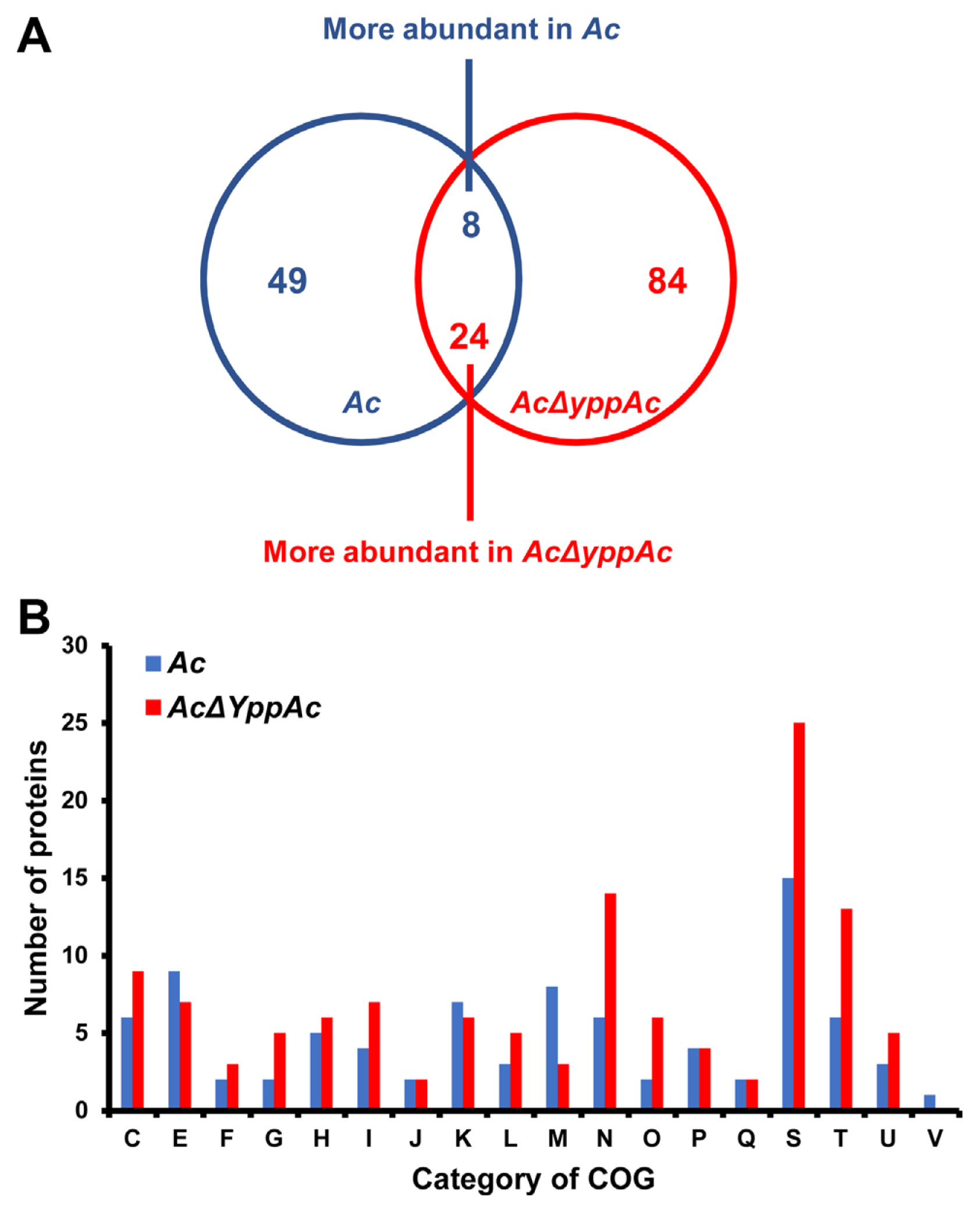
Comparative proteomic analysis in Acidovorax citrulli (Ac) and AcΔyppAc. (A) Venn diagram of differentially expressed proteins. (B) Differentially abundant protein classifications (2 > fold) by clusters of orthologous groups (COG). C, Energy production and conversion; D, Cell cycle control and mitosis; E, Amino acid metabolism and transport; F, Nucleotide metabolism and transport; G, Carbohydrate metabolism and transport; H, Coenzyme metabolism; I, Lipid metabolism; J, Translation; K, Transcription; L, Replication and repair; M, Cell wall/membrane/envelop biogenesis; N, Cell motility; O, Post-translational modification, protein turnover, chaperone functions; P, Inorganic ion transport and metabolism; Q, Secondary structure; S, Function unknown; T, Signal transduction; U, Intracellular trafficking and secretion; V, Defense mechanisms.
Among these classified proteins, we found diverse virulence-related proteins, including type VI secretion system tip proteins, two-component system response regulators, type VI secretion system protein TssL, type VI secretion system ATPase TssH, type VI secretion system baseplate subunit TssK, type VI secretion system baseplate subunit TssF, and LysR family transcriptional regulators). In particular, we detected abundant motility-associated proteins (type VI secretion system TssL, motility MotB, chemotaxis CheD, methyl-accepting chemotaxis, type II secretion system F family, type IV pili twitching motility PilT, and type IV pilus biogenesis/stability PilW).
We also noted proteins affecting various phenotypes such as biofilm formation and external environment tolerance (type I glyceraldehyde-3-phosphate dehydrogenase, phosphoenolpyruvate-protein phosphotransferase, PTS fructose transporter subunit EIIBC, trehalose-phosphatase, NAD-dependent dehydratase, disulfide isomerase, penicillin-binding protein 1A, D-3-hydroxybutyrate oligomer hydrolase, aspartate/glutamate racemase family protein, phospholipase C, phosphocholine-specific, efflux RND transporter periplasmic adaptor subunit, and glycosyltransferase family 2 protein). This comparative proteomic data suggest that YppAc is involved in diverse phenotype regulation, including biofilm formation and bacterial movement.
AcΔyppAc reduced biofilm formation
Bacterial biofilms are vital virulence factors that protect themselves from external or environmental stresses (Davey and O’toole, 2000). In comparative proteomics analysis, there were substantial changes in proteins involved in polysaccharide metabolism, a biofilm component, and cell membranes or walls. Thus, we evaluated Ac strains’ biofilm formation, confirming that it AcΔyppAc(EV)’s biofilm formation was significantly more reduced (over 1.3-fold) compared to Ac(EV). In addition, AcΔyppAc(YppAc) and Ac(EV) revealed similar biofilm formation levels (Fig. 5A). These data confirm YppAc’s association with Ac biofilm formation.
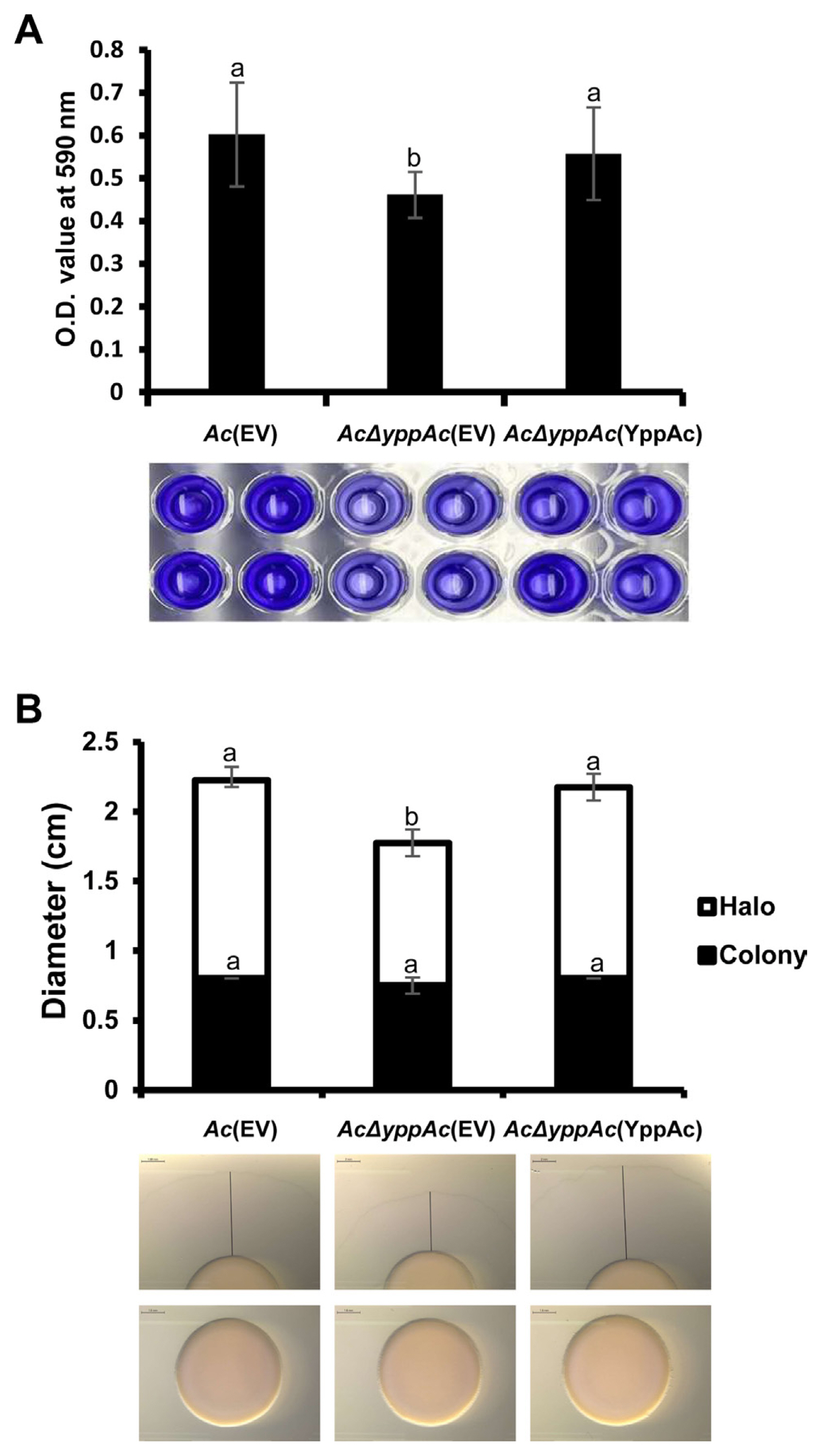
Biofilm formation and twitching motility halo production in Ac(EV), AcΔyppAc(EV), and AcΔyppAc(YppAc). (A) Biofilm formation was evaluated on a polyvinyl chloride (PVC) 96-well plate at O.D. 590 nm with a spectrophotometer 2 days post-incubation. (B) Twitching motility was observed on a TSA (tryptic soy broth with 1.5% agar) medium containing 0.5% agar 2 days post-incubation. Scale bars = 2 mm. The black lines indicate sizes of halos. Different letters on the bar graph indicate statistical differences determined through ANOVA (P < 0.05) with Turkey HSDab.
AcΔyppAc decreased twitching motility
Pili are how Ac maneuvers with twitching motility (Bahar et al., 2010). In the comparative proteomic analysis, type 4 pili-related gene expression differences were involved in twitching motility and other proteins related to bacterial movement. Therefore, twitching motility was measured in three strains: Ac(EV), AcΔyppAc(EV), and AcΔyppAc(YppAc) (Fig. 5B). The colony sizes produced from the three strains were similar. However, AcΔyppAc(EV)’s twitching halo was 1.78 cm, which was markedly reduced compared to Ac(EV)’s 2.23 cm. AcΔyppAc(YppAc)’s twitching halo was 2.18 cm, which was recovered to a level similar to that of Ac(EV). These results convey that YppAc is associated with twitching motility.
Discussion
YggS family pyridoxal phosphate-dependent proteins are involved in various metabolic processes and display YggS-related pleiotropic effects in many organisms (Ito et al., 2019). When the YggS protein was deleted in E. coli, the mutant was sensitive to PN, and the growth of the mutant was suppressed by isoleucine, threonine, and leucine (Ito et al., 2019; Prunetti et al., 2016); however, these features’ mechanisms have not been elucidated, although it is presumed to be due to an internal PN 5′-phosphate concentration increase (Prunetti et al., 2016). Wild-type and AcΔyppAc(EV) expressed no PN resistance differences, and homoserine suppressed mutant growth. These differences suggest YppAc is involved in more diverse mechanisms than YggS family proteins in other bacteria. Therefore, we postulate that YppAc possesses distinct functions.
A previous study concluded that a putative pyridoxal phosphate synthase was involved in Actinobacillus pleuropneumoniae’s pathogenicity (Xie et al., 2017). In addition, when the vitamin B6 biosynthesis pathway containing PLP synthase was deleted, Streptococcus pneumoniae expressed reduced virulence (El Qaidi et al., 2013). However, the YggS family pyridoxal phosphate-dependent enzyme’s association with plant and animal pathogen virulence is poorly documented. Recently, Wang et al. reported that theYggS family pyridoxal phosphate-dependent enzyme’s involvement with Ac’s group I strain in melons (Wang et al., 2022). This study corroborates this as we used two virulence assays to demonstrate that YppAc in the Ac strain belonging to group II was indispensable for pathogenicity. Additionally, there was no growth rate difference between Ac(EV) and AcΔyppAc(EV) in TSB and M9, indicating that reduced mutant virulence is not due to growth and YppAc is a pathogenicity factor.
Current research has yet to theorize that bacteria propagate well in a minimal liquid medium but not a minimal solid medium. Thus, this study demonstrated that AcΔyppAc(EV) could not grow well in the M9 solid medium but did in the M9 liquid medium and rich media TSA and TSB. This finding indicates that nutrient-minimal and solid conditions attenuate mutant growth; however, the exact mechanisms related to this abnormal growth and YppAc are still unknown. Growth-related protein expressions, including isovaleryl-CoA dehydrogenase, peptide chain release factor, phosphomethylpyrimidine synthase, and VgrG (ATG96540, ATG93713, ATG95464, and ATG93564, respectively) were significantly altered in our comparative proteomic analysis. Further molecular and biochemical study on their associated mechanisms is needed to establish whether these proteins’ different abundance cause abnormal phenotypes. At this point, it can only be predicted by the phenotype.
Biofilm is known as one of the virulence factors (Davey and O’toole, 2000). In this study, AcΔyppAc(EV)’s biofilm formation was remarkably decreased compared to Ac(EV), and the comparative proteomic analysis identified group G (carbohydrate metabolism and transport) and M (cell wall/membrane/envelope biosynthesis) diverse proteins closely related to biofilm formation. Among these differentially abundant proteins, the wild-type uniquely expressed disulfide isomerase (ATG92779) and glycosyltransferase (ATG94052), proteins involved in E. coli and Klebsiella pneumonia biofilm formation (Lee et al., 2008; Pal et al., 2019), respectively. Type 4 pili-mediated twitching motility is required for Ac virulence (Bahar et al., 2010). In agreement with this study, AcΔyppAc(EV) displayed reduced twitching motility and could not infect the watermelon.
Proteomic analysis revealed that YppAc altered type 4 pili-related protein abundance (ATG96977, ATG95626, ATG94949) and that disulfide isomerase (ATG92779) and the peptide chain release factor (ATG93713) are involved in E. coli and Pseudomonas aeruginosa motility, respectively (Lee et al., 2008; Pustelny et al., 2013). In addition to these proteins, group N’s motility-related protein expression levels were significantly altered (ATG96505, ATG96536, ATG92975, ATG93157, ATG93438, ATG93503), indicating YppAc regulation. Therefore, YppAc’s involvement in biofilm formation and twitching motility likely contributes to Ac virulence.
This study concluded that YppAc, a putative YggS family pyridoxal phosphate-dependent enzyme, is indispensable for Ac pathogenicity. We further verified that disrupting YppAc with homoserine pleiotropically affected growth, colony development on minimal solid mediums, biofilm formation, and twitching motility. Therefore, these results indicate that YppAc in group II Ac strains is a virulence factor and a potential target for anti-virulence agents to control BFB in watermelon.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
We thank J. Kim for their technical help at the BT research facility center, Chung-Ang University. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. NRF-2020R1A2C1013040), Republic of Korea. This research was also supported by the Chung-Ang University Graduate Research Scholarship in 2022 (awarded to Yongmin Cho).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).