Alemu, K. 2014. Real-time PCR and its application in plant disease diagnostics. Adv. Life Sci. Technol 27:39-49.
Alruwaili, M., Siddiqi, M. H., Khan, A., Azad, M., Khan, A. and Alanazi, S. 2022. RTF-RCNN: an architecture for real-time tomato plant leaf diseases detection in video streaming using Faster-RCNN.
Bioengineering 9:565.



Bhat, I. A., Bhat, R. A., Shrivastava, M. and Sharma, D. M. 2018. In: Universal dependency parsing for Hindi-English code-switching, In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; 1:eds. by In : M. Walkter, H. Ji and A. Stent, pp. 987-998. Association for Computational Linguistics, New Orleans, LA, USA.
Bhujel, A., Kim, N.-E., Arulmozhi, E., Basak, J. K. and Kim, H.-T. 2022. A lightweight attention-based convolutional neural networks for tomato leaf disease classification.
Agriculture 12:228.

Caldeira, R. F., Santiago, W. E. and Teruel, B. 2021. Identification of cotton leaf lesions using deep learning techniques.
Sensors (Basel) 21:3169.



Chang, Y. K., Zaman, Q. U., Schumann, A. W., Percival, D. C., Esau, T. J. and Ayalew, G. 2012. Development of color co-occurrence matrix based machine vision algorithms for wild blueberry fields.
Appl. Eng. Agric 28:315-323.

Chen, K., Wang, J., Pang, J., Cao, Y., Xiong, Y., Li, X., Sun, S., Feng, W., Liu, Z., Xu, J., Zhang, Z., Cheng, D., Zhu, C., Cheng, T., Zhao, Q., Li, B., Lu, X., Zhu, R., Wu, Y., Dai, J., Wang, J., Shi, J., Ouyang, W., Loy, C. C. and Lin, D. 2019 MMDetection: open MMLab detection toolbox and benchmark Preprint at
https://doi.org/10.48550/arXiv.1906.07155.
Cynthia, S. T., Hossain, K. M. S., Hasan, M. N., Asaduzzaman, M. and Das, A. K. 2019. Automated detection of plant diseases using image processing and Faster R-CNN algorithm. In: 2019 International Conference on Sustainable Technologies for Industry 4.0 (STI); pp 1-5. Institute of Electrical and Electronics Engineers, Piscataway, NJ, USA.

Dai, J., Li, Y., He, K. and Sun, J. 2016. R-FCN: object detection via region-based fully convolutional networks. In: Proceedings of the 30th International Conference on Neural Information Processing Systems, eds. by D. D. Lee, M. Sugiyama, U. Luxburg, I. Guyon and R. Garnett, pp. 379-387. Curran Associates Inc, Red Hook, NY, USA.
Deng, R., Tao, M., Xing, H., Yang, X., Liu, C., Liao, K. and Qi, L. 2021. Automatic diagnosis of rice diseases using deep learning.
Front. Plant Sci 12:701038.



Dubey, S. R. and Jalal, A. S. 2012. Detection and classification of apple fruit diseases using complete local binary patterns. In: 2012 Third International Conference on Computer and Communication Technology; pp 346-351. IEEE Computer Society, Washington, DC, USA.

Fernández-Campos, M., Huang, Y. T., Jahanshahi, M. R., Wang, T., Jin, J., Telenko, D. E. P., Góngora-Canul, C. and Cruz, C. D. 2021. Wheat spike blast image classification using deep convolutional neural networks.
Front. Plant Sci 12:673505.



Girshick, R. 2015. Fast R-CNN. In: Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV); pp 1440-1448. IEEE Computer Society, Washington, DC, USA.

Girshick, R., Donahue, J., Darrell, T. and Malik, J. 2014. Rich feature hierarchies for accurate object detection and semantic segmentation. In: Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); pp 580-587. IEEE Computer Society, Washington, DC, USA.

He, K., Gkioxari, G., Dollár, P. and Girshick, R. 2017. Mask R-CNN. In: Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV); pp 2980-2988. IEEE Computer Society, Washington, DC, USA.

He, K., Zhang, X., Ren, S. and Sun, J. 2015. Spatial pyramid pooling in deep convolutional networks for visual recognition.
IEEE Trans. Pattern Anal. Mach. Intell 37:1904-1916.


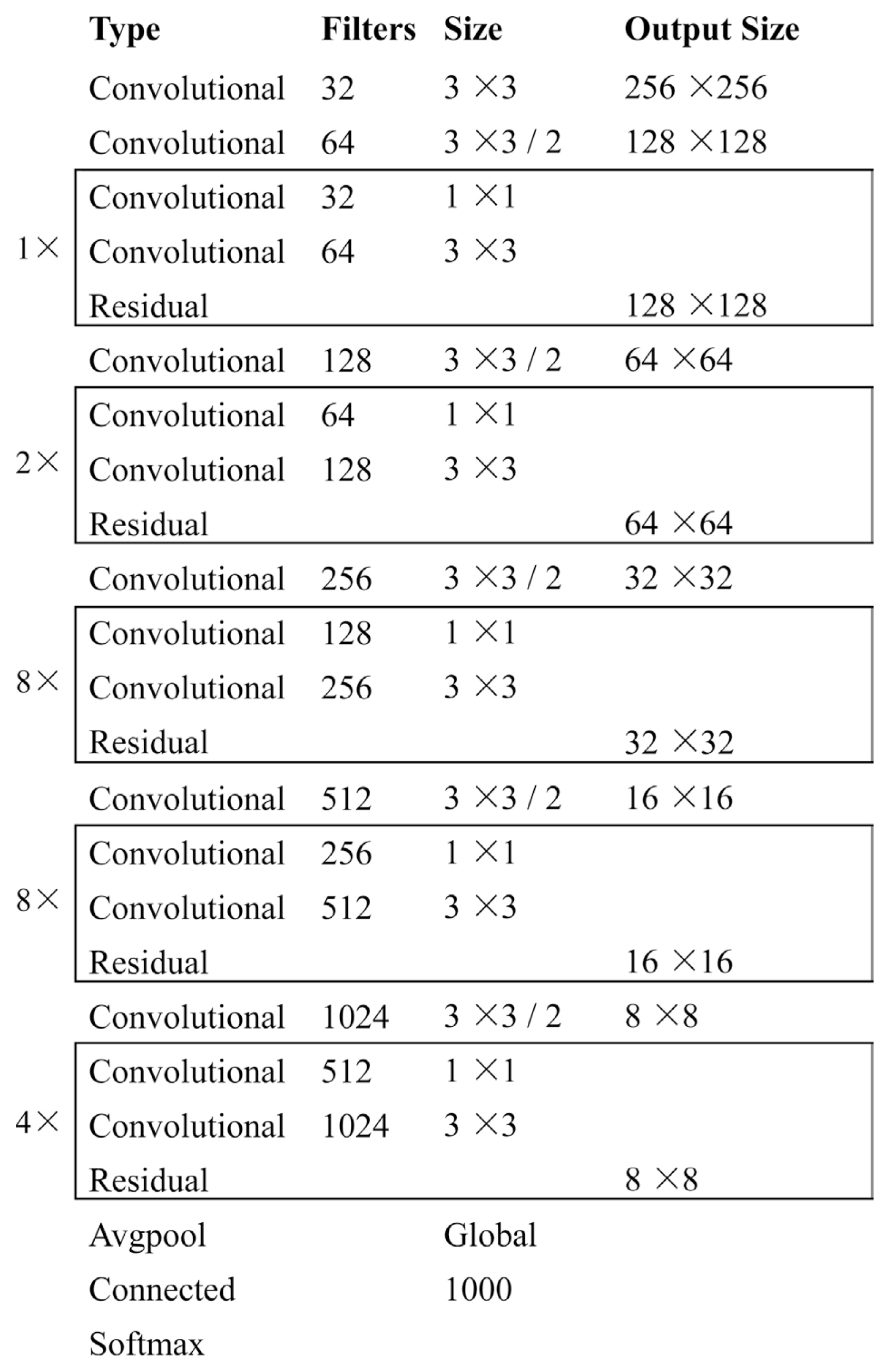
He, K., Zhang, X., Ren, S. and Sun, J. 2016. Deep residual learning for image recognition. In: Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); pp 770-778. IEEE Computer Society, Washington, DC, USA.

Hlaing, C. S. and Maung Zaw, S. M. 2018. Tomato plant diseases classification using statistical texture feature and color feature. In: Proceedings of the 2018 IEEE/ACIS 17th International Conference on Computer and Information Science (ICIS); pp 439-444. IEEE Computer Society, Los Alamitos, CA, USA.

Hou, J., Yang, C., He, Y. and Hou, B. 2023. Detecting diseases in apple tree leaves using FPN-ISResNet-Faster RCNN.
Eur. J. Remote Sens 56:2186955.

Jia, Y., Zhang, Q., Zhang, W. and Wang, X. 2019. In: CommunityGAN: community detection with generative adversarial nets, In: The Web Conference 2019; eds. by In : L. Liu and R. White, pp. 784-794. Association for Computing Machinery, New York, NY, USA.
Jos, J. and Venkatesh, K. A. 2020. Pseudo color region features for plant disease detection. In: 2020 IEEE International Conference for Innovation in Technology (INOCON 2020); pp 1-5. Institute of Electrical and Electronics Engineers, Piscataway, NJ, USA.

Karadağ, K., Tenekeci, M. E., Taşaltın, R. and Bilgili, A. 2020. Detection of pepper fusarium disease using machine learning algorithms based on spectral reflectance.
Sustain. Comput. Inform. Syst 28:100299.

Krizhevsky, A., Sutskever, I. and Hinton, G. E. 2012. ImageNet classification with deep convolutional neural networks.
Adv. Neural Inf. Process Syst 25:1097-1105.

Li, J.-H., Lin, L.-J. and Tian, K. 2020a. Detection of leaf diseases of balsam pear in the field based on improved Faster R-CNN. Trans. Chin. Soc. Agric. Eng 36:179-185 (in Chinese).
Li, Z., Guo, R., Li, M., Chen, Y. and Li, G. 2020b. A review of computer vision technologies for plant phenotyping.
Comput. Electron. Agric 176:105672.

Lin, T.-Y., Dollár, P., Girshick, R., He, K., Hariharan, B. and Belongie, S. 2017. Feature pyramid networks for object detection. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); pp 936-944. Institute of Electrical and Electronics Engineers, Piscataway, NJ, USA.

Liu, J. and Wang, X. 2020. Tomato diseases and pests detection based on improved Yolo V3 convolutional neural network.
Front. Plant Sci 11:898.



Liu, W., Anguelov, D., Erhan, D., Szegedy, C., Reed, S., Fu, C.-Y. and Berg, A. C. 2016. SSD: single shot MultiBox detector. In:
Computer Vision - ECCV 2016, eds. by B. Leibe, J. Matas, N. Sebe and M. Welling, pp. 21-37. Springer, Cham, Switzerland.

Mathew, M. P. and Mahesh, T. Y. 2022. Leaf-based disease detection in bell pepper plant using YOLO v5.
Signal Image Video Process 16:841-847.


Nandhini, N. and Bhavani, R. 2020. Feature extraction for diseased leaf image classification using machine learning. In: 2020 International Conference on Computer Communication and Informatics (ICCCI); pp 1-4. Institute of Electrical and Electronics Engineers, Piscataway, NJ, USA.

Nawaz, M., Nazir, T., Javed, A., Masood, M., Rashid, J., Kim, J. and Hussain, A. 2022. A robust deep learning approach for tomato plant leaf disease localization and classification.
Sci. Rep 12:18568.




Pradhan, P., Kumar, B. and Mohan, S. 2022. Comparison of various deep convolutional neural network models to discriminate apple leaf diseases using transfer learning.
J. Plant Dis. Protec 129:1461-1473.


Pushpa, B. R., Shree Hari, A. V. and Adarsh, A. 2021. In: Diseased leaf segmentation from complex background using indices based histogram, In: Computational Experimental Simulations in Engineering: Proceedings of ICCES 2020; 1:eds. by In : S. N. Atluri and I. Vušanović, pp. 1502-1507. Springer, Cham, Switzerland.
Redmon, J., Divvala, S. K., Girshick, R. B. and Farhadi, A. 2016. You only look once: unified, real-time object detection. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); pp 779-788. IEEE Computer Society, Washington, DC, USA.

Redmon, J. and Farhadi, A. 2017. YOLO9000: better, faster, stronger. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); pp 6517-6525. IEEE Computer Society, Washington, DC, USA.

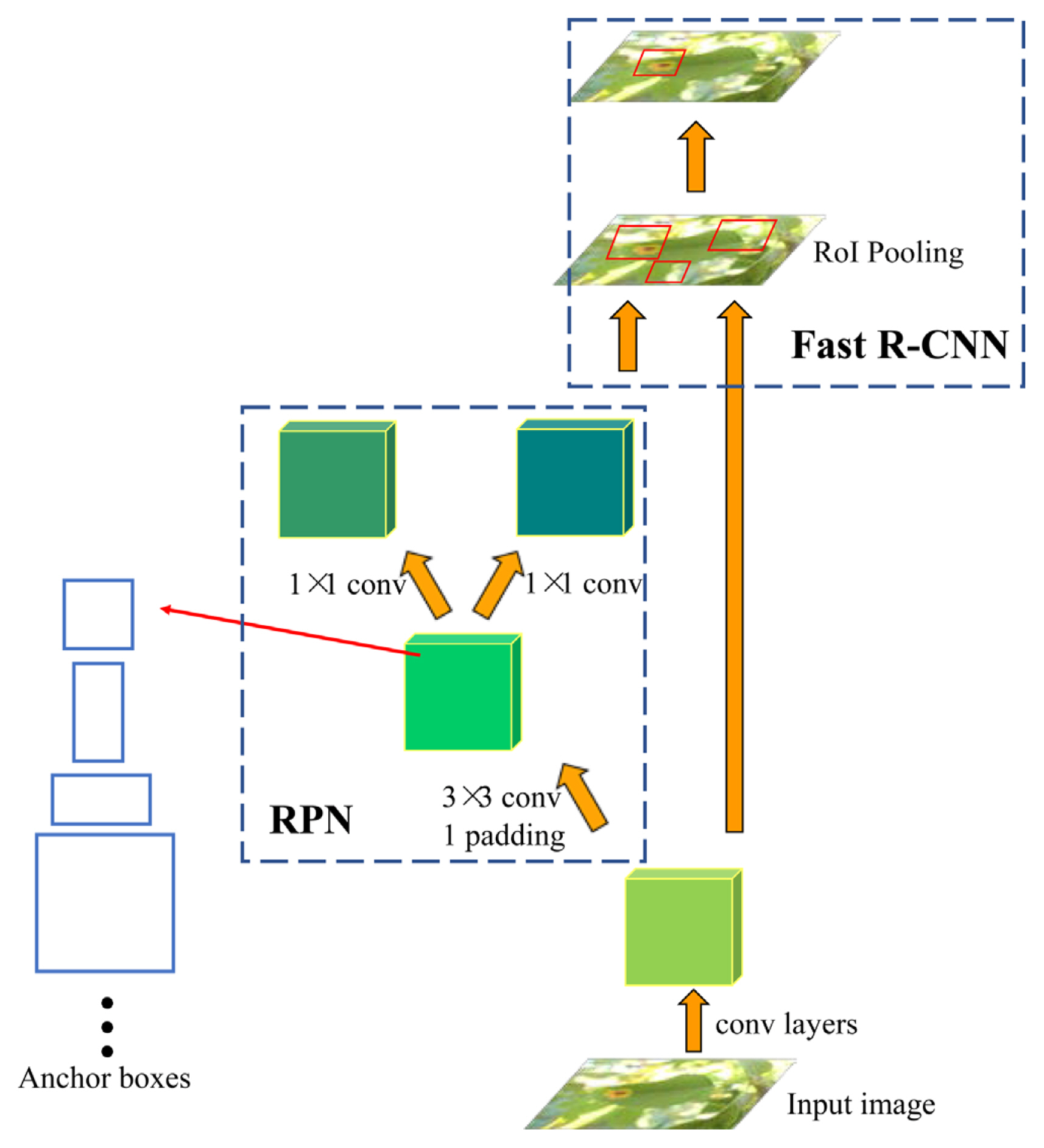
Ren, S., He, K., Girshick, R. and Sun, J. 2017. Faster R-CNN: towards real-time object detection with region proposal networks.
IEEE Trans. Pattern Anal. Mach. Intell 39:1137-1149.


Sardoğan, M., Özen, Y. and Tuncer, A. 2020. Detection of apple leaf diseases using Faster R-CNN.
Düzce Univ. J. Sci. Technol 8:1110-1117.

Shrivastava, V. K. and Pradhan, M. K. 2021. Rice plant disease classification using color features: a machine learning paradigm.
J. Plant Pathol 103:17-26.


Simon, M., Milz, S., Amende, K. and Gross, H.-M. 2019. Complex-YOLO: an Euler-Region-Proposal for real-time 3D object detection on point clouds. In:
Computer Vision - ECCV 2018, eds. by L. Leal-Taixé and S. Roth, pp. 197-209. Springer, Cham, Switzerland.

Srivastava, A., Jha, D., Chanda, S., Pal, U., Johansen, H. D., Johansen, D., Riegler, M. A., Ali, S. and Halvorsen, P. 2021. MSRF-Net: a multi-scale residual fusion network for biomedical image segmentation.
IEEE J. Biomed. Health Inform 26:2252-2263.

Sterling, A. and Melgarejo, L. M. 2020. Leaf spectral reflectance of
Hevea brasiliensis in response to
Pseudocercospora ulei.
Eur. J. Plant Pathol 156:1063-1076.


Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., Vanhoucke, V. and Rabinovich, A. 2015. Going deeper with convolutions. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); pp 1-9. IEEE Computer Society, Washington, DC, USA.

Tamuli, P. 2020. The polymerase chain reaction (PCR) and plant disease diagnosis: a review. Plant Arch 20:3495-3498.
Taohidul Islam, S. M., Masud, M. A., Ur Rahaman, M. A. and Hasan Rabbi, M. M. 2019. Plant leaf disease detection using mean value of pixels and canny edge detector. In: 2019 International Conference on Sustainable Technologies for Industry 4.0 (STI); pp 1-6. Institute of Electrical and Electronics Engineers, Piscataway, NJ, USA.

Tian, Y., Yang, G., Wang, Z., Li, E. and Liang, Z. 2019. Detection of apple lesions in orchards based on deep learning methods of CycleGAN and YOLOV3-Dense.
J. Sens 2019:7630926.


Tugrul, B., Elfatimi, E. and Eryigit, R. 2022. Convolutional neural networks in detection of plant leaf diseases: a review.
Agriculture 12:1192.

Waheed, A., Goyal, M., Gupta, D., Khanna, A., Hassanien, A. E. and Pandey, H. M. 2020. An optimized dense convolutional neural network model for disease recognition and classification in corn leaf.
Comput. Electron. Agric 175:105456.

Wang, Q. and Qi, F. 2019. Tomato diseases recognition based on Faster RCNN. In: 2019 10th International Conference on Information Technology in MedicineEducation (ITME 2019); pp 772-776. Institute of Electrical and Electronics Engineers, Piscataway, NJ, USA.

Wang, X. and Liu, J. 2021. Tomato anomalies detection in greenhouse scenarios based on YOLO-Dense.
Front. Plant Sci 12:634103.



Wang, Y., Wang, Y. and Zhao, J. 2022. MGA-YOLO: a lightweight one-stage network for apple leaf disease detection.
Front. Plant Sci 13:927424.



Wani, J. A., Sharma, S., Muzamil, M., Ahmed, S., Sharma, S. and Singh, S. 2022. Machine learning and deep learning based computational techniques in automatic agricultural diseases detection: methodologies, applications, and challenges.
Arch. Comput. Methods Eng 29:641-677.


Yan, Q., Yang, B., Wang, W., Wang, B., Chen, P. and Zhang, J. 2020. Apple leaf diseases recognition based on an improved convolutional neural network.
Sensors (Basel) 20:3535.



Zhang, K., Xu, Z., Dong, S., Cen, C. and Wu, Q. 2019. Identification of peach leaf disease infected by Xanthomonas campestris with deep learning.
Eng. Agric. Environ. Food 12:388-396.

Zhang, Y., Qin, J., Park, D. S., Han, W., Chiu, C.-C., Pang, R., Le, Q. V. and Wu, Y. 2020 Pushing the limits of semi-supervised learning for automatic speech recognition Preprint at
https://doi.org/10.48550/arXiv.2010.10504.
Zhou, C., Zhou, S., Xing, J. and Song, J. 2021. Tomato leaf disease identification by restructured deep residual dense network.
IEEE Access 9:28822-28831.







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print






