 |
 |
| Plant Pathol J > Volume 39(5); 2023 > Article |
|
Abstract
Ralstonia solanacearum species complex (RSSC) is a soil borne plant pathogen causing bacterial wilt on various important crops, including Solanaceae plants. The bacterial pathogens within the RSSC produce exopolysaccharide (EPS), a highly complicated nitrogen-containing heteropolymeric polysaccharide, as a major virulence factor. However, the biosynthetic pathway of the EPS in the RSSC has not been fully characterized. To identify genes in EPS production beyond the EPS biosynthetic gene operon, we selected the EPS-defective mutants of R. pseudosolanacearum strain SL341 from Tn5-inserted mutant pool. Among several EPS-defective mutants, we identified a mutant, SL341P4, with a Tn5-insertion in a gene encoding a putative NDP-sugar epimerase, a putative membrane protein with sugar-modifying moiety, in a reverse orientation to EPS biosynthesis gene cluster. This protein showed similar to other NDP-sugar epimerases involved in EPS biosynthesis in many phytopathogens. Mutation of the NDP-sugar epimerase gene reduced EPS production and biofilm formation in R. pseudosolanacearum. Additionally, the SL341P4 mutant exhibited reduced disease severity and incidence of bacterial wilt in tomato plants compared to the wild-type SL341 without alteration of bacterial multiplication. These results indicate that the NDP-sugar epimerase gene is required for EPS production and bacterial virulence in R. pseudosolanacearum.
Ralstonia solanacearum species complex (RSSC) causes bacterial wilt (BW) in over 450 of plants species belonging to 50 families worldwide occurring in tropical, subtropical and temperate climates region (Fegan and Prior 2005; Hayward, 1991, 1994). Bacterial pathogens within RSSC infect various economically important crops of Solanaceae family, including tomato, pepper, tobacco and potato, as well as banana, sesame and plants of the Cucurbitaceae family, leading to lethal wilt disease (Wicker et al., 2007). Most strains of the RSSC are known to survive for more than 40 years in dry soil and can also endure low temperature in the plant roots and residues. As a result, these overwintering pathogens can re-infect host plants, making effective management of BW challenging (Balatero et al., 2005).
The whole genome of various RSSC strains have been analyzed, and its virulence mechanism has been extensively investigated. RSSC strains invade the host plants through wounds or natural openings in the plant roots, suppress the innate immunity of the host plant and multiply in the intercellular space (Arlat et al., 1992; Genin and Boucher, 2002; Jacobs et al., 2012). Subsequently, the bacteria move to the xylem vessels of the plants to express their virulence factors. A unique quorum sensing system by the autoinducer molecule 3-hydroxypalmitic acid methyl ester (3-OH PAME) or (R)-methyl 3-hydroxymyristate methyl ester (3-OH MAME) regulates the expression of RSSC virulence functions (Clough et al., 1997; Kai et al., 2015; Saile et al., 1997; Schell, 2000). Exopolysaccharide (EPS), biofilm and cell-wall degradation enzymes (CWDE) such as pectinase and cellulase are known as the major virulence factors expressed through quorum sensing in RSSC (Hikichi et al., 2007). Bacterial biofilm formation with EPS and subsequent CWDE production interfere with water transport in the xylem vessels of the host plant, causing lethal wilt symptoms (Genin and Boucher, 2002; Saile et al., 1997; Schell 2000). Despite of well-known virulence factors, the pathogenicity of RSSC strains against host plant is controlled by complicated interactions among various regulatory genes and their regulation network (de Pedro-Jové et al., 2021; Hikichi et al., 2017; Schell, 2000). Despite extensive effort in studying the virulence of RSSC strains, the complete understanding of the virulence mechanism remains elusive.
The EPS is a high-molecular-weight polymer produced as a slime layer or capsular polysaccharide in various bacteria, including Gram-positive and Gram-negative bacteria as well as RSSC strains (Botta et al., 1994; Reeves et al., 1996). It is typically composed of monosaccharide units and contains non-carboxyl substituents such as acetate, pyruvate, succinate, and phosphate (Kumar and Mody, 2009). EPS plays a crucial role in the bacterial attachment to their habitats and acts as a nutrient trap, promoting bacterial growth (Harimawan and Ting, 2016). It also provides protection to bacteria against stresses in extreme environments through the EPS matrix (Harimawan and Ting, 2016). The bacterial cells in the EPS are organized into a multicellular community (Costerton et al., 1995). Due to these characteristics, EPS is known to be contribute to the colonization of bacteria on plant roots and rhizosphere, which are nutrient-rich compartments in soil ecosystems. Consequently, EPS plays a vital role in successful invasions and infections into the host plants (Ghosh and Maiti, 2016).
In RSSC strains, EPS I is a major virulence factor and it is composed highly heterogeneous acidic polymer consisting of N-acetyl-galactosamine and N-acetylated amino sugars (Denny and Baek, 1991; Orgambide et al., 1991). The genes responsible for EPS biosynthesis are clustered in RSSC strains (Huang and Schell, 1995) and regulation of this gene cluster expression has been characterized (Chapman and Kao, 1998). However, the biosynthetic pathway of EPS in RSSC strains was not elucidated.
In this study, we characterized a mutant strain of R. pseudosolanacearum SL341, SL341P4, which carries a transposon insertion in a putative gene encoding NDP-sugar epimerase like protein. The NDP-sugar epimerase gene appears to be transcribed separately from the EPS biosynthesis gene cluster in SL341, as it is located downstream of the EPS gene cluster in the reverse orientation. The SL341P4 mutant exhibited a defect in EPS production and showed a reduction of virulence in virulence, depending on the tomato cultivar, without the alteration in bacterial growth in plant. This study suggests that the putative NDP-sugar epimerase gene may play a crucial role in the full activation of R. pseudosolanacearum SL341 virulence in tomato plants by facilitating EPS production and biofilm formation.
Bacterial strains and plasmids used in this study are listed in Supplementary Table 1. Escherichia coli strains were incubated in lysogeny broth (LB) at 37°C with shaking 200 rpm for 24 h, or grown statically condition on LB agar at 37°C for 24 h. R. pseudosolanacearum strains were grown in CPG broth (Schaad et al., 2001) at 30°C with shaking at 200 rpm for 24 h, or incubated statically on TZC agar (CPG agar supplemented with 2,3,5-triphenyltatrazolium chloride [TZC]) and mannitol-glutamate (MG) agar (1% mannitol, 0.2% L-glutamic acid, 0.05% KH2PO4, 0.02% NaCl, 0.02% MgSO4, and 1.5% agar) at 30°C for 48 h. Antibiotics concentrations used for the incubation of R. pseudosolanacearum strains and E. coli strains were as follows: tetracycline, 20 μg/ml; kanamycin, 50 μg/ml; ampicillin, 100 μg/ml. Triparental mating between R. pseudosolanacearum strains and E. coil was performed on YDC medium (1% yeast extract, 2% dextrose, 2% CaCO3 and 1.5% agar).
Plasmid preparation, restriction endonuclease digestion, DNA ligation, agarose gel electrophoresis and other standard DNA recombinant techniques were performed following the protocols as described previously (Sambrook et al., 1989). Total genomic DNA of R. pseudosolanacearum SL341 and its mutants was extracted using standard DNA purification kit (Elpis-Biotech, Daejeon, Korea).
For transposon mutagenesis of R. pseudosolanacearum SL341, random Tn5 insertion mutants were generated using the EZTn5<KAN-2>Tnp Transposome kit (Epicentre, Madison, WI, USA) according to the manufacturer’s instructions. The transposon-inserted mutants were selected on CPG agar supplemented with kanamycin, and all of the mutants were stored in 40% glycerol at −80°C until phenotype screening (Wu et al., 2015).
To identify the Tn5-insertion sites in the selected mutants, genomic DNA was extracted and subjected to DNA sequencing using transposon primers KAN-2 FP-1 and KAN-2 RP-1. Additionally, the genomic DNA fragment carrying the Tn5 transposon was cloned into pUC119 to confirm the insertion site. Genomic DNA from the SL341P4 was digested with EcoRI, ligated into EcoRI digested pUC119 and transformed into E. coli. and Transformants carrying the plasmid with Tn-inserted genomic DNA fragments were selected on LB agar supplemented with kanamycin. The recombinant plasmid was subjected to DNA sequence analysis.
To complement the NDP-sugar epimerase gene mutation in SL341P4, the disrupted target gene, NDP-sugar epimerase gene, was amplified from wild-type R. pseudosolanacearum SL341 genomic DNA as a template using oligonucleotides 453F2 and 453R2. These primers were designed based on the Tn-inserted DNA sequences of SL341P4 to amplify a 2-kb DNA fragment. Polymerase chain reaction (PCR) amplification was performed with the following program: an initial denaturation step at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 66°C for 30 s, and extension at 72°C for 2 min, and a final extension step at 72°C for 5 min. The amplified PCR product was initially cloned into pGEM-T Easy, resulting in the generation of pF2R2. The DNA sequences of NDP-sugar epimerase gene were verified in pF2R2. Next, the NDP-sugar epimerase gene in pF2R2 was digested with NotI, treated with S1 nuclease (Promega, Madison, WI, USA) to obtain blunt ends, and inserted into the pRK415 digested with SmaI. This resulted in the subcloning of the NDP-sugar epimerase gene behind the lac promotor of pRK415. The resulting shuttle plasmid was designated pTDP415. Plasmids pTDP415 and pRK415 were introduced into the mutant strain SL341P4 by triparental mating, generating the complementation strain SL341P4C6 and vector control strain SL341P4V2, respectively.
EPS production of R. pseudosolanacearum strains used in this study was quantified using the biochemical Elson-Morgan assay (Peyraud et al., 2017). The R. pseudosolanacearum strains were grown in CPG broth supplemented with appropriate antibiotics at 30°C with shaking at 200 rpm for 60 h. The bacterial cell density was examined by dilution plating on TZC medium. Cell-free culture filtrates were obtained by filtrating the liquid culture through a 0.2 μm pore size membrane filter. The EPS fraction was precipitated, and the reaction with Ehrlich’s reagent was performed following the previously described method (Gatt and Berman, 1966). D-(+)-galactosamine was used as a reference compound to generate standard curve. The amount of EPS produced by R. pseudosolanacearum strains was quantified as the amount of galactosamine per 109 bacterial cells using the standard curve.
Biofilm production of R. pseudosolanacearum strains was assessed using crystal violet staining, following a modified version of the method described by O’Toole and Kolter (1998). In briefly, each R. pseudosolanacearum strains was cultured in 5 ml of CPG broth supplemented with appropriate antibiotics (tetracycline, 20 μg/ml; kanamycin, 50 μg/ml) overnight. Bacterial cells were harvested by centrifugation, resuspended in 30% glycerol, and adjusted to an optical density of 1.0 at 600 nm (OD600). Then, 100 μl of bacterial suspension was transferred to 96-well polyvinylchloride (PVC) microplate (Corning, New York, NY, USA), while 30% glycerol solution was used as a negative control. The PVC microplate was covered with Parafilm and statically incubated at 30°C for 24 and 72 h.
After incubation, the bacterial suspension was stained with 25 μl of 0.1% crystal violet solution for 15 min at 30°C. Subsequently, the planktonic cells were carefully removed, and the microplate was washed three times with sterile distilled water before being air dried. The cell-bound crystal violet was dissolved in 200 μl of dimethyl sulfoxide twice and then 400 μl of the bacterial suspension was mixed with 600 μl of sterile distilled water. Finally, the OD600 was measured using spectrophotometer (Beckman Coulter, Brea, CA, USA) to quantify the biofilm production.
All NDP-sugar epimerase homologous amino acid sequences from diverse phytopathogens were obtained through the NCBI (https://www.ncbi.nlm.nih.gov/) and INRA database (https://iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cgi) (Table 1). The phylogenetic analysis was performed using the neighbor-joining method with MEGA-X software (https://www.megasoftware.net/). Conserved motifs of NDP-sugar epimerase were identified using the MEME suite tool (http://meme-suite.org/tools/meme) with the other default parameters and a maximum of 15 motifs. Functional domains analysis of the NDP-sugar epimerase was conducted using KEGG MOTIF search (https://www.genome.jp/tools/motif/) and Pfam database (https://pfam.xfam.org/).
To compare the growth of wild-type R. pseudosolanacearum SL341 and the mutant SL341P4, both strains were cultured at 30°C in different media, including CPG, MG, and M9 broth supplemented with 0.4% glucose. Bacterial cell density was assessed by measuring the OD600 using spectrophotometer (Beckman Coulter), and the viable colony counts were determined by plating serial dilutions of the cultures od TZC agar medium.
For the virulence teste of R. pseudosolanacearum strains, the commercial cultivar Zuiken, which is susceptible to BW, and the resistant cultivar Hawaii 7996 were used. Tomato seeds were surface sterilized, and the tomato plants were grown in pots following established method (Kwak et al., 2018). The pots contained commercial horticulture nursery-media soil (Punong Co. Ltd., Gyeongju, Korea), and the plants were maintained in a growth room for 3 weeks at 28°C with a light period of 14 h followed dark period of 10 h. Prior to inoculation, the tomato plants were transferred to a growth chamber and kept at a 28°C with light for 14 h and in the dark for 10 h.
The virulence of R. pseudosolanacearum strains were assessed by inoculating 3-4-week-old tomato plants using either the soil-soaking or petiole-injection method, as described by Lee et al. (2011). Bacterial cells were cultured in CPG broth and subsequently centrifuged at 6,000 ×g. The resulting pellets were washed twice with sterile distilled water and resuspended in sterile distilled water to an OD600 of 0.4 (ca. 4-5 × 108 cfu/ml). For bacterial inoculation, each tomato plant was exposed to bacterial suspension at a final concentration of approximately 107 cfu/g of soil by soil-soaking or 105 cfu/plant for petiole-injection. Each strain was inoculated on 10 plants, including non-inoculated controls. The inoculation experiments were repeated three times. Following inoculation, all plants were incubated in a growth chamber under a 14 h light and 10 h dark cycle at 28°C. Plants were monitored for disease progress over time after inoculation, and BW disease severity was scored using the following method: (number of wilted leaves/total number of leaves) × 100%.
To compare the bacterial multiplication of SL341 and SL341P4, tomato plants were inoculated by soil-soaking method, and samples were collected at 7- and 14-days post-inoculation (dpi). Tomato midstems were removed, weighed, and cut into small pieces. After vigorous vortex mixing, the samples were maintained in sterile water for 10 min. subsequently, a series of dilution samples were plated on SMSA medium (Denny and Hayward, 2001) to determine the cfu per gram of stem fresh weight. Additionally, rhizosphere soils from tomato plants at14 dpi were sampled. Soils loosely attached to the plant roots were removed by gentle shaking, and soils tightly associated with plant roots were separated by vigorous vortexing in a sterilized saline (0.75% NaCl). Dilution samples were then plated on SMSA medium. Statistical significance among treatments was evaluated using analysis of variance (ANOVA) followed by a least significant difference post hoc test.
To investigate the CWDE activity and motility of bacterial strains, the bacterial cells were cultured in CPG broth with appropriated antibiotics. After cultivation, the bacterial cells were harvested by centrifugation and resuspended in phosphate buffered saline (pH 7.4). Cellulase, pectinase, and polygalacturonase assays were conducted using MG agar containing 1% carboxyl methyl cellulose (CMC) for pectinase activity and pectin for pectinase activity, and Hildebrand’s polypectate medium for polygalacturonase activity. Each medium was inoculated with 5 μl of bacterial suspension (OD600 = 1.0) and statically incubated at 30°C for 3 days. After incubation, MG agar plates containing CMC or pectin were stained with 0.1% Congo Red and washed three times with 1 M NaCl. The cellulase, pectinase and polygalacturonase activities were evaluated by measuring the diameter of clear zone on the CMC and pectin medium, and the well size around the bacterial colonies on the polypectate medium, respectively. Motility assay was performed by stab-inoculating the bacterial strains into the center of separate test tube MG media containing 0.3% agar, inoculating to half the depth. The medium was then statically incubated at 30°C, and the swimming motility of bacterial strains was observed at 14 dpi.
Previously, we generated 3,900 transposon-inserted mutants of R. pseudosolanacearum SL341 (Wu et al., 2015). Among these mutants, 17 EPS-defective mutants were selected. Through sequencing of transposon insertion sites in Tn5-inserted mutants, we identified genes involved in EPS production, including phcA, xpsR, vsrA, epsD, and gdhA (Wu et al., 2015). Among these mutants, we identified a mutant strain, SL341P4, which harbored a transposon insertion in the gene encoding a putative NDP-sugar epimerase and dTDP-glucose 4,6-dehydratase protein (Fig. 1A). In comparison to the wild-type SL341 strain, SL341P4 exhibited reduced EPS production and displayed a small colony shape on TZC agar medium. However, the production of EPS was partially restored in the complementation strain, SL341P4C6 (Fig. 1B).
To quantify EPS production in each R. pseudosolanacearum strains, the total amount of hexosamine was measured in TZC broth medium using the Elson-Morgan biochemical assay (Fig. 1C). Compared with SL341, the amount of galactosamine per 109 cells was reduced by 75.9- and 54.4-fold in SL341P4 and a vector control strain SL341P4V2 (SL341P4 mutant with pRK415), respectively (Fig. 1C). However, the production of galactosamine in SL341P4C6 were increased by 19.0- and 15.3-fold compared with SL341P4 and SL341P4V2, respectively. Additionally, we investigated impact of disruption of the putative NDP-sugar epimerase gene on biofilm formation in vitro (Supplementary Fig. 1). At 1 dpi, biofilm production of SL341P4 was 3.08-fold higher than that of wild type strain SL341. However, at 3 dpi, biofilm formation of SL341P4 was reduced by 1.46-fold compared to SL341. These results demonstrate that the putative NDP-sugar epimerase contributes to EPS production and biofilm formation in R. pseudosolanacearum SL341.
To compare the sequence of NDP-sugar epimerase in SL341 with that of other bacterial pathogens, we found the 19 homologous amino acid sequences from diverse pathogenic bacteria including other RSSC strains, Burkholderiales bacterium, Pseudomonas syringae, Rhizobium leguminosarum, Agrobacterium sp., and Xathomonas campestris (Table 1). Among 19 homologous amino acid sequences of NDP-sugar epimerase, 14 sequences from diverse RSSC showed more than 93% identity with the sequence of SL341 (Table 1). However, five sequences from other pathogens displayed low identity with the SL341 sequence, ranging from 37.87% to 53.26% (Table 1). In the phylogenetic analysis, the sequences from RSSC strains were distinct from the five sequences of other phytopathogens (Supplementary Fig. 2). Furthermore, each sequence from the RSSC strains was further divided based on the phylotype of the RSSC in the neighbor-joining trees (Supplementary Fig. 2).
To identify the conserved domains or motifs of NDP-sugar epimerase, 20 homologous amino acid sequences from diverse pathogenic bacteria were submitted to the MEME suite tool (Table 2, Fig. 2, Supplementary Fig. 3). The analysis revealed the presence of 15 common motifs in the diverse NDP-sugar epimerase sequences (Table 2, Fig. 2, Supplementary Fig. 3). Motif 1, 2, 4, 7, 10, and 13 formed a fundamental structural combination that was conserved across all NDP-sugar epimerases. Additionally, among 15 motifs, motif 1, 2, 3, 4, 10, 11, and 13 were predicted to belong “Polysacc_synt_2” motif family, which is associated with putative polysaccharide biosynthesis proteins (Table 2, Fig. 2). Notably, the transposon was inserted in motif 1, which is the most conserved motif, in SL341P4 mutant (Supplementary Fig. 3). Meanwhile, motif 5 and 6 were predicted to be part of the “CoA_binding” motif, which is associated with coenzyme A biding domain (Table 2, Fig. 2). All RSSC strains possess both motifs, whereas none of the other pathogens, except for B. bacterium, exhibit these motifs. B. bacterium only has motif 6 and lacks motif 5 (Fig. 2). These results indicate that NDP-sugar epimerase is conserved among diverse phytopathogens and may play a role in polysaccharide biosynthesis.
Because the colony size of SL341P4 was smaller than that of SL341 wild type on TZC medium (Fig. 1B), we investigated whether the mutation in the putative NDP-sugar epimerase gene affects bacterial growth in SL341. To address this, the bacterial growth of SL341P4 was investigated in CPG, MG, and M9 broth media (Fig. 3). Unexpectedly, the growth of SL341 and SL341P4 mutant was comparable in all three media (Fig. 3). The growth rate of both strains was faster in rich CPG medium compared to the minimal media M9 and MG broth (Fig. 3). In CPG broth, the population of SL341 and SL341P4 were reached approximately 109 cfu/ml at 12 hours post-inoculation (hpi) and were saturated after 24 hpi (Fig. 3B). The OD600 of SL341 and SL341P4 mutant was exceeded 1.5 at 48 hpi in CPG broth (Fig. 3A). In the case of MG and M9 broth, the population of SL341 and SL341P4 were reached approximately 109 cfu/ml after 24 hpi, and the OD600 of both strains was exceeded 1.0 after 72- and 96-hpi, respectively (Fig. 3A and B).
Since SL341P4 exhibited attenuated virulence characteristics such as reduced EPS production and biofilm formation (Fig. 1, Supplementary Fig. 1), we investigated whether the mutation in the putative NDP-sugar epimerase gene of SL341 affect its virulence in tomato plants. To assess virulence of SL341P4, we inoculated cell suspensions of SL341 and SL341P4 into the root system of commercial tomato BW-susceptible cultivar Zuiken (Fig. 4A). Tomato plants inoculated with SL341P4 mutant showed significantly delayed and decreased disease progression compared to SL341-inoculated plants (Fig. 4A). At 21 dpi, the disease severity of BW caused by SL341P4 and SL341P4V2 were reduced by 2.17- and 2.60-fold, respectively, compared to SL341-inoculated plants. Meanwhile, the SL341P4C6 exhibited the partially recovered virulent activity in the tomato cultivar (Fig. 4A). The disease severity in SL341P4C6-inoculated plant were increased by 1.52- and 1.82-fold, compared to SL341P4 and SL341P4V2 (Fig. 4A), respectively. Interestingly, the SL341P4-inoculated plant showed not only low disease severity but also low disease incidence. At 7 and 14 dpi, 57.1% and 92.9% of SL341-inoculated plant exhibited BW symptom, while the 96.4% and 71.4% of SL341P4-inoculated plant did not show any symptom of BW at 7 and 14 dpi (data not shown).
To evaluate the bacterial multiplication of R. pseudosolanacearum in tomato plants, the population density of SL341P4 were measured in the midstem tissue and rhizosphere soil of tomato cultivar Zuiken (Supplementary Fig. 4). At 14 dpi, there was no significant difference in population density between SL341 and SL341P4 in both the midstem tissue and rhizosphere soil (Supplementary Fig. 4). Taken together, these results indicated that the putative NDP-sugar epimerase gene plays a role for the virulence of R. pseudosolanacearum SL341 in tomato plants, without affecting bacterial multiplication in the susceptible tomato cultivar Zuiken.
We further examined the impact of SL341P4 mutant on virulence in BW-resistant cultivar Hawaii 7996 (Fig. 4B). Although Hawaii 7996 is BW-resistant cultivar, SL341 showed high virulence to Hawaii 7996 (Choi et al., 2020; Jeong et al., 2007; Jung et al., 2014; Kwak et al., 2018; Murugaiyan et al., 2011; Thoquet et al., 1996). When cell suspension of SL341 and mutant strains were treated using soil-soaking method, the virulence of SL341P4 was reduced in Hawaii 7996 compared to Zuiken (Fig. 4B). The disease severity in SL341P4-inoculated plant were reduced by 6.6-fold at 21 dpi in Hawaii 7996 (Fig. 4B). Similarly, SL341P4V2 and SL341P4C6 also exhibited significant decrease in virulence in Hawaii 7996 (Fig. 4B). Meanwhile, when the petiole injection method employed, SL341 and SL341P4 showed enhanced virulence in Hawaii 7996 compared to the soil-soaking method (Supplementary Fig. 5). All Hawaii 7996 plants inoculated with SL341 displayed 100% disease severity at 12 dpi. Notably, the disease severity caused by SL341P4 was reduced by 2.29-fold at 13 dpi compared to SL341 (Supplementary Fig. 5). These results indicated that the mutation of the putative NDP-sugar epimerase led to reduction in the virulence of R. pseudosolanacearum.
In addition to EPS production and biofilm formation, CWDE production and cell motility are also known as a virulence factors of R. pseudosolanacearum (Liu et al., 2005; Tans-Kersten et al., 2001). We investigated whether the mutation of the putative NDP-sugar epimerase gene in SL341 affects the production of other virulence factors. First, we assessed the activity of CWDE such as cellulase, polygalacturonase and pectinase in R. pseudosolanacearum strains SL341, SL341P4, SL341P4C6, and SL341P4V2 (Supplementary Fig. 6A). However, these four strains showed similar activity levels of cellulase, polygalacturonase, and pectinase activity (Supplementary Fig. 6A). Additionally, swimming motility of SL341, SL341P4, SL341P4C6, and SL341P4V2 was tested in MG medium with low agar concentration (Supplementary Fig. 6B). However, the swimming motility among four strains was not different. These results suggested that the mutation of putative NDP-sugar epimerase gene did not affect the CWDE production and cell motility in R. pseudosolanacearum SL341.
In this study, we identified the putative NDP-sugar epimerase gene of R. pseudosolanacearum SL341, which is involved in EPS production. The NDP-sugar epimerase gene family has been reported in diverse bacterial species, but its functional roles, such as the virulence of phytopathogens, remain largely unknown. Here, we demonstrated that the mutation of the putative NDP-sugar epimerase gene caused the reduction in virulence in R. pseudosolanacearum SL341, as evidenced by the alteration of EPS and biofilm production.
NDP-sugar epimerase including dTDP-glucose 4,6-dehydratase, plays a crucial role in the biosynthesis of deoxy and amino sugars. These sugars are involved in a diverse biological functions, including polysaccharide biosynthesis, cell envelop biosynthesis, and virulence activity in various bacteria such as Rhizobium, Agrobacterium, R. solanacearum, Xanthomonas campestris, Bacillus subtilis, Pseudomonas putida, and E. coli (Becker et al., 1995; Capela et al., 2017; Czolkoss et al., 2021; Grangeasse et al., 2003; Huang and Schell, 1995; Koller and Lassak, 2021; Li et al., 2014; Mazur et al., 2002; Mijakovic et al., 2003; Soldo et al., 2003; Vogel et al., 2022; Wang et al., 2022). The putative NDP-sugar epimerase of SL341 is highly conserved among diverse phytopathogen and RSSC, especially GMI1000 (Table 1, Fig. 2). The putative NDP-sugar epimerase Rsp1004 in GMI1000 is known to be involved in EPS biosynthesis and virulence (Capela et al., 2017; Wang et al., 2022). Additionally, the homologues of putative NDP-sugar epimerase in SL341 and other phytopathogen contain “Polysacc_synt_2” motif domain, which is found in diverse bacterial polysaccharide biosynthesis proteins, including the ExoT protein from Rhizobium meliloti, the AmsL protein from Erwinia amylovora, the CapD protein from Staphylococcus aureus, the WalL protein from Vibrio cholerae, and several putative epimerases (Becker et al., 1993; Fallarino et al., 1997; Klee et al., 2020; Lin et al., 1994). Thus, our results suggested that the putative NDP-sugar epimerase is a novel virulence gene regulating EPS biosynthesis in SL341.
The similar efficacy of multiplication in vitro and in planta of SL341P4 indicates that the putative NDP-sugar epimerase gene is not involved in bacterial growth in RSSC (Fig. 3, Supplementary Fig. 4) (Wu et al., 2015). Thus, the differential virulence of SL341 and SL341P4 appears to be primarily determined by EPS production and biofilm formation. Bacterial EPS is one of the main pathogenicity determinants in RSSC (Denny and Back, 1991) and contributes to the occlusion of vascular system in the host plant after root invasion (Dalsing and Allen, 2014; Wallis and Truter, 1978). Generally, EPS-deficient mutants exhibit poor movement to the upper portion of the stem in infected plant (Araud-Razou et al., 1998; Denny and Baek, 1991; Kao et al., 1992; Saile et al., 1997). The SL341P4 mutant displayed the invasion into roots and midstems of tomato plants but did not completely kill the host plant (Fig. 4A, Supplementary Fig. 4A). This suggests that the reduction of EPS production in SL341P4 mutant might influence the expression of wilt symptoms caused by R. pseudosolanacearum after invasion into the tomato vascular tissue (Planas-Marquès et al., 2019).
The EPS-defective mutation was partially restored by providing the original NDP-sugar epimerase gene in trans under lac promoter. The complementation strain SL341P4C6 showed partial restoration of virulence in a susceptible cultivar Zuiken but not in a resistant cultivar Hawaii 7996 (Fig. 4). Although EPS production was partially restored in SL341P4C6, it still impaired the formation of biofilm (Fig. 1, Supplementary Fig. 1). Biofilm formation plays important role in the attachment to root surface and root invasion in R. pseudosolanacearum (Denny and Baek, 1991; Liu et al., 2005; Yao and Allen, 2007). Specifically, the attachment and root invasion of R. pseudosolanacearum were more reduced in the vascular cylinder of Hawaii 7996 than in the susceptible cultivar (Caldwell et al., 2017). Meanwhile, when the SL341P4 strain was directly injected into the tomato petiole, SL341P4 exhibited similar levels of virulence to susceptible cultivars, even in resistant ones, unlike soil-soaking inoculation (Supplementary Fig. 5). This indicates that the virulence of R. pseudosolanacearum SL341 is not solely determined by EPS production in the resistant tomato cultivar. Since SL341 wild-type, SL341P4, and SL341P4C6 did not show any difference in CWDE activity and cell motility (Supplementary Fig. 6), the disruption of NDP-sugar epimerase gene might affect other virulence factors of SL341, such as type III secretion system or quorum sensing, which are involved infecting the resistant cultivar. Further investigation of multiple virulence pathways controlled by the putative NDP-sugar epimerase in R. pseudosolanacearum SL341 should be required.
Acknowledgments
This work was supported by National Research Foundation of Korea (NRF) grants to S.-W. L. (No. 2020R1A2C3005453 and 2020R1A6A1A03047729) and Green Fusion Technology Program funded by the Korea government (MSIT, MOE, ME), Republic of Korea.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
The mutation of the putative NDP-sugar epimerase gene and exopolysaccharide (EPS) production in Ralstonia pseudosolanacearum SL341. (A) Genomic position and surrounding regions of the transposon insertion site of R. pseudosolanacearum SL341P4 mutant. Map of a 28 kb fragment of R. pseudosolanacearum SL341 strain carrying 21 open reading frames (ORFs) encoding function of genes including transposon inserted gene (dark arrow). Circle at the top of the map indicates the transposon insertion site located at C-terminus region of the putative NDP-sugar epimerase protein. (B) Colony morphologies of R. pseudosolanacearum strains of SL341 (wild-type), and the NDP-sugar epimerase gene mutant SL341P4 and a complementation strain SL341P4C6 were taken at 2 days post inoculation on TZC agar medium at 30°C. (C) Biochemical quantification of EPS hexosamine from R. pseudosolanacearum strains. Total amount of hexosamine was quantified from bacterial culture grown in CPG broth medium. The amount of hexosamine was normalized to the 109 bacterial cells. Different letters above the bar represent the significant difference in Tukey means comparison (P = 0.05).
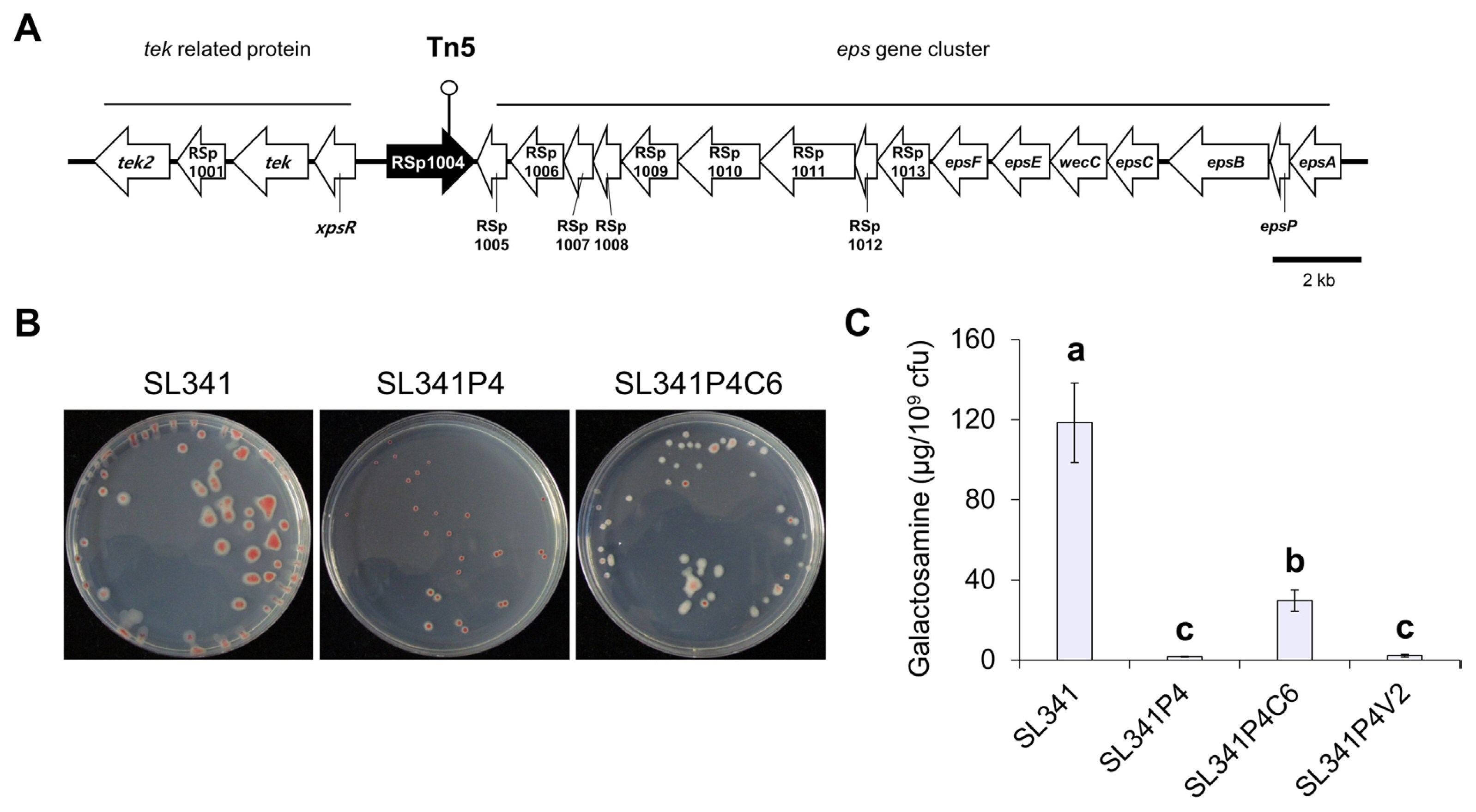
Fig. 2
Alignment of conserved domain of the putative NDP-sugar epimerase of Ralstonia pseudosolanacearum SL341 and its homologs from diverse phytopathogen. Different colored boxes representing different motifs with different conservative sequences. The serial number of boxes corresponds to the sequences of conserved motifs in Supplementary Fig. 3. There are two functional domains in NDP-sugar epimerase, CoA-binding domain and Polysaccharide biosynthesis domain. The functional domains above boxes corresponds to the list in Table 2. RSSC, Ralstonia solanacearum species complex.
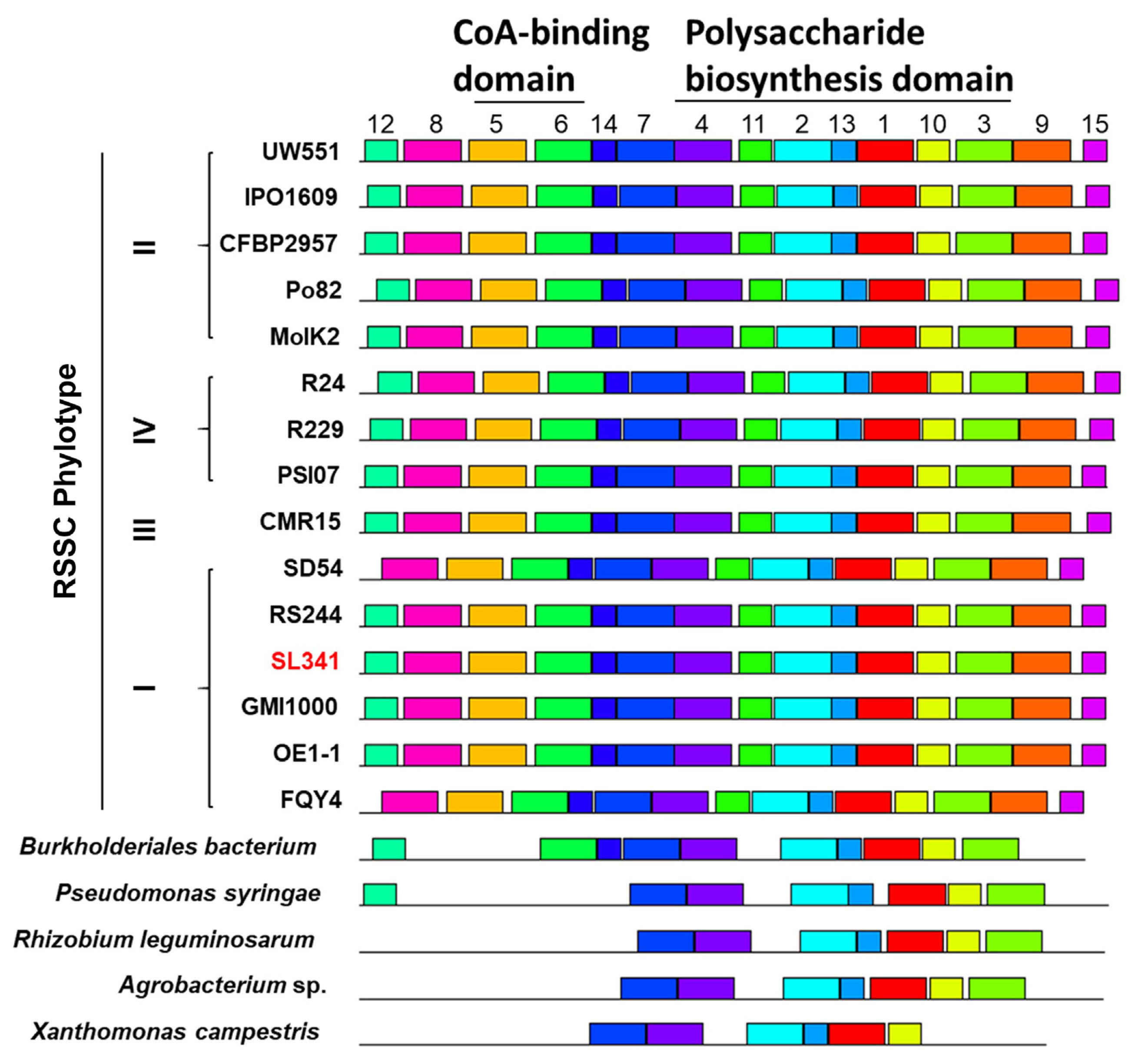
Fig. 3
The assessment of bacteria growth of Ralstonia pseudosolanacearum SL341 and SL341P4. R. pseudosolanacearum SL341 and SL341P4 were cultured in CPG, MG, and M9 broth medium by shaking at 200 rpm over time. The closed and opened symbols represent the wild-type strain SL341 and the mutant strain SL341P4, respectively. The bacterial growth of the wild-type strain SL341 and the mutant strain SL341P4 were assessed by measuring the optical density (OD600) using by spectrophotometer (A) and bacterial cell count on SMSA medium (B). ■, SL341 in CPG broth; ♦, SL341 in MG broth; ▲, SL341 in M9 broth; □, SL341P4 in CPG broth ⋄, SL341P4 in MG broth; △, SL341P4 in M9 broth. Vertical bars indicate the standard deviations from 3 replications.
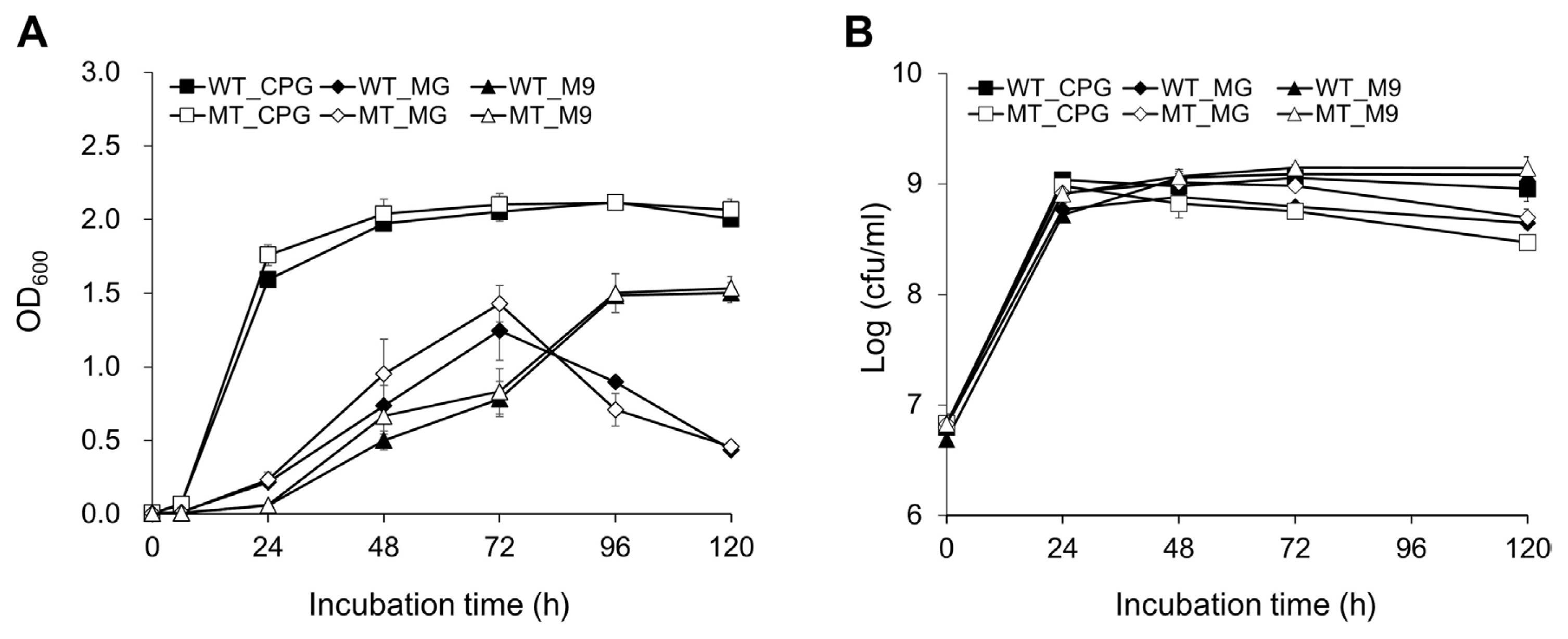
Fig. 4
Disease responses on tomato plants Zuiken (A) and Hawaii 7996 (B) inoculated with Ralstonia pseudosolanacearum strain SL341, SL341P4, SL341P4C6, and SL341P4V2 using soil-soaking inoculations. Disease severity of bacterial wilt was investigated for 21 days post inoculation of R. pseudosolanacearum strains. Disease severities were calculated using followed method: (number of wilted leaves/total number of leaves) × 100 (%). ▲, inoculation of SL341 wildtype; ■, inoculation of SL341P4 mutant; △, inoculation of SL341P4C6 mutant; □, inoculation of SL341P4V2 mutant. Values are the average of three replicates (each replication with 10 plants, n = 30 for each treatment) and vertical bars represent standard error. Significant difference was noticed by repeated measures ANOVA (***P < 0.001).
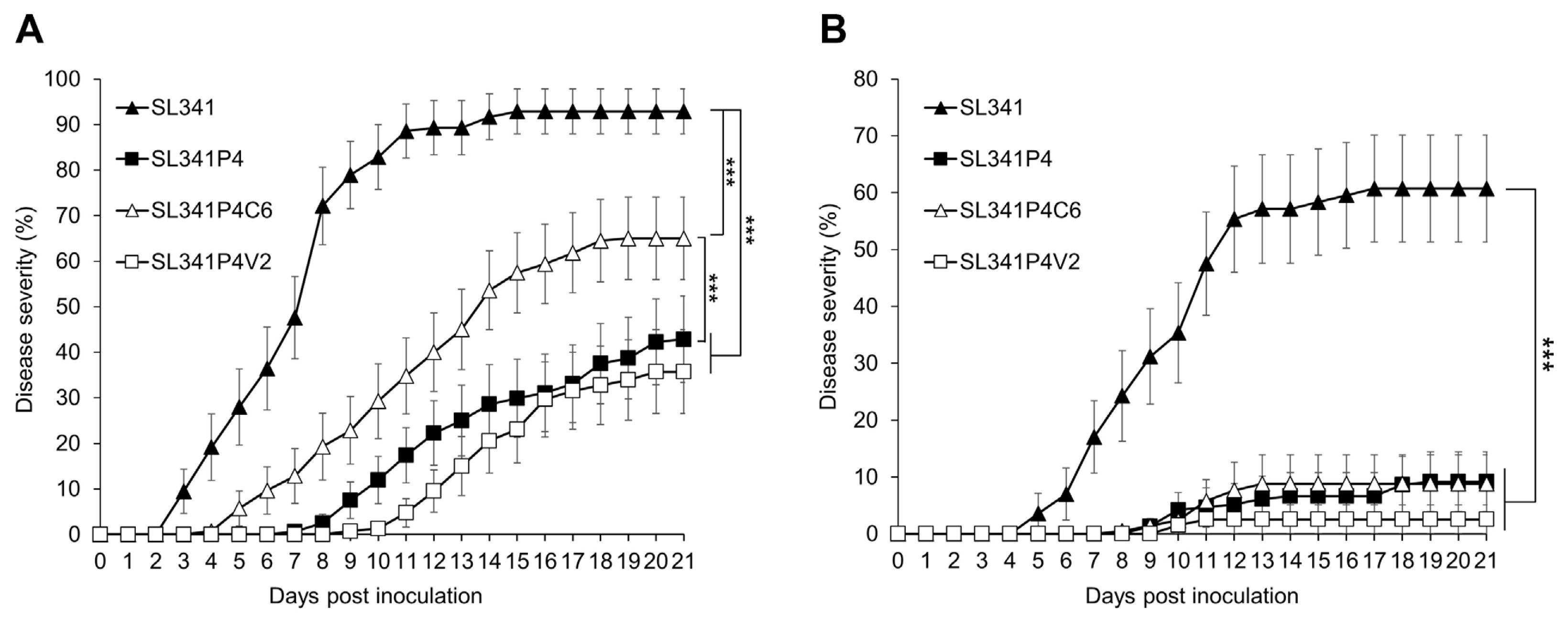
Table 1
List of homologous amino acid sequence of NDP-sugar epimerase in other 19 bacterial phytopathogens
The homologous sequences were retrieved from INRA (https://iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cgi) and NCBI (https://www.ncbi.nlm.nih.gov/) database.
Table 2
List of conserved motifs of NDP-sugar epimerase
There are 15 conserved motifs in NDP-sugar epimerase amino acid sequences in Fig. 2. Each motif sequence was classified by Pfam database.
References
Araud-Razou, I., Vasse, J., Montrozier, H., Etchebar, C. and Trigalet, A. 1998. Detection and visualization of the major acidic exopolysaccharide of Ralstonia solanacearum and its role in tomato root infection and vascular colonization. Eur. J. Plant Pathol 104:795-809.
Arlat, M., Gough, C. L., Zischek, C., Barberis, P. A., Trigalet, A. and Boucher, C. A. 1992. Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol. Plant-Microbe Interact 5:187-193.


Balatero, C. H., Hautea, D. M., Narciso, J. O. and Hanson, P. M. 2005. QTL mapping for bacterial wilt resistance in Hawaii. 7996. using AFLP, RGA, and SSR markers. In: Bacterial wilt disease and the Ralstonia solanacearum species complex, eds. by C. Allen, P. Prior and A. C. Hayward, pp. 301-307. APS Press, St. Paul, MN, USA.
Becker, A., Kleickmann, A., Küster, H., Keller, M., Arnold, W. and Pühler, A. 1993. Analysis of the Rhizobium meliloti genes exoU, exoV, exoW, exoT, and exoI involved in exopolysaccharide biosynthesis and nodule invasion: exoU and exoW probably encode glucosyltransferases. Mol. Plant-Microbe Interact 6:735-744.

Becker, A., Niehaus, K. and Pühler, A. 1995. Low-molecular-weight succinoglycan is predominantly produced by Rhizobium meliloti strains carrying a mutated ExoP protein characterized by a periplasmic N-terminal domain and a missing C-terminal domain. Mol. Microbiol 16:191-203.


Botta, G. A., Arzese, A., Minisini, R. and Trani, G. 1994. Role of structural and extracellular virulence factors in gram-negative anaerobic bacteria. Clin. Infect. Dis 18:Suppl 4. S260-S264.


Caldwell, D., Kim, B.-S. and Iyer-Pascuzzi, A. S. 2017. Ralstonia solanacearum differentially colonizes roots of resistant and susceptible tomato plants. Phytopathology 107:528-536.


Capela, D., Marchetti, M., Clérissi, C., Perrier, A., Guetta, D., Gris, C., Valls, M., Jauneau, A., Cruveiller, S., Rocha, E. P. C. and Masson-Boivin, C. 2017. Recruitment of a lineage-specific virulence regulatory pathway promotes intracellular infection by a plant pathogen experimentally evolved into a legume symbiont. Mol. Biol. Evol 34:2503-2521.


Chapman, M. R. and Kao, C. C. 1998. EpsR modulates production of extracellular polysaccharides in the bacterial wilt pathogen Ralstonia (Pseudomonas) solanacearum. J. Bacteriol 180:27-34.




Choi, K., Choi, J., Lee, P. A., Roy, N., Khan, R., Lee, H. J., Weon, H. Y., Kong, H. G. and Lee, S.-W. 2020. Alteration of bacterial wilt resistance in tomato plant by microbiota transplant. Front. Plant Sci 11:1186.



Clough, S. J., Lee, K. E., Schell, M. A. and Denny, T. P. 1997. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J. Bacteriol 179:3639-3648.




Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R. and Lappin-Scott, H. M. 1995. Microbial biofilms. Annu. Rev. Microbiol 49:711-745.


Czolkoss, S., Safronov, X., Rexroth, S., Knoke, L. R., Aktas, M. and Narberhaus, F. 2021. Agrobacterium tumefaciens type IV and type VI secretion systems reside in detergent-resistant membranes. Front. Microbiol 12:754486.



Dalsing, B. L. and Allen, C. 2014. Nitrate assimilation contributes to Ralstonia solanacearum root attachment, stem colonization, and virulence. J. Bacteriol 196:949-960.



de Pedro-Jové, R., Puigvert, M., Sebastià, P., Macho, A. P., Monteiro, J. S., Coll, N. S., Setúbal, J. C. and Valls, M. 2021. Dynamic expression of Ralstonia solanacearum virulence factors and metabolism-controlling genes during plant infection. BMC Genomics 22:170.




Denny, T. P. and Baek, S.-R. 1991. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol. Plant-Microbe Interact 4:198-206.

Denny, T. P. and Hayward, A. C. 2001. Gram-negative bacteria: Ralstonia. In: Laboratory guide for identification of plant pathogenic bacteria, eds. by N. W. Schaad, J. B. Jones and W. Chun, 3rd ed. pp. 151-174. APS Press, St. Paul, MN, USA.
Fallarino, A., Mavrangelos, C., Stroeher, U. H. and Manning, P. A. 1997. Identification of additional genes required for O-antigen biosynthesis in Vibrio cholerae O1. J. Bacteriol 179:2147-2153.




Fegan, M. and Prior, P. 2005. How complex is the Ralstonia solanacearum species complex. In: Bacterial wilt disease and the Ralstonia solanacearum species complex, eds. by C. Allen, P. Prior and A. C. Hayward, pp. 449-461. APS Press, St. Paul, MN, USA.
Gatt, R. and Berman, E. R. 1966. A rapid procedure for the estimation of amino sugars on a micro scale. Anal. Biochem 15:167-171.


Genin, S. and Boucher, C. 2002. Ralstonia solanacearum: secrets of a major pathogen unveiled by analysis of its genome. Mol. Plant Pathol 3:111-118.



Ghosh, P. K. and Maiti, T. K. 2016. Structure of extracellular polysaccharides (EPS) produced by rhizobia and their functions in legume-bacteria symbiosis: a review. Achiev. Life Sci 10:136-143.

Grangeasse, C., Obadia, B., Mijakovic, I., Deutscher, J., Cozzone, A. J. and Doublet, P. 2003. Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J. Biol. Chem 278:39323-39329.


Harimawan, A. and Ting, Y.-P. 2016. Investigation of extracellular polymeric substances (EPS) properties of P. aeruginosa and B. subtilis and their role in bacterial adhesion. Colloid Surf. B Biointerfaces 146:459-467.

Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol 29:65-87.


Hayward, A. C. 1994. Systematics and phylogeny of Pseudomonas solanacearum and related bacteria. In: Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum, eds. by A. C. Hayward and G. L. Hartman, pp. 123-135. CAB International, Wallingford, UK.
Hikichi, Y., Mori, Y., Ishikawa, S., Hayashi, K., Ohnishi, K., Kiba, A. and Kai, K. 2017. Regulation involved in colonization of intercellular spaces of host plants in Ralstonia solanacearum. Front. Plant Sci 8:967.



Hikichi, Y., Yoshimochi, T., Tsujimoto, S., Shinohara, R., Nakaho, K., Kanda, A., Kiba, A. and Ohnishi, K. 2007. Global regulation of pathogenicity mechanism of Ralstonia solanacearum. Plant Biotechnol 24:149-154.

Huang, J. and Schell, M. 1995. Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol. Microbiol 16:977-989.


Jacobs, J. M., Babujee, L., Meng, F., Milling, A. and Allen, C. 2012. The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. MBio 3:e00114-12.




Jeong, Y., Kim, J., Kang, Y., Lee, S. and Hwang, I. 2007. Genetic diversity and distribution of Korean isolates of Ralstonia solanacearum. Plant Dis 91:1277-1287.


Jung, E. J., Joo, H. J., Choi, S. Y., Lee, S. Y., Jung, Y. H., Lee, M. H., Kong, H. G. and Lee, S.-W. 2014. Resistance evaluation of tomato germplasm against bacterial wilt by Ralstonia solanacearum. Res. Plant Dis 20:253-258 (in Korean).

Kai, K., Ohnishi, H., Shimatani, M., Ishikawa, S., Mori, Y., Kiba, A., Ohnishi, K., Tabuchi, M. and Hikichi, Y. 2015. Methyl 3-hydroxymyristate, a diffusible signal mediating phc quorum sensing in Ralstonia solanacearum. ChemBioChem 16:2309-2318.


Kao, C. C., Barlow, E. and Sequeira, L. 1992. Extracellular polysaccharide is required for wild-type virulence of Pseudomonas solanacearum. J. Bacteriol 174:1068-1071.




Klee, S. M., Sinn, J. P., Christian, E., Holmes, A. C., Zhao, K., Lehman, B. L., Peter, K. A., Rosa, C. and McNellis, T. W. 2020. Virulence genetics of an Erwinia amylovora putative polysaccharide transporter family member. J. Bacteriol 202:e00390-20.



Koller, F. and Lassak, J. 2021. Two RmlC homologs catalyze dTDP-4-keto-6-deoxy-D-glucose epimerization in Pseudomonas putida KT2440. Sci. Rep 11:11991.




Kumar, A. S. and Mody, K. 2009. Microbial exopolysaccharides: variety and potential applications. In: Microbial production of biopolymers and polymer precursors: applications and perspectives, eds. by B. H. A. Rehm, pp. 229-254. Academic Press, Norfolk, UK.
Kwak, M.-J., Kong, H. G., Choi, K., Kwon, S.-K., Song, J. Y., Lee, J., Lee, P. A., Choi, S. Y., Seo, M., Lee, H. J., Jung, E. J., Park, H., Roy, N., Kim, H., Lee, M. M., Rubin, E. M., Lee, S.-W. and Kim, J. F. 2018. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol 36:1100-1109.


Lee, H. J., Jo, E. J., Kim, N. H., Chae, Y. and Lee, S.-W. 2011. Disease responses of tomato pure lines against Ralstonia solanacearum strains from Korea and susceptibility at high temperature. Res. Plant Dis 17:326-333 (in Korean).

Li, C.-T., Liao, C.-T., Du, S.-C., Hsiao, Y.-P., Lo, H.-H. and Hsiao, Y.-M. 2014. Functional characterization and transcriptional analysis of galE gene encoding a UDP-galactose 4-epimerase in Xanthomonas campestris pvcampestris. Microbiol. Res 169:441-452.


Lin, W. S., Cunneen, T. and Lee, C. Y. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol 176:7005-7016.




Liu, H., Zhang, S., Schell, M. A. and Denny, T. P. 2005. Pyramiding unmarked deletions in Ralstonia solanacearum shows that secreted proteins in addition to plant cell-wall-degrading enzymes contribute to virulence. Mol. Plant-Microbe Interact 18:1296-1305.


Mazur, A., Król, J. E., Wielbo, J., Urbanik-Sypniewska, T. and Skorupska, A. 2002. Rhizobium leguminosarum bv. trifolii PssP protein is required for exopolysaccharide biosynthesis and polymerization. Mol. Plant-Microbe Interact 15:388-397.


Mijakovic, I., Poncet, S., Boël, G., Mazé, A., Gillet, S., Jamet, E., Decottignies, P., Grangeasse, C., Doublet, P., Le Maréchal, P. and Deutscher, J. 2003. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J 22:4709-4718.



Murugaiyan, S., Bae, J. Y., Wu, J., Lee, S. D., Um, H. Y., Choi, H. K., Chung, E., Lee, J.-H. and Lee, S.-W. 2011. Characterization of filamentous bacteriophage PE226 infecting Ralstonia solanacearum strains. J. Appl. Microbiol 110:296-303.


Orgambide, G., Montrozier, H., Servin, P., Roussel, J., Trigalet-Demery, D. and Trigalet, A. 1991. High heterogeneity of the exopolysaccharides of Pseudomonas solanacearum strain GMI 1000 and the complete structure of the major polysaccharide. J. Biol. Chem 266:8312-8321.


O’Toole, G. A. and Kolter, R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol 28:449-461.



Peyraud, R., Denny, T. P. and Genin, S. 2017. Exopolysaccharide quantification for the plant pathogen Ralstonia solanacearum. Bio Protoc 7:e2289.



Planas-Marquès, M., Kressin, J. P., Kashyap, A., Panthee, D. R., Louws, F. J., Coll, N. S. and Valls, M. 2019. Four bottlenecks restrict colonization and invasion by the pathogen Ralstonia solanacearum in resistant tomato. J. Exp. Bot 71:2157-2171.



Reeves, P. R., Hobbs, M., Valvano, M. A., Skurnik, M., Whitfield, C., Coplin, D., Kido, N., Klena, J., Maskell, D., Raetz, C. R. and Rick, P. D. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol 4:495-503.


Saile, E., McGarvey, J. A., Schell, M. A. and Denny, T. P. 1997. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 87:1264-1271.


Sambrook, J., Fritsch, E. F. and Maniatis, T. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Habor Laboratory Press, Cold Spring Habor, NY, USA. pp. 1546.
Schaad, N. W., Jones, J. B. and Chun, W. 2001. Laboratory guide for the identification of plant pathogenic bacteria. APS Press, St. Paul, MN, USA. pp. 373.
Schell, M. A. 2000. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol 38:263-292.


Soldo, B., Scotti, C., Karamata, D. and Lazarevic, V. 2003. The Bacillus subtilis Gne (GneA, GalE) protein can catalyse UDP-glucose as well as UDP-N-acetylglucosamine 4-epimerisation. Gene 319:65-69.


Tans-Kersten, J., Huang, H. and Allen, C. 2001. Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol 183:3597-3605.




Thoquet, P., Olivier, J., Sperisen, C., Rogowsky, P., Laterrot, H. and Grimsley, N. 1996. Quantitative trait loci determining resistance to bacterial wilt in tomato cultivar Hawaii7996. Mol. Plant-Microbe Interact 9:826-836.

Vogel, U., Beerens, K. and Desmet, T. 2022. Nucleotide sugar dehydratases: structure, mechanism, substrate specificity, and application potential. J. Biol. Chem 298:101809.



Wallis, F. M. and Truter, S. J. 1978. Histopathology of tomato plants infected with Pseudomonas solanacearum, with emphasis on ultrastructure. Physiol. Plant Pathol 13:307-310.

Wang, L., Gao, Y., Jiang, N., Yan, J., Lin, W. and Cai, K. 2022. Silicon controls bacterial wilt disease in tomato plants and inhibits the virulence-related gene expression of Ralstonia solanacearum. Int. J. Mol. Sci 23:6965.



Wicker, E., Grassart, L., Coranson-Beaudu, R., Mian, D., Guilbaud, C., Fegan, M. and Prior, P. 2007. Ralstonia solanacearum strains from Martinique (french west indies) exhibiting a new pathogenic potential. Appl. Environ. Microbiol 73:6790-6801.




- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 2,074 View
- 164 Download
- ORCID iDs
-
Seon-Woo Lee

https://orcid.org/0000-0001-6142-7612 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print



