Evaluation and Genome Mining of Bacillus stercoris Isolate B.PNR1 as Potential Agent for Fusarium Wilt Control and Growth Promotion of Tomato
Article information
Abstract
Recently, strategies for controlling Fusarium oxysporum f. sp. lycopersici (Fol), the causal agent of Fusarium wilt of tomato, focus on using effective biocontrol agents. In this study, an analysis of the biocontrol and plant growth promoting (PGP) attributes of 11 isolates of loamy soil Bacillus spp. has been conducted. Among them, the isolates B.PNR1 and B.PNR2 inhibited the mycelial growth of Fol by inducing abnormal fungal cell wall structures and cell wall collapse. Moreover, broad-spectrum activity against four other plant pathogenic fungi, F. oxysporum f. sp. cubense race 1 (Foc), Sclerotium rolfsii, Colletotrichum musae, and C. gloeosporioides were noted for these isolates. These two Bacillus isolates produced indole acetic acid, phosphate solubilization enzymes, and amylolytic and cellulolytic enzymes. In the pot experiment, the culture filtrate from B.PNR1 showed greater inhibition of the fungal pathogens and significantly promoted the growth of tomato plants more than those of the other treatments. Isolate B.PNR1, the best biocontrol and PGP, was identified as Bacillus stercoris by its 16S rRNA gene sequence and whole genome sequencing analysis (WGS). The WGS, through genome mining, confirmed that the B.PNR1 genome contained genes/gene cluster of a nonribosomal peptide synthetase/polyketide synthase, such as fengycin, surfactin, bacillaene, subtilosin A, bacilysin, and bacillibactin, which are involved in antagonistic and PGP activities. Therefore, our finding demonstrates the effectiveness of B. stercoris strain B.PNR1 as an antagonist and for plant growth promotion, highlighting the use of this microorganism as a biocontrol agent against the Fusarium wilt pathogen and PGP abilities in tomatoes.
Tomato (Solanum lycopersicum L.) is one of the most significant economic vegetable crops. Recently, the total area of tomato cultivation has been growing based on the demand for and nutritional value of tomatoes. However, there are significant crop losses in tomatoes due to several plant pathogens, including bacteria, viruses, and fungi (Ajilogba and Babalola, 2013). Fusarium wilt disease is one of the most severe in tomato plants that is caused by Fusarium oxysporum f. sp. lycopersici (Fol). The Fol pathogen is a severe threat to tomato production because it affects tomato plants worldwide by reducing yields in both open fields and greenhouses (McGovern, 2015). Moreover, this disease is difficult to control because the pathogen can overseason for many years in the soil and crop residues due to chlamydospore production, and it then enters the tomato root through wounds, invading the vascular tissue (McGovern, 2015; Srinivas et al., 2019).
Disease management approaches, including resistant variety utilization (Chen et al., 2019; Zuo et al., 2018), cultural control practices (Abdallah et al., 2018), fungicide application (Amini and Sidovich, 2010; Bauer et al., 2016), and biological control by plant growth-promoting bacteria (Castaldi et al., 2021), have been used to control this pathogen. However, the development of resistant tomato varieties is a time-consuming process (Du et al., 2022). Fungicides can pose risks to human health, as well as cause harm to non-target organisms and the environment (Fatima and Anjum, 2017; Vurukonda et al., 2018). Additionally, their use can induce the emergence of fungicide-resistant pathogens (Kanini et al., 2013; Reis et al., 2005). Thus, the use of chemical fungicides in sustainable agriculture should be restricted (Babalola and Glick, 2012; López-Aranda et al., 2016; Wisniewski et al., 2016). Therefore, alternative methods, such as the utilization of antagonistic microorganisms as a biocontrol strategy to control the Fol pathogen, represent a new choice in sustainable agriculture (Fatima and Anjum, 2017). This approach is considered more environmentally friendly (Albayrak, 2019). Among the antagonistic biocontrol agents, bacteria belonging to the Bacillus species have been widely used because of their effectiveness and wide-ranging ability to control pathogens, and their resistance to adverse environmental conditions (Albayrak, 2019).
Bacillus spp. are one of the predominant bacterial genera found in soil (Saxena et al., 2020), and they are well-known for their various advantageous characteristics that directly or indirectly benefit plants. These characteristics include nutrient uptake, siderophore production, nitrogen fixation ability, phytohormone production, and nutrient solubilization (Albayrak, 2019; Vejan et al., 2016), as well as protection from phytopathogens and other abiotic stressors (Fan et al., 2017; Saxena et al., 2020). Bacillus spp. can directly and indirectly control plant pathogens through different mechanisms. Direct mechanisms involve competition for space and nutrients with pathogens, as well as the secretion of extracellular metabolites such as antibiotics, antifungal compounds, lipopeptides, and cell wall degrading compounds (Albayrak, 2019; Lastochkina et al., 2019). Indirect mechanisms include the ability of certain strains to enhance systemic resistance or stress tolerance in their plant hosts by inducing the expression of stress-response genes and stress-related metabolites (Hashem et al., 2019; Radhakrishnan et al., 2017). For example, Elanchezhiyan et al. (2018) reported that the endophytic Bacillus strain FZB24, isolated from different parts of tomato plants, exhibited maximum inhibition of Fol and promoted plant growth in vitro. Furthermore, the genome of the FZB24 strain contained antibiotic biosynthesis genes encoding iturin A, iturin C, surfactin, bacillomycin A, and bacillomycin D.
Therefore, a good understanding of antimicrobial or secondary metabolite profiles from antagonistic potential Bacillus spp. may help develop a suitable strategy to control the fungal pathogen. Currently, whole genome sequencing has been applied and used to study the biosynthetic properties of biosynthesis gene clusters (BGCs) from potential Bacillus isolates (Xia et al., 2022). The analyzed data will help the identification of the biocontrol agent and PGP abilities of Bacillus to reveal their potential for crop protection and environmental sustainability. For example, the phylogenomic analysis through genome mining has been used to identify the 13 BGCs encoding secondary metabolites with biocontrol functions as well as genes/gene clusters involved in plant colonization, plant growth promotion, and induced systemic resistance in the genome of B. velezensis LM2303 (Chen et al., 2018). The results from whole genome sequencing provide a mechanistic understanding of B. velezensis LM2303 for control of F. graminearum pathogen and increase the potential for biocontrol against this pathogen (Chen et al., 2018). Therefore, the objective of this study was to isolate potential strains of Bacillus from soil and investigate their antagonistic activities, including their mechanisms for inhibiting the fungal pathogen Fol. In addition, we aimed to evaluate the broad-spectrum antagonistic activity of the isolated strains against other fungal pathogens, addressing the potential application of these strains as biocontrol agents for various plant diseases. Furthermore, the study aimed to assess the plant growth-promoting properties of the isolated strains. The investigation was carried out through a combination of in vitro and pot experiments. The outstanding isolate was preliminarily identified using the 16S rRNA gene. The whole genome sequencing with genome data mining of the best strain was performed to identify the bacterial species and to find the genes/gene clusters encoding bioactive compound substances and plant growth-promoting traits. The findings may provide valuable data to support the antagonistic activity of the best strain against the Fusarium wilt pathogen and PGP abilities in tomatoes for further use in sustainable agriculture.
Materials and Methods
Isolation of microorganisms
Bacillus isolates
Bacillus spp. were isolated from the soil near three separate extinct volcanoes, Phanom Rung, Plai Bat, and Khao Khok, in Buriram Province, Thailand. The isolation procedure was performed with the modified method described by Boottanun et al. (2017). Briefly, 100 g of each soil sample was collected and air-dried at ambient temperature for 2–3 days. One gram of each soil sample was suspended in 99 ml sterile distilled water and boiled at 100°C for 5 min; then, soil suspensions were diluted 10-fold serially. The diluted soil suspensions were spread on nutrient agar (NA) and incubated at 37°C for 24–48 h. Pure culture of Bacillus was isolated and kept on an NA slant at 4°C.
Plant pathogenic fungi
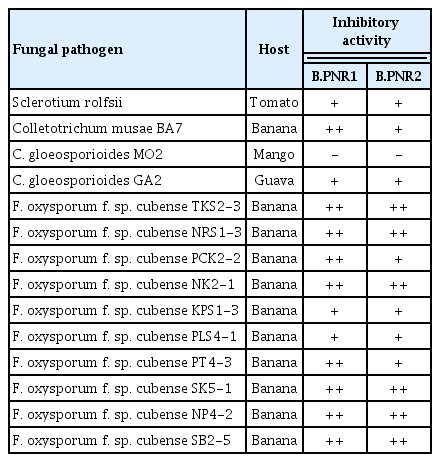
The isolate TFPK401 of Fol race 1, which is a reference isolate for Fol race 1-resistant tomato screening (Kawicha et al., 2023b) and disease control assessments (Kawicha et al., 2023a; Saman et al., 2022) and nine other isolates of Fol were used to evaluate the antagonistic efficacy of the isolated Bacillus. The other fungal pathogens, including 10 isolates of F. oxysporum f. sp. cubense race 1 (Foc), one isolate of S. rolfsii, one isolate of Colletotrichum musae, and two isolates of C. gloeosporioides were used to determine broad spectrum antagonistic activity of the Bacillus spp. The Fol and Foc isolates were provided by the Plant Pest and Biocontrol Research Unit, Department of Agriculture and Resources, Faculty of Natural Resources and Agro-Industry, Kasetsart University Chalermphrakiat Sakon Nakhon Province Campus, Sakon Nakhon. S. rolfsii was provided by the Entomology and Plant Pathology Section, Faculty of Agriculture, Khon Kaen University. The C. musae and two isolates of C. gloeosporioides were isolated from banana, mango, and guava using a tissue transplanting technique. Before being used, all fungal pathogen isolates’ pathogenicity was confirmed in strict conformity to Koch’s postulates. The conidia morphological characteristics and the internal transcribed spacer DNA sequencing were used to confirm and identify the fungal species before being used for further study.
Preliminary screening of antagonistic Bacillus under in vitro condition
Eleven isolates of Bacillus were tested for antifungal activity against 10 isolates of Fol. A 6-mm-diameter agar plug with Fol mycelium was placed in the center of a potato dextrose agar (PDA) plate, and Bacillus isolates were spotted on the plate at two locations, 3 cm from the agar plug. The plates were incubated at room temperature for seven days before mycelial growth was measured. The antagonistic ability was reported as the percentage inhibition of radial growth (PIRG). The PIRG was calculated using the formula:
, where C is the radius of the fungal growth of a pathogen colony (mm) in the control plates, and T is the radius of fungal growth of a pathogen colony interacting with Bacillus (mm) (Devi et al., 2022). The experiment was conducted using a completely randomized design (CRD) with triplicates and performed twice on the same objects under identical conditions. ANOVA was used to evaluate the data, and Tukey’s range test was used to compare means (at P < 0.05). The potential isolates were selected with the criteria of a PIRG of more than 50% and used for further analysis of the broad-spectrum antagonistic ability of Bacillus spp. against 10 isolates of Foc, one isolate of S. rolfsii, one isolate of C. musae, and two isolates of C. gloeosporioides with the same procedure described above.
Evaluation of antifungal activity of Bacillus culture filtrate
Bacillus culture filtrate preparation
Isolates of Bacillus were cultured in 50 ml of nutrient broth (NB) and incubated in an incubator shaker at 150 rpm, 30°C for 48 h. Bacterial cells were collected by centrifugation at 10,000 rpm for 10 min, and then the supernatant was filtered with a 0.22 μm syringe filter. The filtered supernatant was used as the culture filtrate for further experiments.
Effect of Bacillus culture filtrate from selected isolates on hyphal growth and conidial germination on culture medium
The effects of the culture filtrates from selected Bacillus spp. against mycelial growth and conidial germination of 10 isolates of Fol was determined on PDA plates containing 10% sterile culture filtrate. For hyphal growth, a Fol colony (6 mm in diameter) was inoculated onto the center of the PDA plates and incubated at room temperature for seven days. Then, the mycelial growth of Fol was measured compared with the control plate. The percentage of mycelial growth reduction was calculated using the formula described by Li et al. (2015).
For conidial germination, a Fol conidial suspension was adjusted to 106 conidia/ml and was spread onto PDA and PDA plus 10% sterile culture filtrate. The plates were incubated at room temperature for 24 h. After incubation, 100 conidia were investigated under a light microscope. The inhibition rate of conidial germination was calculated as described by Kgosi et al. (2022). The experiment used CRD with triplicates and was performed twice on the same objects under identical conditions.
Effect of Bacillus culture filtrate from selected isolates on fungal hyphal under scanning electron microscope and transmission electron microscopy observation
The mechanism of the culture filtrate of selected Bacillus against Fol TFPK401 were investigated using a scanning electron microscope (SEM) and transmission electron microscopy (TEM). In the experiments, the fungal pathogen Fol isolate TFPK 401 was cultured in potato dextrose broth containing 10% culture filtrate of selected Bacillus. At seven days after inoculation, the hyphae of Fol TFPK401 were collected and washed with 1× phosphate buffer saline (PBS) three times to remove the excess medium.
For the SEM examination, the fungal hyphae were fixed in 2.5% glutaraldehyde at 4°C overnight before being rinsed with 1× PBS three times (5 min each time). Then, the samples were dried using a critical point drier with CO2 after being dehydrated in an ethanol series (25%, 50%, 70%, 95%, and 100%). After that, the samples were cut into small pieces, coated with gold, and examined with an SEM (Model Auriga FESEM, Carl Zeiss, Oberkochen, Germany).
For TEM observation, fungal hyphae were fixed overnight in 2.5% glutaraldehyde at 4°C. After that, the samples were post-fixed in 1% osmium tetroxide at 4°C for 3 h and washed with distilled water three times for 15 min. Then, the samples were dehydrated in a graded series of acetone (20%, 40%, 60%, 80%, 100%, 100%, v/v) with 15 min per change and then embedded in pure Epon 812. The embedded samples were sectioned on an ultramicrotome (MTX, RMC Products, Tucson, MZ, USA) with a diamond knife. Then, the sections were placed on formvar-coated copper grids. These sections were sequentially stained with 2% uranyl acetate and 4% lead citrate for 15 min and washed two times with distilled water before being examined with a TEM (FEI, Model TECNAI G2 20 S-TWIN, FEI Company, Hillsboro, OR, USA).
Evaluation of biocontrol activity under in vivo conditions of Fusarium wilt disease
The biological control ability of selected antagonistic Bacillus was assessed against the Fol isolate TFPK401. Tomato seedlings were grown in sterilized loamy soil for 30 days before the disease control ability of culture filtrates and cell suspensions of the selected Bacillus was examined. Two approaches to Bacillus application were compared between applying Bacillus 48 h before inoculating Fol (approach A) and applying Bacillus 48 h after inoculating Fol (approach B). Then, for both approaches, continue applying Bacillus every week for 4 weeks.
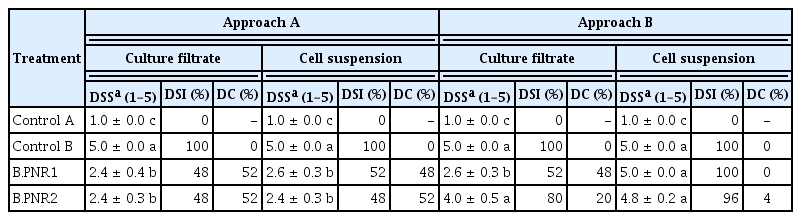
The plants were treated with 10 ml of culture filtrate or cell suspension, 107 cfu/ml, of selected Bacillus by drenching them near the root crown. Fol isolates TFPK401 were inoculated into the plants according to Kawicha et al. (2023a). Briefly, 10 ml of Fol conidia suspension, 106 conidia/ml, were drenched on four sites of wounds on tomato roots created using a cutting blade, 18 mm wide blade, 5 cm below the soil surface, and 5 cm from the tomato stem. At 35 days after Fol inoculation, the disease severity score (DSS) and disease severity index (DSI) were assessed and compared with the positive control (inoculated with only Fol isolate TFPK401) and negative control (mock inoculation with no culture filtrate treatment). The disease was rated on a 1–5 scale according to a modified method described by Marlatt et al. (1996). The DSS was scored using a five-grade severity scale, with (1) symptomless, (2) chlorotic plants, (3) chlorotic plants and wilting, (4) wilting plants, and (5) plant death. DSI was calculated using the formula:
, where Si is the DSS, Nj is the number of tested tomatoes with Si severity score, S is the highest DSS, and Nt is the total number of tested tomatoes. Disease control efficacy (DC) was calculated using a formula modified from Torguet et al. (2022):
, where DSIT is the DSI in a given treatment and DSIUTC is the DSI of untreated control. The experiment was performed using Factorial in CRD with five replicates and performed twice.
Evaluation of antagonistic properties and plant growth-promoting traits of selected Bacillus under in vitro conditions
Antagonistic activities, including cellulolytic and amylase activity of selected Bacillus were tested in vitro. Indole acetic acid (IAA), phosphate solubilization, and siderophore production were also evaluated as the plant growth-promoting activities of selected Bacillus. The isolates were grown on NA medium for two days before being tested. All experiments were performed with three independent replicates. The production of amylase and cellulase was determined by the method of Smibert and Krieg (1994). Siderophore production was assayed on Chrome Azurol S blue agar (Schwyn and Neilands, 1987). IAA was assayed on L-tryptophan NB (Glickmann and Dessaux, 1995). Phosphate solubilization activity was assayed on Pikovskaya agar medium (Pikovskaya, 1948). The experiment used CRD with triplicates and was performed twice on the same objects under identical conditions.
Evaluation of plant growth promotion activity of Bacillus in pot experiment
Tomato seedlings were grown in sterilized loamy soil for 30 days. Afterward, the tomato plants were drenched with 10 ml of the culture filtrate or cell suspension, 107 cfu/ml, of the selected Bacillus isolate at 7, 14, 21, and 28 days. At day 35, the growth parameters, including total length, shoot length, root length, and dry weight of shoot and root, were measured, and compared with the control (mock inoculation and no culture filtrate treatment). The experiment was performed using CRD with five replicates and performed twice.
Preliminary characterization of Bacillus
The selected Bacillus was subjected to further identification according to Bergey’s Manual of Systematic Bacteriology (Vos et al., 2011) and sequence analysis of the 16S rRNA gene. The selected Bacillus was grown in NA medium at 28°C and 37°C for 24 h. The bacterial Gram stain and endospore characteristics were examined under a compound microscope. For molecular characterization, the selected isolates of Bacillus were cultured in NB at 28°C for 12 h. The cells were harvested, and genomic DNA was extracted using a bacterial DNA extraction kit (Vivantis, Selangor, Malaysia). The 16S rRNA gene was amplified using a universal primer set of fD1 and rP2 (Weisburg et al., 1991). Thermal cycle profiles were performed according to Sangdee et al. (2016). Polymerase chain reaction products were subjected to DNA sequencing at Macrogen Inc. Seoul, South Korea, and the partial sequence of 16S rDNA was compared with the sequences in the National Center for Biotechnology Information (NCBI) database. A phylogenetic tree was constructed using MEGA 11: Molecular Evolutionary Genetics Analysis version 11 (Tamura et al., 2021), and the neighbor-joining tree was created with 1,000 bootstrap replicates using the Kimura 2-parameter model (Nishimaki and Sato, 2019).
Whole genome sequencing of Bacillus sp. isolate B.PNR1
Whole genome sequencing
The genomic DNA of Bacillus sp. isolate B.PNR1 was extracted. Then, the whole genome sequencing was performed using an Illumina platform by the next-generation sequencing service from Macrogen (Seoul, Korea). Qualified reads were used for a de novo assembly, and then the assembled genome was validated using the self-mapping strategy and BUSCO analysis. Genome annotation was done using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). The data available from the NCBI database included the Bioproject number (PRJNA875222), Biosample number (SAMN30608759), and accession number (JANYMN000000000).
For genome comparison, reference genome sequences belonging to Bacillus were obtained from the NCBI database and compared using the Type Strain Genome Server (TYGS) (Makuwa and Serepa-Dlamini, 2019). Genome BLAST Distance Phylogeny (GBDP) distances derived from genome sequences were processed by FastME 2.1.6.1 (Lefort et al. 2015) to construct the phylogenetic tree. The branch lengths were scaled in terms of the GBDP distance formula d4.
Digital DNA-DNA hybridization (dDDH) values between Bacillus sp. isolate B.PNR1 and closely related strains, including Bacillus stercoris LJBS06, B. stercoris D7XPN1, B. subtilis KCTC 1028= ATCC6051a, B. subtilis NCIB 3610, B. subtilis subsp. spizizenii TU-B-10, B. inaquosorum KCTC 13429, B. cabrialesii TE3, B. tequilensis EA-CB0015, and B. tequilensis NCTC13306 were calculated using the Genome-to-Genome Distance Calculator (GGDC) (Meier-Kolthoff et al. 2013). ANIBLAST (ANIb), ANIMUMmer (ANIm), and Tetra-nucleotide signature correlation index algorithm analyses were performed using the free software from the JspeciesWS web service.
In silico comparison of genomes and prediction of biosynthetic gene clusters
The antiSMASH tool version 6.1.1 (https://antismash.secondarymetabolites.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) on RAST (https://rast.nmpdr.org/rast.cgi) were used to analyze predicted BGCs, genes related to antibiotics, and genes that promote plant growth in Bacillus sp. isolate B.PNR1 genomes.
Results
Isolation and antagonistic ability of Bacillus under in vitro condition
Eleven isolates of Bacillus were obtained from soil samples collected from Buriram Province, Thailand. The result showed that all isolates had activity to inhibit the mycelial growth of the 10 isolates of Fol by dual culture method. Two isolates, B.PNR1 and B.PNR2, showed the highest percentage of mycelial growth reduction. Bacillus sp. isolate B.PNR1 could inhibit the growth of all tested Fol isolates with an inhibition percentage over 50% followed by isolate B.PNR2. The rest of the Bacillus isolates showed low efficacy in inhibiting Fol mycelial growth (Fig. 1). Bacillus isolates B.PNR1 and B.PNR2 showed broad-spectrum activity against 10 isolates of Foc, one isolate of Sclerotium rolfsii, one isolate of Colletotrichum musae, and one isolate of C. gloeosporioides (Table 1). Therefore, Bacillus isolates B.PNR1 and B.PNR2 were selected to be examined in further experiments.

Efficiency of selected Bacillus isolates against Fusarium oxysporum f. sp. lycopersici (Fol) isolates, causal agent of Fusarium wilt disease in tomato. Different lower-case letters above mean bars indicate significant differences according to Tukey’s range test (P < 0.05).
Effect of culture filtrates from selected Bacillus isolates on hyphal growth and conidial germination
The effect of the culture filtrates of Bacillus isolates B.PNR1 and B.PNR2 on the fungal mycelial growth and conidia germination of 10 isolates of Fol was determined on PDA plates containing 10% sterile culture filtrate. The results showed that these isolates could inhibit fungal mycelial growth and conidia germination with various percentages of mycelial growth reduction and inhibition rates of conidial germination (Table 2). Isolate B.PNR1 showed more activity inhibiting the fungal mycelial growth and reducing conidial germination than isolate B.PNR2 (Table 2). During the SEM and TEM observations, significant alterations were observed in the cell wall structures, and morphology of Fol mycelial cells when exposed to the culture filtrate of Bacillus isolates B.PNR1 and B.PNR2 (Fig. 2). Under SEM, treated mycelial cells exhibited abnormal cell wall structures and rough cell walls, contrasting the normal morphology observed in the control samples (Fig. 2B and C vs. Fig. 2A). Furthermore, TEM analysis revealed that when exposed to the culture filtrate, Fol exhibited noticeable effects on its cell walls and cell membranes. Specifically, the cell walls of Fol were observed to shrink, and the cell membranes showed signs of alteration or damage (Fig. 2E and F). In contrast, the control samples exhibited uniform and regularly shaped cell membranes, cytoplasm, and nucleus, with intact and regular ultrastructure of the mycelial cell wall (Fig. 2D).
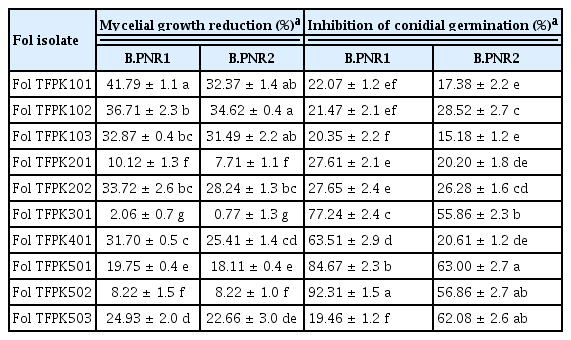
Antagonistic activity of culture filtrates of Bacillus B.PNR1 and B.PNR2 on inhibiting mycelial growth and conidial germination of Fol isolates
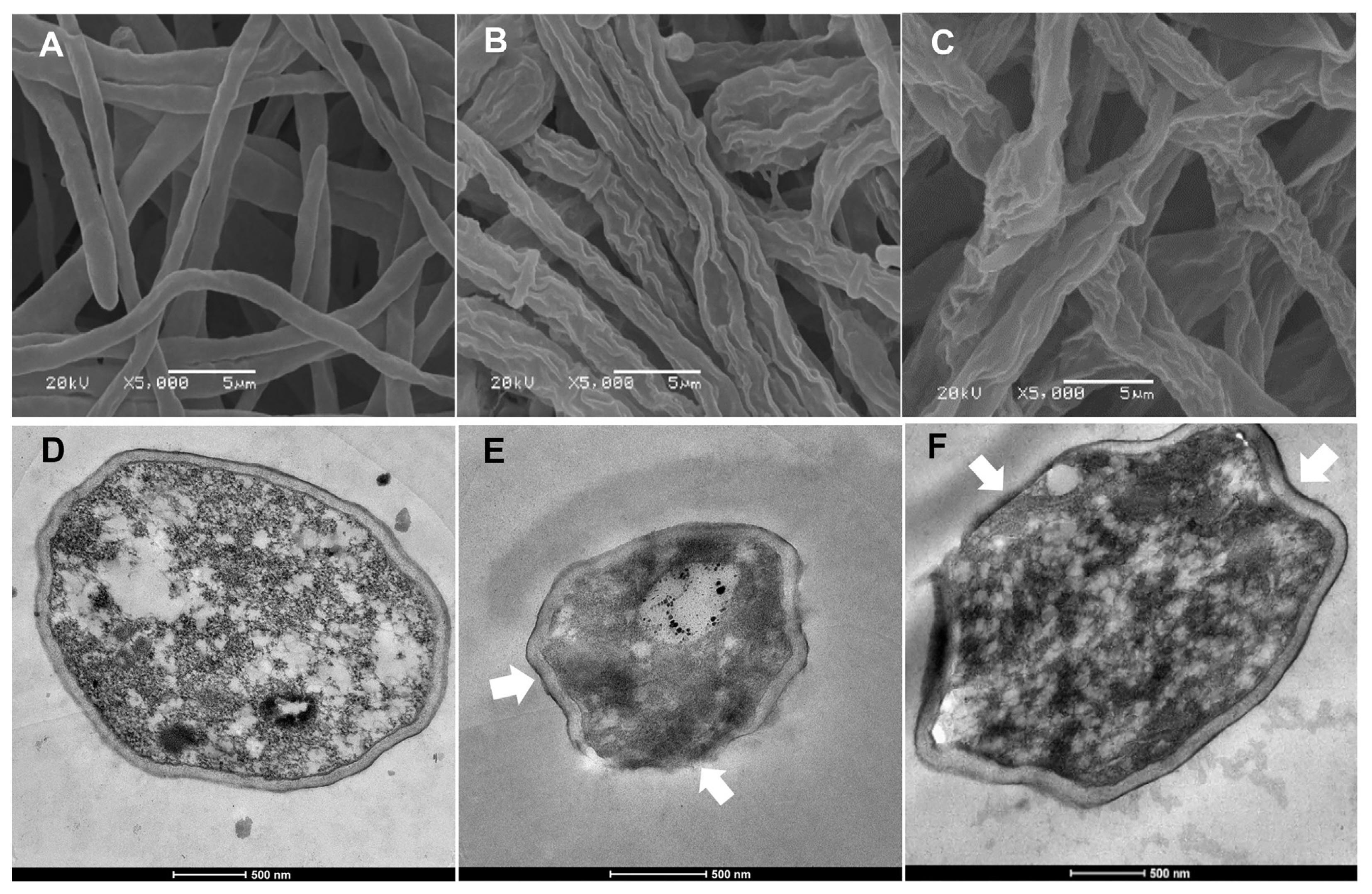
Scanning (A–C) and transmission (D–F) electron micrographs of Fusarium oxysporum f. sp. lycopersici (Fol) isolate TFPK 401 treated with culture filtrates of Bacillus sp. isolates B.PNR1 and B.PNR2. (A, D) Mycelial of Fol TFPK 401 without applying Bacillus culture filtrate (control). (B, E) Mycelial of Fol TFPK 401 treated with culture filtrate of B.PNR1. (C, F) Mycelial of Fol TFPK 401 treated with culture filtrate of B.PNR2. Arrows indicate abnormal cell wall shape and structure.
Evaluation of biocontrol activities of selected Bacillus isolates under in vivo conditions
The culture filtrates and cell suspensions of Bacillus isolates B.PNR1 and B.PNR2 were evaluated for their efficacy in controlling Fusarium wilt disease under pot experiments. The results showed that the application of the culture filtrate and cell suspension of both Bacillus isolates according to approach A, applying biocontrol agents before Fol infection, showed a low DSS and index resulting in higher DC than the control treatment (control B: Fol TFPK 401 inoculation without Bacillus application) as shown in Table 3 and Fig. 3. In approach B, applying the biocontrol agents after Fol infection, only the application of the culture filtrate of Bacillus isolate B.PNR1 could significantly reduce the Fusarium wilt disease. Bacillus isolate B.PNR2 could not reduce the disease according to approach B (Table 3). The results indicated that approach A significantly reduced the Fusarium wilt disease and that isolate B.PNR1 had a more robust antagonistic activity than isolate B.PNR2.

Fusarium wilt symptoms on tomatoes treated with culture filtrates and cell suspensions of Bacillus isolates by approaches A and B. (A) Approach A: apply Bacillus 48 h before inoculating Fusarium oxysporum f. sp. lycopersici (Fol), and then continuously apply Bacillus every week for 4 weeks. (B) Approach B: apply Bacillus 48 h after inoculating Fol, and then continuously apply Bacillus every week for 4 weeks. 1: Fol TFPK 401 inoculation without Bacillus application (control B), 2: Mock inoculation without Bacillus application (control A), 3–4: Fol inoculated plants and treated with Bacillus isolates, 3: B.PNR1, 4: B.PNR2.
Evaluation of antagonistic properties and plant growth promoting traits of selected Bacillus under in vitro conditions
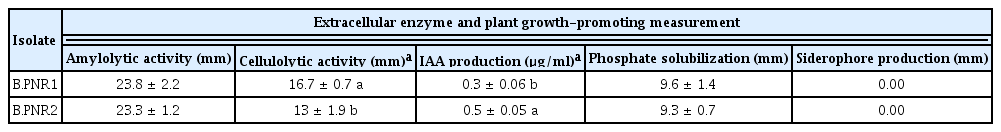
The selected Bacillus isolates B.PNR1 and B.PNR2 were tested for antagonistic properties involving extracellular enzyme production and plant growth promotion traits, such as IAA production, phosphate solubilization, and siderophores production. The results showed that both bacterial isolates could produce extracellular enzymes, including amylolytic and cellulolytic. Bacillus isolate B.PNR1 produced significantly higher cellulolytic activity than isolate B.PNR2. In addition, both isolates had the activity to produce IAA and phosphate solubilization. Bacillus isolate B.PNR2 produced significantly higher IAA than isolate B.PNR1. Siderophore production was not present in either isolate in the tested conditions (Table 4).
Evaluation of plant growth promotion activity of Bacillus in pot experiment
The selected Bacillus isolates B.PNR1 and B.PNR2 were assessed for their potential to promote tomato growth in a pot experiment. The results showed that the selected Bacillus’ culture filtrates and cell suspensions affected the growth parameters. The culture filtrate of Bacillus isolate B.PNR1 showed the highest shoot dry weight and total dry weight compared with the other Bacillus isolate B.PNR2 and the control treatments (Table 5). The cell suspension of the selected Bacillus isolates did not affect the growth parameters evaluated (Table 5). This indicates that the culture filtrates have better activity than the cell suspensions.
Preliminary characterization of Bacillus
The bacterial colonies of the two selected Bacillus isolates, B.PNR1 and B.PNR2, when cultured on NA for 24 h, exhibited a similar colony appearance. When incubated at 28°C, the colonies appeared as opaque white circles with a raised elevation and an even edge. At 37°C, the typical colonies displayed an irregular-wrinkled colony (Fig. 4B and D). Under microscopic observation, the bacterial cells were Gram-positive and displayed a rod-shaped morphology, with dimensions of approximately 0.5–0.7 μm in width and 3.5–4 μm in length. Additionally, these cells could produce central endospores (Fig. 4C and E).

Phylogenetic relationship of Bacillus isolates B.PNR1 (B, C) and B.PNR2 (D, E) based on partial 16S rRNA gene sequence using neighbor-joining algorithm. Bootstrap values (%) based on 1,000 replications were given at nodes. The scale bar indicates the number of bases substitutions per site, computed using the Kimura 2-parameter method (A). The colony morphology of Bacillus sp. isolate B.PNR1 on nutrient agar (NA) (B) and its endospore characteristic (C), and B.PNR2 on NA (D) and its endospore characteristic (E).
The 16S rRNA gene sequences of both strains showed high homology with previous sequences of the member species of Bacillus from the GenBank database. Isolate B.PNR1 (accession no. OP592212) was closely related to B. stercoris D7XPN1 and B. stercoris LJBS06, with 96.27% and 96.23% identity values respectively. Isolate B.PNR2 (accession no. OP592213) was closely related to B. stercoris D7XPN1 and B. stercoris LJBS06, with its identity value at 98.95% and 99.11%, respectively. The phylogenetic tree based on partial sequence of 16S rRNA gene showed that isolates B.PNR1 and B.PNR2 were in the same clade as B. stercoris LJBS06, B. stercoris D7XPN1, B. halotolerans NRRL B-41617, B. tequilensis NCTC13306, B. vallismortis, B. subtilis ATCC6051 and B. amyloliquefaciens GKT04 (Fig. 4). Therefore, these two selected isolates were preliminarily assigned to be Gram-positive bacteria in the genus Bacillus.
Whole-genome sequencing of best-selected isolate of Bacillus.
Whole genome sequence analysis was employed to determine the bacterial species of isolate B.PNR1, which exhibited remarkable antifungal and plant growth-promoting activities. The draft genome of isolate B.PNR1 had a size of 4,161,238 bp, with a G + C content of 43.83%. It consisted of eight contigs with an N50 value of 2,141,500 bases. The genome contained 4,051 protein-coding sequences, 58 tRNAs, and 8 rRNAs (5S, 16S, 23S: 2, 4, 2).
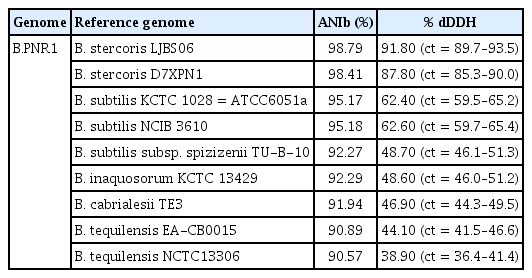
Phylogenetic analysis based on the TYGS database revealed that isolate B.PNR1 formed a distinct cluster with B. stercoris D7XPN1 and B. stercoris LJBS06 (Fig. 5), indicating their close relationship. Moreover, analysis of ANIb and dDDH values demonstrated a high similarity between isolate B.PNR1 and B. stercoris LJBS06 B. with ANIb and dDDH scores of 98.79% and 91.80%, respectively (Table 6).

Phylogenomic tree based on Type Strain Genome Server (TYGS) results showing relationship between Bacillus sp. isolate B.PNR1 and related type strain. Matching square colors indicate same species and subspecies clusters as defined by each parameter indicated at top of squares.
In silico analysis of BGCs prediction
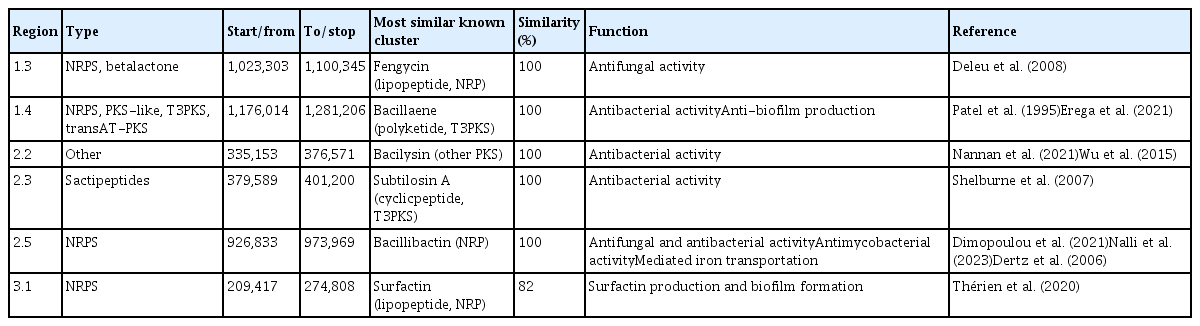
A draft genome of Bacillus sp. isolate B.PNR1 was analyzed for predicting the genes/gene clusters related to BGCs, including antibiotics and plant growth promotion, using the antiSMASH program and KEGG pathway analysis. The results showed that the draft genome contained genes involved in the production of antibiotics, such as the nonribosomal peptide synthetase (NRPS)/polyketide synthase (PKS) cluster. The NRPS clusters for fengycin (100% similarity), surfactin (82% similarity), and polyketide type-3 polyketide synthase clusters for bacillaene (100% similarity), sactipeptide clusters for subtilosin A (100% similarity), and other clusters for bacilysin (100% similarity) were noted from the B.PNR1 genome. In addition, Bacillus isolate B.PNR1 contained a gene encoding for bacillibactin (100% similarity), involved in producing plant growth promoters, as shown in Tables 7 and 8. The core genes and related genes of the gene clusters mentioned above are shown in Fig. 6. In the Fengicin gene cluster, the core biosynthetic genes include fenA, fenB, fenC, fenD, and fenE. The bacillaene gene cluster contains core biosynthetic genes such as baeN, baeM, baeL, baeJ, baeH, baeG, baeE, baeD, baeB, and baeC. The Bacilycin gene cluster includes the core biosynthetic gene bacC, while the subtilosin gene cluster contains the core biosynthetic gene ywiA. The core biosynthetic genes in the bacillibactin gene cluster are dhbA, dhbC, and dhbF. Lastly, the surfactin gene cluster consists of core biosynthetic genes srfAD, srfAC, srfAB, and srfAA. Additional genes can be observed in Fig. 6.

Genes involved in antifungal and plant growth promoting properties of Bacillus sp. isolate B.PNR1 identified by antiSMASH
Discussion
Bacillus spp. are well-characterized and exhibit multiple beneficial properties in plant nutrition and antimicrobial activity against phytopathogens (Fan et al., 2017; Saxena et al., 2020). Among them, B. amyloliquefaciens, B. subtilis, and B. tequilensis have been reported to produce IAA, extracellular enzymes (cellulase, lipase, and protease), and antibiotics (surfactins, iturins, fengycins, macrolactins, and bacillomycin D) that promote plant growth and inhibit the growth of phytopathogens with various mechanisms (Aloo et al., 2019; Shahid et al., 2021). For example, the endophytic B. amyloliquefaciens has been reported to produce some antibiotic substances (iturin A, iturin C, surfactin, bacillomycin A, and bacillomycin D) and extracellular enzymes (protease, cellulase, pectinase, and alpha-amylase) that exhibited control of Fusarium wilt disease in tomato (Elanchezhiyan et al., 2018). Moreover, they also have the activity of siderophore production, ammonia production, and ZnCO3 solubilization that enhances tomato growth (Devi et al., 2022; Elanchezhiyan et al., 2018). Therefore, in this study, 11 Bacillus isolates were isolated from the soil near three separate extinct volcanoes and evaluated for their antagonistic potential against the fungal Fol pathogen, and their plant growth promoting (PGP) potential in tomatoes was also evaluated under in vitro and in vivo conditions. Among them, the selected Bacillus isolates B.PNR1 and B.PNR2 showed robust antifungal activity against the Fol pathogen. Moreover, these isolates showed good antagonistic activity against the other nine isolates of Fol, 10 isolates of Foc, one isolate of S. rolfsii, one isolate of C. musae, and one isolate of C. gloeosporioides. The findings presented in this research align with a study conducted by Wang et al. (2021) that emphasized the biocontrol potential of B. stercoris LJBS06 against a specific pathogen. However, in this current research, the effects of B.PNR1 culture filtrate on the morphology and structure of Fol mycelium were investigated through SEM and TEM studies. The antagonistic activity involves the extracellular hydrolytic enzymes such as amylase and cellulase produced from isolates B.PNR1 and B.PRN2. These enzymes may help to control fungal mycelial growth by degrading the polysaccharide in the fungal cell wall (Haddoudi et al., 2021; Khan et al., 2018; Shahid et al., 2021; Singh et al., 2008; You et al., 2021). Cellulases may play a dual role in antibiosis and parasitism processes (Ghasemi et al., 2020). Extracellular amylase limits the availability of space and nutritional resources for plant pathogens, inhibiting their growth and preventing plant diseases. Additionally, amylase enzymes can promote plant growth by breaking down complex polysaccharides such as starch into simple sugars or glucose, which plants easily absorb (Choubane et al., 2016; Huang et al., 2022). For example, Choub et al. (2021) reported that B. velezensis CE 100 produced the enzymes chitinase, protease, and 1,3-glucanase, which play a key role in deleting the cell wall and inhibiting hyphal growth and spore germination of C. gloeosporioides. Ali et al. (2020) reported that B. siamensis S3 and B. tequilensis S5 exhibited the activity to inhibit the phytopathogen Pestalotiopsis versicolor XJ27.
In addition, SEM and TEM observations confirmed the antifungal activities in this study. The observations indicated that the culture filtrates induced damage to hyphae, resulting in abnormal cell wall structures and ultimately leading to cell death. This was correlated with Sha et al. (2020), who found that the substances from B. pumilus strain S9 and B. amyloliquefaciens strain S170 have effects on the cells of Magnaporthe oryzae P131 by inducing abnormal fungal cell morphology, such as cell wall degrading, cell membrane destruction, and damaging cellular organelles. These findings suggest that the culture filtrate of Bacillus isolates B.PNR1 and B.PNR2 can potentially disrupt the integrity of Fol cell walls and membranes. However, further investigations, such as assessing the leakage of cellular contents, are needed to substantiate the claim regarding the breakdown of cell walls and cell membranes.
In this study, the pot experiment was set up to observe and evaluate the antagonistic activity of the culture filtrates and cell suspensions of isolates B.PNR1 and B.PNR2 against the fungal Fol isolate TFPK401. Based on the disease control efficacy, the application of the culture filtrates and cell suspensions of B.PNR1 and B.PNR2 before the infection of Fol was more effective against Fol isolate TFPK 401 than the application after the Fol infection. Different degrees of disease control in tomato plants may depend on antagonistic isolates, the number of secondary metabolites and/or secretion of cell wall degrading enzymes (Balderas-Ruíz et al., 2021; Shafi et al., 2017; Yoshida et al., 2001). Moreover, in this study, the culture filtrates more effectively control Fusarium wilt disease than the cell suspensions because the culture filtrates contained active substances that could directly fight the fungal pathogen. Conversely, cell suspensions required several weeks to colonize and survive in the root system (Weller, 1988).
The evaluation of PGP activities is an important criterion when selecting a promising biocontrol agent (Deketelaere et al., 2017). Biocontrol agents with PGP activities enhance efficacy and support plant growth while suppressing pathogens. IAA production and phosphate solubilization in the treatments may explain the difference in tomato plant growth (Kang et al., 2015; Zhao et al., 2020). IAA improves plant growth by promoting seed germination, root elongation, and root dry weight (El-Tarabily et al., 2009; Khamna et al., 2010), while inorganic phosphate solubilization activity enhances plant growth (Hamdali et al., 2008). Bacillus isolate B.PNR1 exhibited superior antagonistic and PGP activities compared to other isolates, as demonstrated by its culture filtrate and cell suspensions promoting specific growth parameters. Bacillus isolate B.PNR1 was identified by 16S RNA gene sequencing and whole genome analysis. The analysis confirmed its high homology with Bacillus stercoris LJBS06 (Wang et al., 2021). The classification of isolate B.PNR1 as Bacillus stercoris was supported by the dDDH and ANI values exceeding the species level cut-offs of 70% for dDDH and 95–96% for ANI (Chun et al., 2018; Meier-Kolthof et al., 2013; Richter and Rosselló-Móra, 2009).
In this study, the BGCs involving antibiotic and plant growth-promoting properties in the genome of B. stercoris isolate B.PNR1 were predicted using the antiSMASH tool. The B. stercoris isolate B.PNR1 genome contains a variety of BGCs related to antibiotics, such as the NRPS/PKS cluster, which was related to the BGCs produced from B. stercoris LJBS06 as reported by Wang et al. (2021). It has been reported that the NRPS/PKS cluster produced by Bacillus plays an essential role in plant growth-promoting and antimicrobial activities against bacterial and fungal pathogens (Ongena and Jacques, 2008; Raaijmakers et al., 2010). Wu et al. (2018) reported that Bacillus sp. GFP-2 isolated from Chiloscyllium plagiosum (white-spotted bamboo shark) can inhibit Gram-positive and Gram-negative bacteria by inducing clusters for secondary metabolites, such as difficidin, bacillibactin, bacilysin, surfactin, butirosin, macrolactin, bacillaene, fengycin, lanthipeptide, and bacteriocins. Andrić et al. (2020) reported that Bacillus spp. have the activity to produce cyclic lipopeptides or polyketides, which play essential roles in responses against fungal pathogens. Xu et al. (2022) reported that B. subtilis YB-15 from wheat rhizosphere soil significantly inhibited Fusarium crown rot caused by Fusarium pseudograminearum by producing a variety of antimicrobial compounds (bacillaene, fengycin, bacillibactin, surfactin, subtilosinA, bacilysin, and paenibacterin), hydrolytic enzymes (β-1,3-glucanase, amylase, protease, and cellulase), and plant growth promoting traits. Chen et al. (2018) reported that the B. velezensis strain LM2303 genome contained genes encoding antifungal metabolites (fengycin B, iturin A, and surfactin A) and exhibited a directly antagonistic effect against F. graminearum. These antifungal metabolites are recognized as potential antimicrobial agents against many phytopathogenic fungi by penetrating cell membranes and forming ion pore channels, which results in membrane osmotic imbalance and even cell death (Ongena and Jacques, 2008).
Moreover, a draft genome of B. stercoris isolate B.PNR1 contained gene clusters for bacillibactin siderophore production and iron acquisition that may support the plant growth promoting properties. Surfactin molecules play an essential role in enhancing biocontrol agent colonization in plant roots and inducing systemic acquired resistance in plants (Bais et al., 2004; García-Gutiérrez et al., 2013; Ongena et al., 2007). Based on the results of this experiment and literature reviews, the plant growth-promoting activity of B. stercoris isolate B.PNR1 in tomato plants may be supported. In addition, this evidence is also in agreement with the opinion of Olanrewaju et al. (2021), who reported that the genomes of B. subtilis A1, B. velezensis A3, and B. subtilis A29 contained genes involving plant growth-promoting activities, such as growth hormone production, volatile compound production, siderophore production, and nitrogen, phosphorous, and sulfur metabolism. Haddoudi et al. (2021) reported that B. cereus, B. mojavensis, B. velezensis, B. subtilis, and B. amyloliquefaciens contained gene clusters related to plant growth-promoting and antifungal activities.
In conclusion, this study provides strong scientific evidence for the biocontrol effects of B. stercoris strain B.PNR1 against Fusarium wilt in tomatoes. B.PNR1 exhibits activities such as producing hydrolytic enzymes, antimicrobial compounds, and the potential for disease control under greenhouse conditions. Additionally, it shows capabilities in IAA production and phosphate solubilization, contributing to enhanced plant growth. These observations provide scientific evidence of the disruptive effects of B.PNR1 on the pathogen. Additionally, we assessed the efficacy of B.PNR1 in controlling Fusarium wilt and promoting growth in tomatoes under greenhouse conditions, which adds practical relevance to our findings. The combined evidence from SEM, TEM, and greenhouse experiments proves the potential of B.PNR1 as a biocontrol agent. Genome sequence analysis reveals candidate genes involved in biocontrol and plant growth promotion. Future research should focus on validating the functionality of these genes, including those associated with plant colonization and plant-Bacillus interactions. This knowledge can be applied to develop targeted strategies for sustainable agriculture and effective management of Fusarium wilt.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This research project was financially supported by Mahasarakham University. The authors acknowledge the Faculty of Science, Mahasarakham University, and the Faculty of Natural Resources and Agro-Industry, Kasetsart University Chalermphrakiat Sakon Nakhon Province Campus, for facilitating this project. Rattana Pengproh acknowledges the Ministry of Higher Education, Science, Research, and Innovation for the Ph.D. scholarship.