Physiology and Gene Expression Analysis of Tomato (Solanum lycopersicum L.) Exposed to Combined-Virus and Drought Stresses
Article information
Abstract
Crop productivity can be obstructed by various biotic and abiotic stresses and thus these stresses are a threat to universal food security. The information on the use of viruses providing efficacy to plants facing growth challenges owing to stress is lacking. The role of induction of pathogen-related genes by microbes is also colossal in drought-endurance acquisition. Studies put forward the importance of viruses as sustainable means for defending plants against dual stress. A fundamental part of research focuses on a positive interplay between viruses and plants. Notably, the tomato yellow leaf curl virus (TYLCV) and tomato chlorosis virus (ToCV) possess the capacity to safeguard tomato host plants against severe drought conditions. This study aims to explore the combined effects of TYLCV, ToCV, and drought stress on two tomato cultivars, Money Maker (MK, UK) and Shalala (SH, Azerbaijan). The expression of pathogen-related four cellulose synthase gene families (CesA/Csl) which have been implicated in drought and virus resistance based on gene expression analysis, was assessed using the quantitative real-time polymerase chain reaction method. The molecular tests revealed significant upregulation of Ces-A2, Csl-D3,2, and Csl-D3,1 genes in TYLCV and ToCV-infected tomato plants. CesA/Csl genes, responsible for biosynthesis within the MK and SH tomato cultivars, play a role in defending against TYLCV and ToCV. Additionally, physiological parameters such as “relative water content,” “specific leaf weight,” “leaf area,” and “dry biomass” were measured in dual-stressed tomatoes. Using these features, it might be possible to cultivate TYLCV-resistant plants during seasons characterized by water scarcity.
Tomato (Solanum lycopersicum L.) is a highly economically valuable crop worldwide, with a production of 180.8 million tons in 2019 (Food and Agriculture Organization of the United Nations, 2020). Viral diseases are significant inhibitors of tomato production, causing yield losses (Hančinský et al., 2020; Ong et al., 2020). A comprehensive analysis of databases and literature revealed at least 312 viruses, satellite viruses, or viroid species associated with tomatoes, representing 22 families and 39 genera, which is a record number for any plant species. The financial losses caused by viral infections in tomato crops are challenging to estimate, but yield losses range from approximately 30 to 50 billion US dollars per year (Rivarez et al., 2021). Biotic and abiotic stress factors have a detrimental impact on global agricultural production. DNA and RNA viruses that infect tomato plants are particularly significant biotic stress factors affecting productivity. Whitefly-transmitted virus diseases, including tomato yellow leaf curl virus (TYLCV), are the main pathogens causing infections in tomato plants (Fiallo-Olivé and Navas-Castillo, 2019). TYLCV is a single-stranded DNA virus belonging to the Geminiviridae family in the Begomovirus genus, known for its negative impact on tomato plant production in tropical and subtropical regions (Czosnek, 2021; Desbiez et al., 2019; Verdin et al., 2018). TYLCV has a circular 2800 nucleotide single-stranded DNA genome encapsulated in a structure measuring 25 × 30 nm. Tomato chlorosis virus (ToCV) is another pathogen affecting tomato plants, belonging to the Closteriviridae genus of the Crinivirus species. It is a single-stranded RNA genome virus causing characteristic symptoms of interveinal chlorosis in the lower leaves and chlorotic spots in the apical part of the plant (Orfanidou et al., 2016). ToCV is a phloem-limited virus with a positive-sense single-stranded bipartite RNA genome. Insect vectors, particularly Bemisia tabaci, play a crucial role in transmitting both ToCV and TYLCV viruses (Macedo et al., 2019; Wei et al., 2019).
Drought, exacerbated by global warming, poses a significant threat to many countries (Fullana-Pericàs et al., 2018; Lesk et al., 2016). Rational utilization of natural water resources is becoming a global challenge (Yan and Wang, 2013). Tomato production is severely affected by abiotic stresses, leading to yield losses of approximately 70% depending on the severity and duration of the stress. Drought is recognized as the most detrimental abiotic stress, rendering cultivated tomato plants lacking novel genes for drought stress tolerance highly susceptible (Krishna et al., 2022). Prolonged drought also impacts plant-pathogen interactions, resulting in further reductions in productivity due to the combined effect of biotic (TYLCV, ToCV) and drought stresses (Dai et al., 2017). Developing resistant plant varieties against pathogenic and abiotic stress factors is economically significant.
Recent studies have shown that virus-infected tomato plants exhibit increased tolerance to drought by reducing metabolic activity in leaves and activating defense mechanisms in roots to cope with water scarcity (Mishra et al., 2022). TYLCV and ToCV have the capacity to prevent wilting and crop loss in tomato plants under extreme drought conditions. The virus-resistant tomato plants can be preserved during water scarcity, preventing crop loss during seasons characterized by water deficit, which is of great economic importance.
Drought stress has a more pronounced impact on qualitative and quantitative indicators in plants compared to other abiotic factors (Rukundo et al., 2014). Severe modifications occur in the morphological, physiological, and biochemical processes of plants when water metabolism is impaired due to drought stress (Torres-Ruiz et al., 2015). Understanding the mechanisms of plant responses to biotic and abiotic stresses can help develop strategies to optimize crop productivity in severe environments by studying the expression profiling of genes, proteins, and metabolites (Torres-Ruiz et al., 2015). Drought restricts plant growth by reducing the intensity of photosynthesis in affected plants (Liang et al., 2018). Stomatal closure due to decreased CO2 levels, decreased photosynthetic activity in mesophyll tissue (nonstomatal), or both can contribute to the reduction in photosynthesis intensity (Liang et al., 2018). Osmotic substance accumulation, changes in membrane lipid balance, induction of reactive oxygen species synthesis, increased protein synthesis, and alterations in hormonal regulatory pathways play important roles in the development of drought-resistant plants (Liang et al., 2020). Metabolite profiling studies have shown that virus infection enhances plant tolerance to abiotic stresses by inducing the synthesis of osmoprotectants and antioxidants. Virus infection can also increase the level of salicylic acid in an abscisic acid-independent manner, further contributing to drought tolerance (Aguilar et al., 2017).
The cellulose synthase gene family, which includes the cellulose synthase (CesA) and cellulose synthase-like (Csl) gene families, plays a vital role in cellulose and hemicellulose biosynthesis in plants. These genes are also involved in cell wall biosynthesis, cell elongation, and plant growth (Choe et al., 2021). Some genes associated with the cellulose biosynthesis pathway have been found to show increased resistance to TYLCV infection (Seo et al., 2018). It has been hypothesized that the symptoms caused by TYLCV, which disrupt plant growth, may be directly related to the suppression of cellulose synthesis genes. CesA/Csl genes have also been associated with drought stress, with CesA genes showing increased gene expression under drought-stress conditions (Wang et al., 2023). The expression of CesA/Csl genes may vary during different developmental stages of plants under drought stress (Zhang et al., 2019). However, CesA/Csl genes have not been extensively studied in Solanaceae plants, particularly tomatoes (Song et al., 2019). Therefore, investigating the roles of these genes in response to virus and drought stress in Solanaceae plants, especially in susceptible and/or resistant cultivars, is of great importance for understanding their functions.
The study of host plant-virus relations has been extensively explored, with significant research addressing this topic. However, there exists a notable gap in our understanding of the combined effect of abiotic and biotic stress factors on plants and the resulting outcomes, as well as the economic implications. Dastogeer et al. (2020) and Poudel et al. (2021) have highlighted this limitation in their research. To address this gap, our research focused on investigating the complex interaction between viruses and drought stress on tomato plants. As a result, we uncovered new priorities for enhancing plant tolerance to both viral infections and severe water reduction. This knowledge can contribute to the development of innovative concepts and strategies vital for combatting the global problem of food insecurity, which is exacerbated by climate change. By utilizing the findings from our research, we can propose the use of virus-resistant cultivars that are infected with the virus during the early stages of development. This approach minimizes yield loss when drought stress is subsequently applied. This novel strategy can have a significant impact on mitigating the negative consequences of both viral infections and water scarcity on crop productivity. The objective of this research was to conduct a physiological (“relative water content,” “specific leaf weight,” “leaf area,” and “dry biomass”) and comparative analysis of the expression profiles of the cellulose synthase gene superfamily in highly productive but susceptible cultivars under biotic and abiotic stress factors. In this study, we utilized the Money Maker (MK) cultivar as a susceptible genotype (Milc et al., 2019; Siniga Geroge et al., 2013) to drought and virus, and the Shalala (SH) cultivar, recently grown in Azerbaijan fields, with unknown sensitivity/resistance to virus and drought. We analyzed four CesA and Csl genes using the quantitative real-time polymerase chain reaction (qRT-PCR) method. qRT-PCR analyses were performed at 1 and 25 time points in both virus-infected and combined virus/drought-stressed MK and SH cultivars. In addition to qRT-PCR studies, we measured four important physiological parameters, including relative water content, specific leaf weight, leaf area, and dry biomass, to assess the impact of virus and drought stress on tomato cultivars. The results obtained from this study are expected to contribute to the development of drought- and virus-resistant crop systems and provide insights into the response and adaptation mechanisms of tomato plants to drought stress.
In conclusion, while host plant-virus relationships have been extensively studied, the combined effects of abiotic and biotic stress factors, along with their economic implications, have received limited attention. Our research tackles this knowledge gap by examining the intricate interplay between viral infections and drought stress on tomato plants. The resulting insights offer new avenues to improve plant tolerance and contribute to addressing global food insecurity amidst the challenges posed by climate change.
Materials and Methods
Plant growth and single-leaflet grafting of TYLCV and ToCV
To investigate the response of tomato plants to the combined effects of virus and drought stresses, we used susceptible Solanum lycopersicum L. cv. Money Maker (MK, UK) and Shalala (SH, Azerbaijan) cultivars. The Money Maker seeds were obtained from the Institute of Biotechnology at Ankara University, while the Shalala seeds were obtained from the seed bank of the Research Institute of Crop Husbandry of the Ministry of Agriculture of the Azerbaijan Republic.
The tomato seeds were germinated in plastic vials containing a mixture of peat, perlite, and vermiculite. The tomato (Solanum lycopersicum) cultivars were grown in an insect-free growth chamber with a temperature of 26/20°C (day/night) and a 16-h light/8-h dark cycle, maintaining a relative humidity of 60–70% (Çevik et al., 2019). At the 3–4 true leaf stage, the tomato seedlings (Money Maker and Shalala MK and SH) were used as recipient plants for the single-leaflet grafting method following the procedure described by Lee et al. (2017). ToCV- and TYLCV-infected tomato plants were used as sources for grafting inoculation. Leaf samples for grafting were collected from greenhouses located in the Kumluca region of Antalya, Turkey, in June (Fig. 1). The identification of the viruses in source plants was confirmed using duplex PCR (for TYLCV) and nested PCR (for ToCV) with specific primer pairs (for TYLCV, 5′-CCG GTG TTG TGC GTT GTG TTA G-3′ and 5′-TGA AGG AGC AGT GTY TGY TG-3′; For ToCV, 5′-GG(G/T)TT(A/G)GA(G/T)TT(C/T)GGTACTAC-3′, 5′-CC(G/T) CCACCAAA(A/G)TCGTA-3′, and 5′-GG TTTGGATTTTGGTACTACTAGT-3′, 5′-AAACTGCCTGCATAAAGTCT C-3′) before grafting inoculation. For grafting, a small incision was made on the stem of the recipient seedling by removing the first leaf at the node (Fig. 2). The leaf axil of the identified diseased plant samples was used as inoculum for grafting. The inoculum was applied to the recipient plant by making a cut on the recipient plant with a lancet. Grafting clips were used to ensure successful grafting. The newly grafted tomato seedlings were kept in a shaded, air-conditioned, insect-free greenhouse with temperatures of 26°C during the day and 20°C at night. The grafted leaflets were misted with water multiple times a day to prevent wilting. The grafted plants were maintained under these conditions for four weeks after grafting. Grafting leaflets collected from virus-free healthy tomato plants were included as a negative control (mock). Four independent sets of the grafting inoculation experiment were conducted, and after four weeks, the grafted tissues were examined (Fig. 3). Four weeks after grafting, nucleic acids were extracted from the upper systemic leaves of the successfully grafted recipient tomato plants. These extracts were subjected to duplex polymerase chain reaction (PCR) and nested PCR using the specific primer pairs mentioned above to detect TYLCV and ToCV, respectively. Success rate of grafting was 98.5% for TYLCV and 85.3% for ToCV after 28 dpi (Çevik et al., 2019; Pathirana and McKenzie, 2005). The experiment was performed with three biological and technical replicates.

Symptomatic tomato samples brought from Antlia (Kumluca) for grafting: ToCV-infected (A) and TYLCV-infected (B) tomato samples. ToCV, tomato chlorosis virus; TYLCV, tomato yellow leaf curl virus.
Drought treatment
Tomato seedlings (MK and SH) were transplanted into 2-liter pots filled with standard soil three weeks after sowing. They were initially irrigated with tap water for one week and then water supply was ceased. No additional minerals or fertilizers were applied throughout the experiment. Growth chamber with a temperature range of 20°C (dark period) and 26°C (daylight) were maintained for both uninfected and virus-infected tomato plants.
Subsequently, the plants were divided into four groups: healthy (mock-inoculated) and watered (HW), healthy and exposed to drought (HD), virus-infected and watered (VW), and virus-infected and exposed to drought (VD). These four groups of plants were further categorized into two cultivars (MK and SH) and were infected with TYLCV and ToCV, representing two different virus isolates. The control group consisted of mock-inoculated plants grafted with leaflet samples obtained from healthy tomato plants of the MK and SH cultivars.
VW and VD plants were infected with TYLCV and ToCV, respectively, using the grafting method. At 4 weeks post-infection (wpi) (Çevik et al., 2019), the VD tomatoes were subjected to drought stress along with the HD group. To ensure long-term growth under water deficit conditions and prevent early plant mortality, 50 ml of water was provided to each pot at 10, 17, and 24 days of drought application (25 d) (Mishra et al., 2022). Generally, the drought-applied plants were watered on the 10th, 17th, and 24th days of the experiment to maintain long-term stress conditions. The experiments were repeated three times, each consisting of four groups of plants containing 15 seedlings.
TYLCV and ToCV grafting confirmation with Duplex and nested PCR
Duplex PCR
For the confirmation of TYLCV, the AV632, AC950, and AC1048 primers were utilized to amplify the coat protein (CP) gene of the viruses, following the protocols described by Brown et al. (1996) and Martinéz-Culebras et al. (2001).
Total DNA extraction from 50 mg of fresh leaf tissue was performed using CTAB (cetyltrimethylammonium bromide) solution, following the protocol described by Aboul-Maaty and Oraby (2019). The resulting DNA pellet was resuspended in Dnase-Purity solution. The concentration and purity of the extracted DNA were determined using a NanoDrop1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA).
For the duplex PCR reaction, a total volume of 25 μl was prepared. The reaction mix contained a final concentration of 1× Taq buffer, 2 mM MgCl2, 0.15 mM dNTPs, 0.2 μM of each primer (AV632, AV950, and AC1048), 1.24 U of Taq DNA polymerase, and 100 ng of DNA template (Louro et al., 2000). The duplex PCR reaction was carried out under the following conditions: initial denaturation at 95°C for 1 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 65°C for 1 min, and elongation at 72°C for 1 min. A final elongation step was performed at 72°C for 10 min. Details of the PCR reaction conditions can be found in Table 1.
Nested PCR
Total RNA extraction was performed using the Tri Reagent solution (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s recommendations. Approximately 30–50 mg of fresh leaf tissue was used for RNA extraction. The resulting RNA pellet was resuspended in Rnase-Purity solution, and the concentration and purity of the extracted RNA samples were determined using a NanoDrop1000 spectrophotometer (Thermo Scientific).
To confirm ToCV in grafted tomatoes, a two-step PCR approach was employed, consisting of reverse transcription polymerase chain reaction (RT-PCR) and nested PCR. The first step involved RT-PCR using the BC-36 and BC-37 primers, as well as a primer specific to the highly conserved heat shock protein (HSP 70) region, designed according to the method outlined by Dovas et al. (2002). For the nested PCR, four different specific primer pairs (BC-36 and BC-37) designed by Dovas et al. (2002) were used. Information of the primers can be found in Table 1.
For RT-PCR, 5 μl of RNA samples were mixed with 1 μl of primer and 6 μl of water, and the mixture was incubated at 65°C for 5 min. The samples were then denatured in a PCR machine and kept on ice for 5 min. In a separate tube, 4 μl of 5× reaction buffer (250 mM Tris-HCl pH 8.3, 250 mM KCl, 20 mM MgCl2, 50 mM DTT), 1 μl of RNase inhibitor, 2 μl of 10 mM dNTPs, and 1 μl of reverse transcriptase were added. The reaction mixture was then combined with the denatured RNA samples. The RT-PCR reaction was performed at 25°C for 5 min, followed by 42°C for 60 min, and a final step at 70°C for 5 min. The cDNA obtained from the RT-PCR reaction was used as a template for PCR amplification of the approximately 587 bp region using specific primers targeting the conserved regions of the ToCV HSP70 gene, and Taq DNA polymerase enzyme was used for the PCR reaction.
For the PCR reaction, a mixture was prepared consisting of 2.5 μl of 10× PCR buffer solution (100 mM Tris-HCl pH 8.8, 500 mM KCl), 1.5 μl of 2.5 mM MgCl2, 0.5 μl of 10 mM dNTPs, 4 μl of 20 pmol primer BC36F, 2 μl of 20 pmol primer BC37R, 0.25 μl of Taq DNA Polymerase, and 11.75 μl of water. To this mixture, 22.5 μl of the PCR mixture and 2.5 μl of the DNA template were added to the PCR tube. The PCR reaction conditions included an initial denaturation step at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 43°C for 30 s, and elongation at 72°C for 1 min. A final extension step was performed at 72°C for 5 min. For the nested PCR, a second PCR reaction was performed using specific primers (0.5 μl of 20 pmol primer BC40F and 0.5 μl of 20 pmol primer BC41R) and the same PCR conditions as the first PCR. The amplification products were separated by electrophoresis on a 1% agarose gel, stained with ethidium bromide, and visualized under ultraviolet light using a Gel Documentation imaging device (Uvitek, UK).
The intensity of viral presence within a plant can exhibit variations based on factors such as the plant’s developmental stage, climatic conditions, or the stage of infection. Consequently, in situations where virus symptoms or intensity are minimal, the utilization of nested RT-PCR, a highly sensitive method, is recommended (Dovas et al., 2002). Although the CP gene is not commonly employed for virus identification, the nested RT-PCR technique targeting the HSP70 gene, which is specific to this virus family and possesses a highly conserved sequence, successfully identified the virus. Previous studies (Dovas et al., 2002; Moriones and Navas-Castillo, 2000; Papayiannis et al., 2011) have effectively employed RT-PCR methodologies targeting the HSP70 gene. The present study demonstrates that the HSP70 gene can serve as an alternative to the CP gene for diagnosing ToCV.
For the nested PCR procedure, a mixture was prepared by combining 2.5 μl of 10× PCR buffer solution (comprising 100 mM Tris-HCl [pH 8.8], 500 mM KCl), 1.5 μl of 2.5 mM MgCl2, 0.5 μl of 10 mM dNTPs, 0.5 μl of a mixture containing 20 pmol of primer BC40F and 20 pmol of primer BC41R, and 0.25 μl of Taq DNA polymerase (0.5 U), with a final volume of 18.25 μl of water. Each PCR tube received 24 μl of the prepared mixture and 1 μl of DNA. The PCR amplification protocol commenced with an initial denaturation step at 95°C for 3 min. This was followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. Finally, a 5 min extension step was performed at 72°C. After the amplification, the PCR product corresponding to a 463-base pair (bp) segment of the ToCV HSP70 gene was maintained at 4°C. Subsequently, the PCR products and a 100-bp DNA ladder were separated via electrophoresis on a 1% agarose gel and visualized under ultraviolet light using a Gel Documentation (Uvitek) imaging device after staining with ethidium bromide.
During the RT-PCR testing, symptomatic tomato samples infected with ToCV isolate from the Kumluca region of Antalya were employed as positive controls.
Primer design and quantitative (qRT-PCR) expression analysis
In the primer design, Solanum lycopersicum cellulose synthase-like protein genes (Gene ID, gene number) were identified with the help of National Center for Biotechnology Information (NCBI) and then NCBI Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer). In the primer validation, pre-optimization reactions were performed at different annealing temperatures and PCR conditions, taking into account the primers, forward and reverse annealing temperatures, using the control cDNA, and the amplifications of the primers were checked. Then, standard curves were generated for each gene/primer to show that each primer was valid. The Roche LightCycler 480 instrument (Roche, Mannheim, Germany) was employed for conducting qRT-PCR experiments (Table 2). A standard curve was generated using six serial dilutions (ranging from 1/10 to 1/100,000) of control cDNA (MCD or SCD) for each primer. For each primer, the standard close were prepared (efficiency and slope values were closed to 2.2 and -3.2, respectively). cDNAs obtained in the nested PCR step were used in qRT-PCR reactions.
The qRT-PCR conditions involved an initial pre-incubation of 2 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 56-57-50-52-58°C for 1 min, and extension at 72°C for 1 min. Prior to loading onto the real-time PCR instrument plates, the cDNAs of the samples were covered with Roche LightCycler 480 Sealing Foil. The samples were prepared using Promega GoTaq qPCR Master Mix (Madison, WI, USA) according to the optimized conditions specific to each primer. Three independent biological replicates were used in each experiment.
The optimization of Csl-H1, Ces-A2, Csl-D3,2, Csl-D3,1, and Solyc11 primers was performed using qRT-PCR. A 12 μl reaction mixture was prepared, comprising 3 μl of cDNA (500 ng/μl), 1.8–2 μl of forward and reverse primers (10 pmol), and 5 μl of LightCycler 480 SYBR Green I Master (Roche). The amplification program involved an initial polymerase activation step at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 10 s, annealing at Tm for 10 s, and extension at 72°C for 5 s.
To verify the specificity of the qRT-PCR amplification, melt curve analysis was performed by gradually increasing the temperature from 95°C to Tm following the final cycle. Melting curve analysis confirmed the presence of single peaks with no dimers. Optimal temperatures were selected on the Roche LightCycler-LC 480 instrument, and the amplification reactions were carried out according to the specified PCR program conditions.
Relative gene expression levels were determined by normalizing the expression of Csl-H1, Ces-A2, Csl-D3,2, Csl-D3,1, and Solyc11 genes with the housekeeping gene Solanum lycopersicum actin-7-like gene (ID: LOC101262163) (Klay et al., 2014). As for technical replicates, we used 3 for each biological sample. The abbreviations used in the qRT-PCR analyses are provided in Table 3.
Statistical analysis of quantitative (qRT-PCR) expression analysis
The experimental setup was designed according to a completely randomized design as three replicates each of which contains 4–5 leaflets. According to the Ct (cycle threshold) values, the relative expression levels were calculated by using the 2−ΔΔCT (the delta-delta-Ct or ddCt) algorithm (Livak and Schmittgen, 2001). Reaction efficiency has been considered as 1 (one). The expression of Csl-H1, Ces-A2, Csl-D3,2, Csl-D3,1 genes was normalized according to the Actin-7-like gene housekeeping gene.
Physiological measurements
At the conclusion of the experiment, several physiological parameters were assessed as follows. The physiological parameters were analyzed in four groups: TYLCV and ToCV grafted (virus healthy, VH; virus drought, VD), as well as mock-inoculated (healthy watered, HW) and healthy drought (HD) groups, in each of the two cultivars (MK, SH). All experiments were conducted with three biological replicates.
The relative water content (RWC) of virus-infected and healthy tomato samples was determined using the method described by Tambussi et al. (2005). To measure the dry biomass of tomato leaves, sections of the same size were prepared from both diseased and healthy leaf samples, and their mass was determined using an electronic scale, following the protocol outlined by Grünzweig et al. (1999). Leaf area measurements were taken for infected and healthy plant samples. The length from the part of the leaf attached to the stem to the tip, as well as the diameter of the widest part, were measured using a ruler. The leaf area was calculated geometrically, in accordance with the method described by Grünzweig et al. (1999). To assess water deficit in the leaves, the initial weight of the leaves was recorded, followed by soaking the leaves in water for 60 min, and finally, the wet weight was measured, as outlined by Shackel (1991). Three independent biological replicates were used in each experiment. The physological parameters were determined using three technical replicates.
Statistical analysis of physiological analysis
The significance of differences between the control and experimental groups was evaluated using two-way analysis of variance (ANOVA). Differences in means were considered statistically significant when the P-value was less than 0.05. For each treatment, three different samples were collected and analyzed twice.
Results
Drought treatment
From the onset of drought symptoms, non-inoculated plants exhibited either wilted shoot tips or complete collapse. In order to investigate the impact of virus infection on plant growth and the development of drought symptoms, we compared virus-infected and mock-inoculated tomato cultivars under prolonged drought conditions. The growth rate of virus-susceptible tomatoes was significantly reduced as a result of viral infection. After 15 to 25 days of growth, plants in the virus-infected (VW) group exhibited smaller sizes compared to the healthy (HW) group. It is important to note that the growth of drought-exposed tomatoes in the healthy (HD) group was also restricted. Drought symptoms became apparent in tomatoes of the virus-infected (VD) and healthy (HD) groups after 10 days of drought treatment. However, by day 25, the tomatoes in the HD group had completely wilted and collapsed, unlike the VD group where only clear wilting was observed. Remarkably, the growth inhibition in the VD group plants was significantly delayed compared to the HD group plants. Even after an extended drought period of 20–25 days, no complete destruction of tomato plants was observed in the VD group.
TYLCV and ToCV grafting confirmation with Duplex and nested PCR
After extracting total DNA from MK and SH tomato samples, the duplex PCR method was employed to identify TYLCV in the tomato plants. The samples exhibited bands at 462 bp on an agarose gel when primer triplets (AC950, AV632, and AC1048) were used. The healthy tomato leaf sample, mock-inoculated sample, and negative control (dH2O) did not display any bands. In contrast, the positive control obtained from infected tomato samples from Antalya exhibited a specific TYLCV band at 462 bp (Fig. 4B).
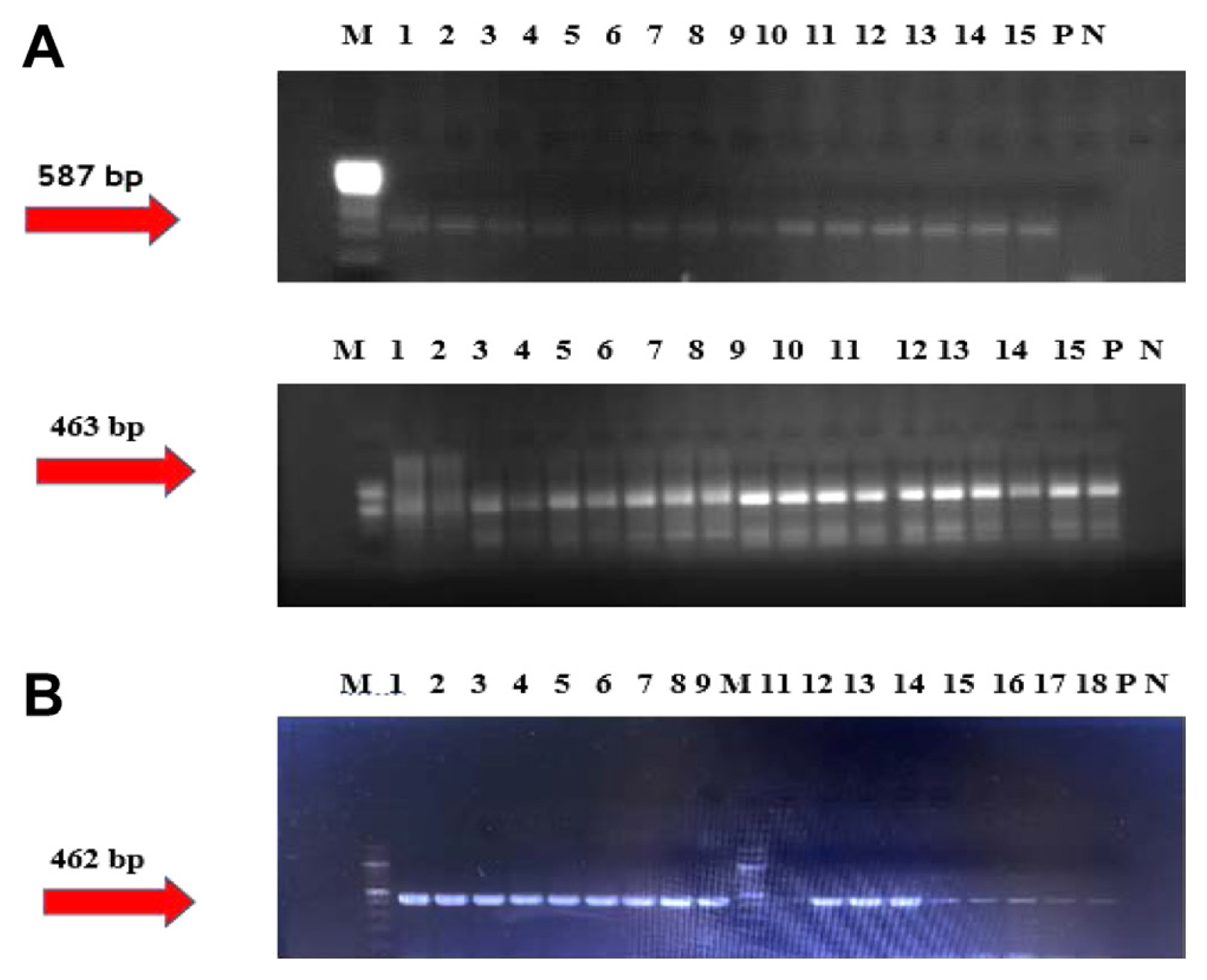
Molecular detection of tomato chlorosis virus by nested polymerase chain reaction (PCR) (A) and tomato yellow leaf curl virus by duplex PCR (B), respectively, from the upper systemic leaves of the single-leaflet grafted tomato plants. PCR amplicons were visualized under ultraviolet light in 1% agarose gel (0.5× TBE buffer) containing 1 μg/ml ethidium bromide. M, 100 bp DNA ladder (Biolabs); N, negative controls of PCR reactions; P, positive controls of PCR reactions. (A) 1–15 test samples. (B) 1–18 test samples.
To detect ToCV, total RNA was isolated from MK and SH tomato samples, and the presence of ToCV was determined using the two-step nested RT-PCR method. Following the nested RT-PCR, a 463 bp fragment of the HSP70 gene was amplified from the positive control sample (Fig. 4A).
Quantitative (qRT-PCR) expression analysis of cellulose synthase genes (CesA/Csl)
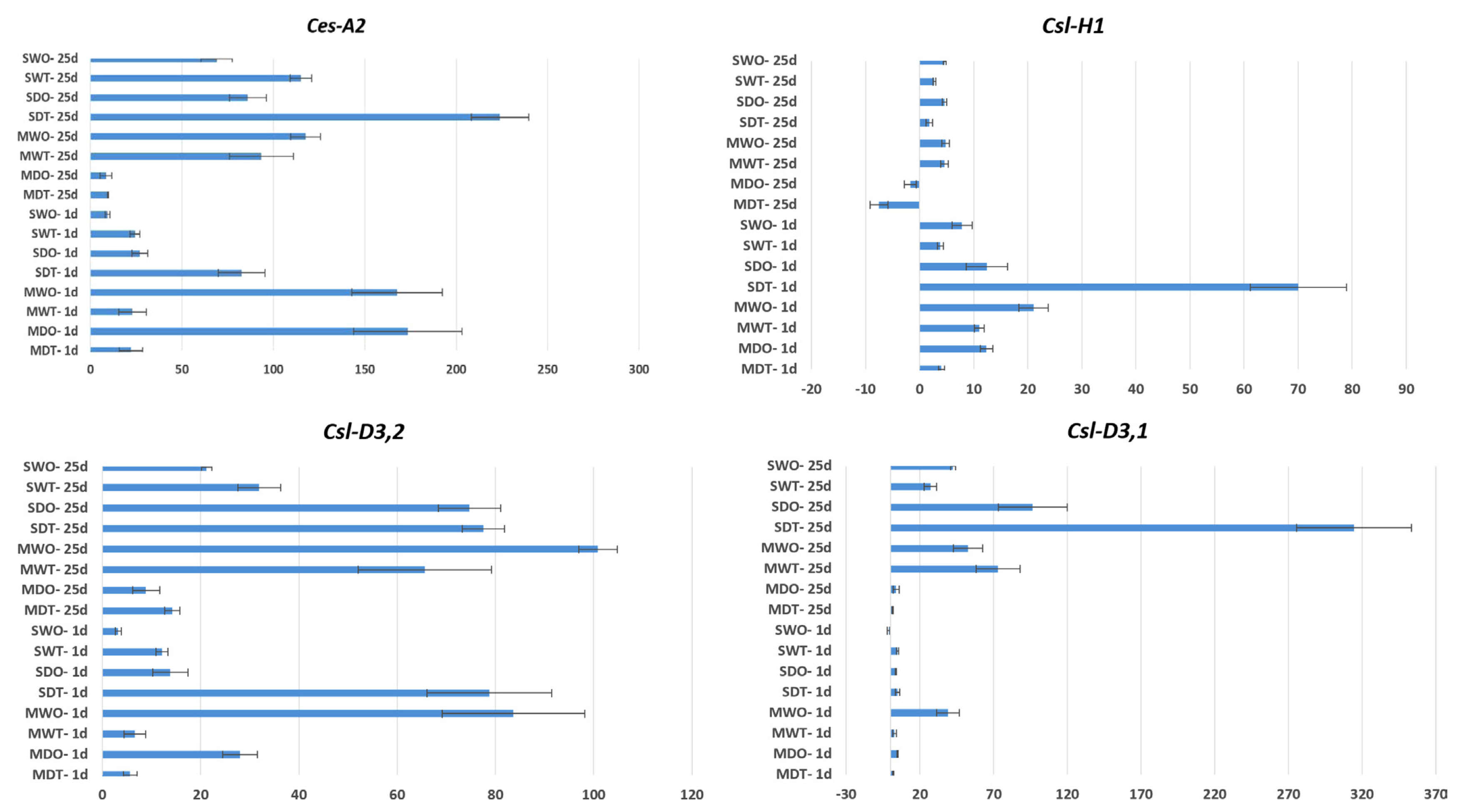
To investigate the involvement of CesA/Csl genes in the response to combined virus and drought stress in tomato cultivars, gene expression analysis was conducted. The transcript levels of CesA and Csl genes exhibited an increase under virus and drought stress conditions, and the extent of this increase varied among different species (Fig. 5).
In the qRT-PCR analyses of the four studied genes (Csl-H1, Csl-D3,1, Csl-D3,2, and Ces-A2), an overall upregulation pattern was observed in both tomato cultivars (MK and SH). Notably, among these genes, Csl-H1 showed relatively lower levels of upregulation or downregulation compared to the other genes when subjected to virus and drought stress on days 1 and 25. It is worth mentioning that the expression of the Csl-H1 gene displayed less variation compared to other CesA/Csl genes in terms of both virus (TYLCV and ToCV) and drought stress.
TYLCV and ToCV virus-infected cultivars
When only TYLCV and ToCV virus-infected (without drought stress application) samples were compared; Significant gene expression differences were determined between day 1 and day 25 in both Money Maker (MK, UK) and Shalala (SH, Azerbaijan) cultivars, especially in TYLCV-infected genotypes. In the Shalala cultivar (SWT), an approximately 3-fold increase in gene expression was observed in the Csl-D3,2 between the 1st day (12 fold change) and the 25th day (31 fold change), approximately 7-fold in the Csl-D3,1 and approximately 4.5-fold in the Ces-A2, respectively (Fig. 5).
Unlike the TYLCV-infected samples, there is no significant difference in the expression increase in the ToCV virus-infected samples, especially in the Money Maker cultivar (MWO), when comparing the Csl-D3,1, Csl-D3,2, and Ces-A2 genes between day 1 and 25. In the ToCV virus-infected Shalala cultivar (SWO), between the 1st and the 25th day, approximately 40 and 6.5-fold increase in gene expression was observed, especially in Csl-D3,1 and Csl-D3,2 genes, respectively (Fig, 5). When evaluated from this point of view, the fact that there are more gene expression changes (between 1st and 25th days) in the ToCV virus-infected Shalala cultivar compared to the Money Maker reveals that the Shalala cultivar may be more sensitive to this virus.
TYLCV and ToCV virus-infected and drought-stressed cultivars
When we look at the samples infected with the virus (TYLCV and ToCV) and also subjected to drought stress, the highest gene expression increase was detected in the Shalala (SDT) (314 fold change) in the Csl-D3,1 gene.
Interestingly, the Money Maker (MDT) genotype, which showed a 4 fold change, upregulation in the Csl-H1 gene on day 1, manifested a 7.5 fold change, downregulation on day 25. According to the results of the analysis, drought stress applications in virus-infected SDO and SDT genotypes, especially in Csl-D3,1, Csl-D3,2 and Ces-A2 genes, significantly increased gene expression levels on the 25 day of drought stress. For example, a 3-fold change, upregulation occurred in the Csl-D3,1 gene on the 1st day of drought-stress treatment in the SDO genotype, while the same genotype had a 96 fold change, upregulation on the 25th day of drought stress. There is a different situation in the Money Maker cultivar. In the Csl-D3,1 gene, there was no significant change in the comparison of virus-infected Money Maker (MDT and MDO) on the 1st day of drought stress with the 25th day of drought stress, and gene expression coefficients were similar. Similarly, in the Csl-D3,2 no significant increase or decrease in gene expression was observed on the 25th day compared to the 1st-day results. For example, the Money Maker (MDO) plants in the Csl-D3,2 gene had 28 fold change, upregulation, while on day 25 there was 8 fold change, upregulation.
Physiological measurements of tomatoes subjected to virus and drought stresses
In the MK and Shalala cultivars, there was an increase in RWC in leaves infected with TYLCV and ToCV compared to mock-inoculated samples. However, RWC in the combined infection of TYLCV and drought (VD) was lower compared to the combined infection of TYLCV and drought (VH). When drought stress was applied to TYLCV-inoculated MK cultivars, the RWC in leaves of the drought treatment group (HD) decreased by 11% compared to the well-watered group (HW), and in the drought treatment and virus-infection group (VW), RWC decreased by 14%. Although the RWC increased by 8–11% in the VD group compared to the other TYLCV and drought-stressed groups, respectively, it dropped by 3% compared to the HW group. Similar results were observed in plants exposed to the combined effect of ToCV and drought, with the RWC in the HD and VW groups showing a reduction of 14% and 12%, respectively, compared to the HW group. In contrast, the VD group showed a 4% increase in RWC. In the Shalala genotype, the RWC percentage changed in response to the dual effects of TYLCV and drought, with RWC values of 64% in the well-watered (HW) group, 56% in the drought treatment (HD) group, 52% in the drought treatment and VW group, and 59% in the drought treatment and combined virus-infection (VD) group. After exposing tomato samples grafted with ToCV to drought stress, the RWC expression decreased by 65% in the well-watered (HW) samples, and 53–51% in the HD and VW groups, respectively. In the VD group, compared to the other stressed groups, there was an increase, with an RWC value of 60% (Fig. 6).
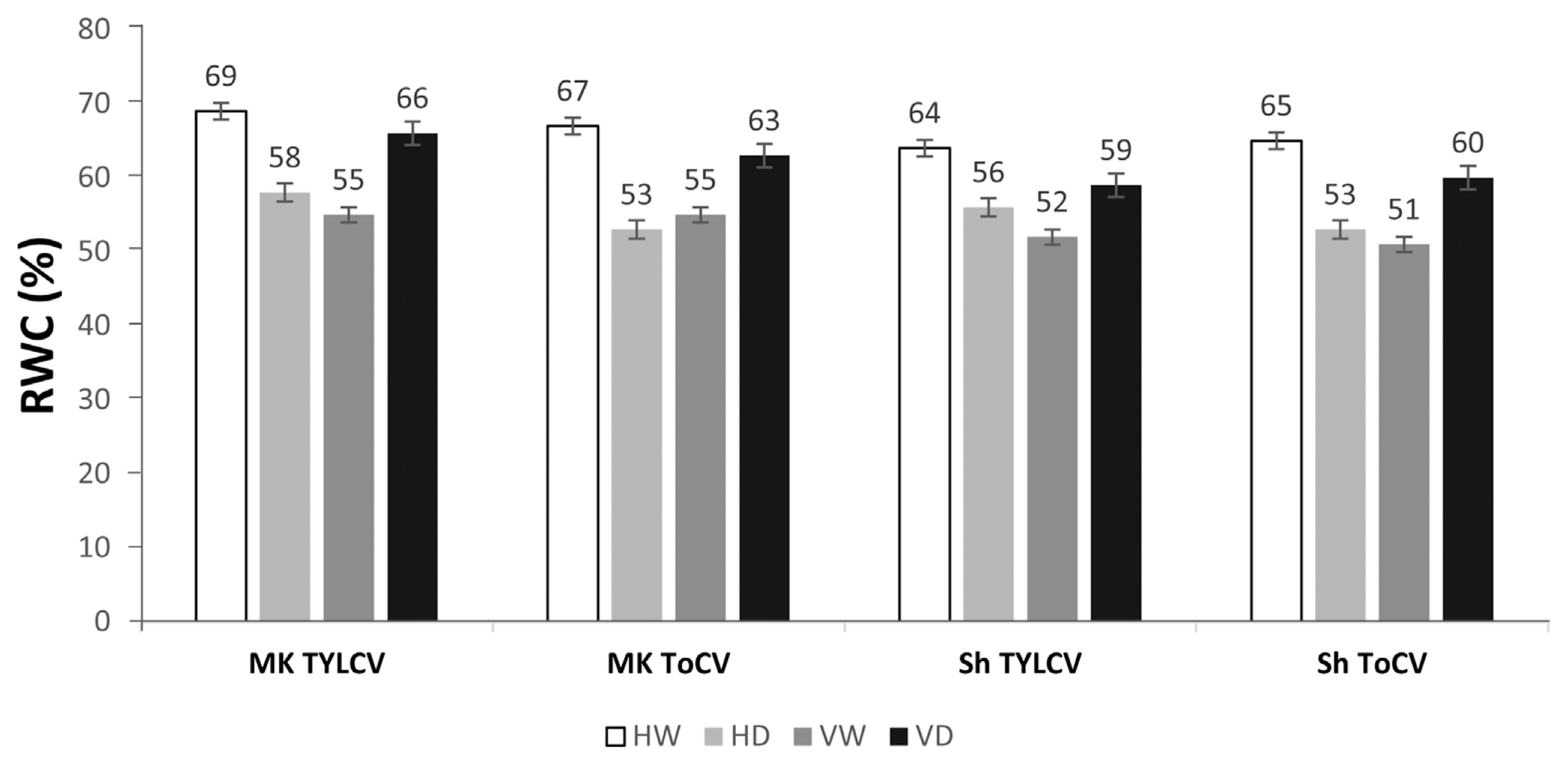
Relative water content (RWC %) in Money Maker (MK) and Shalala (SH) cultivars exposed to combined virus (TYLCV and ToCV) and drought stress. HW, healthy watered; HD, healthy drought; VW, virus watered; VD, virus drought; TYLCV, tomato yellow leaf curl virus; ToCV, tomato chlorosis virus.
During the experiments, the dry biomass percentage of tomato samples belonging to the MK and SH cultivars was measured following the procedure outlined in the Materials and Methods section. In plants exposed to the dual effect of TYLCV and drought, there was an approximately 11% reduction in leaf dry biomass in the HD group compared to the HW group. The biomass of the VW group was approximately half (23%) of that of the HW group. The VD group showed a 14% reduction, representing a ~1.6 times reduction compared to the VW and HD groups. In samples inoculated with ToCV, compared to the HW group, the dry biomass increased 2 times, reaching 24% in the HD group. The dry biomass percentage in the VW samples was 21%. Compared to the mock-inoculated group, there was a decrease of up to 17% in the ToCV + drought variants, with a slight increase of up to 3% compared to the control variant. In the Shalala genotype, the values of dry biomass in plants exposed to the combined effects of the viruses (TYLCV and ToCV) and drought were 12% and 17%, respectively. In mock-inoculated plants, there was an increase of up to 19% and 23% in the HD samples, and up to 17% and 14% in the VW samples. Only slight increases of 2% and 4% were observed in the VD group compared to the HW group, indicating a minimal increase compared to the control variant (Fig. 7).

Dry biomass (%) in Money Maker (MK) and Shalala (SH) cultivars exposed to combined virus (TYLCV and ToCV) and drought stress. HW, healthy watered; HD, healthy drought; VW, virus watered; VD, virus drought; TYLCV, tomato yellow leaf curl virus; ToCV, tomato chlorosis virus.
Water deficit was examined in both cultivars. An increase in water deficit was observed in the VD samples compared to the HW samples, while a significant reduction was detected in the VW samples compared to the VD samples. In the MK cultivar, the water deficit in mock-inoculated leaves was 9%. This index increased to 16% and 20% in the HD group, and to 18% (TYLCV) and 19% (ToCV) in the VW group. In the VD group, water deficit was 13% and 15%, representing 4% and 6% increases compared to the HW group. In the Shalala cultivar, the water content of the HW samples was approximately 8%. There were increases of 13% and 11% in the HD variant compared to the control (HW), and 14% (TYLCV) and 6% (ToCV) in the VW variant of tomato leaves. Compared to the control variant, there was a 4% increase in the TYLCV + drought (VD) variant and a 3% increase in the ToCV + drought (VD) variant in plant leaves exposed to dual stress (Fig. 8).
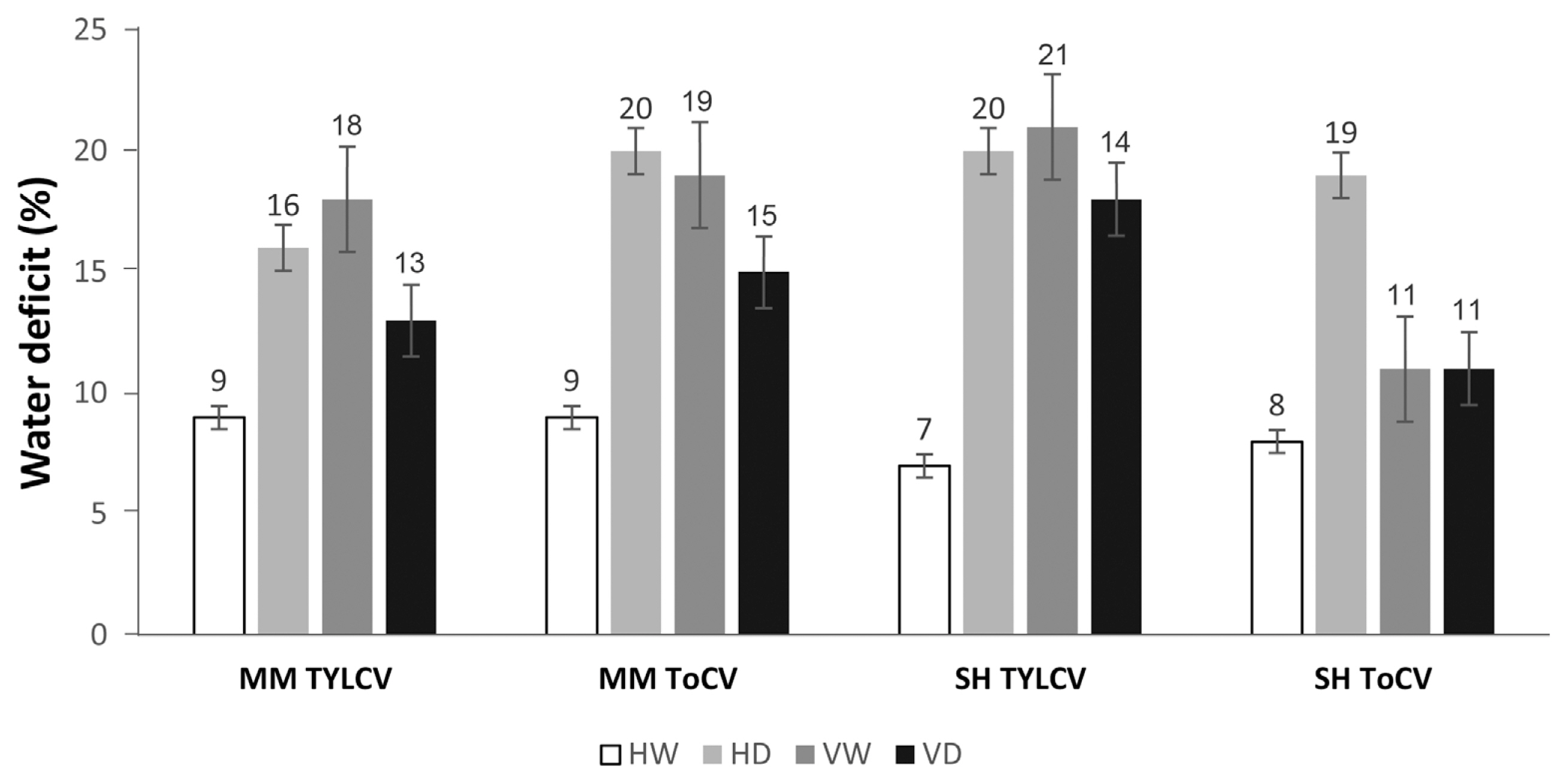
Water deficit (%) in Money Maker (MK) and Shalala (SH) cultivars exposed to combined virus (TYLCV and ToCV) and drought stress. HW, healthy watered; HD, healthy drought; VW, virus watered; VD, virus drought; TYLCV, tomato yellow leaf curl virus; ToCV, tomato chlorosis virus.
In both cultivars, an increase in leaf area was observed in the VW samples compared to the HW samples, while a decrease was observed in the VD samples compared to the VW samples. However, the decrease in leaf area in the VD samples was quite insignificant compared to the other groups of plants (HD, VW). In the MK cultivar, the leaf area was 22 cm2 in the HW group, and 30 cm2 in the HD group, 3 cm2 (TYLCV) and 10 cm2 (ToCV) in the VW group, representing a reduction of approximately 20 cm2 for the HD group and approximately 16 cm2 for the VW group compared to the leaf area of mock-inoculated plants. In the VD group, the leaf area showed a decrease of up to 4 cm2 for both viruses. In the Shalala cultivar, the leaf area changed as follows: 31 cm2 and 29 cm2 in the HW group, 16 cm2 and 18 cm2 in the HD group, 19 cm2 (TYLCV) and 27 cm2 (ToCV) in the VW groups, and 27 cm2 (TYLCV) and 21 cm2 (ToCV) in the VD groups. The reduction in leaf area in the VD leaf samples was minimal compared to the other groups of plants (HD, VW) (Fig. 9).
Discussion
Quantitative (qRT-PCR) expression analysis of cellulose synthase genes (CesA/Csl)
The plant cell wall, composed of polyphenolic structures such as lignin and polysaccharides like cellulose, pectin, and hemicellulose, is crucial for the construction of plant biomass and finds applications in various industries, including food, feed, and fuel (Hu et al., 2018; Somerville, 2006; Wang et al., 2012). In addition to providing structural support, the cell wall plays vital roles in plant growth, determining plant shape and cell size, and protecting against environmental stresses (Landrein and Hamant, 2013; Le Gall et al., 2015; Malinovsky et al., 2014). Understanding the genes involved in cell wall formation and reshaping is crucial as they contribute to plant growth, control water movement, regulate cell differentiation, defend against pathogens and pests, and facilitate intercellular adhesion and communication (Heidari et al., 2019).
The cellulose synthase gene superfamily, which includes the CesA and Csl gene families, plays a vital role in cellulose and hemicellulose biosynthesis in plants. However, the study of these gene families in tomato plants has been limited. Recently, 38 CesA/Csl family members, containing cellulose synthase domain regions, have been identified and analyzed in tomato plants. Through phylogenetic analysis, these genes have been classified into six subfamilies: CesA, CslA, CslB, CslD, CslE, and CslG. Although most CesA/Csl genes in tomato plants are similar to those found in Arabidopsis, they exhibit distinguishable characteristics in terms of gene structure, chromosome distribution and localization, phylogeny, and protein sequence, indicating that they have evolved through various processes (Song et al., 2019).
Understanding the roles and functions of CesA/Csl genes in tomato plants will contribute to our knowledge of cell wall biosynthesis and its regulation. These genes play critical roles in plant growth, cell differentiation, and defense mechanisms against pathogens and pests. By studying the CesA/Csl genes, we can gain insights into the molecular mechanisms underlying cell wall formation and the regulation of cell wall-related processes in tomato plants.
The studies on TYLCV-infected plants have shown that silencing of Ces genes leads to plant growth arrest, reduced leaf size, and leaf shape distortion, similar to the symptoms caused by TYLCV (Burn et al., 2002; Chu et al., 2007). In RNA-Seq analyses of healthy and TYLCV-infected tomato cultivars, it was observed that the cellulose synthase family gene Csl-H1g043390.2.1, a homolog of the AtCESA8 gene, was highly down-regulated in response to TYLCV infection (Seo et al., 2018). This downregulation of cellulose synthase genes may be an important factor triggering stunted growth and leaf curl in TYLCV-infected plants (Li et al., 2019). However, in our experiments using TYLCV-infected cultivars MK and SH, no downregulation was observed in the 4 CesA/Csl genes used, in contrast to the findings of the previous studies.
In the comparison between the 1st and 25th day, it was observed that the Ces-A2 (93 fold change and 117 fold change) and Csl-D3,1 (73 fold change and 3 fold change) genes in the MK genotype, known to be sensitive, and the Ces-A2 gene (115-fold change and 223-fold change) in the SH cultivar showed marked upregulation within 25 days. Transcriptome profiling analyses of TYLCV-infected tomato lines have reported approximately 4-fold upregulation of the Csl gene in TYLCV-resistant breeding lines, and in TYLCV-infected tomato samples, one CesA gene and two Csl genes were significantly upregulated (Chen et al., 2013; Choe et al., 2021). Based on this information, the upregulation of the 4 CesA/Csl genes used in our study, albeit at different levels, in the susceptible MK cultivar with TYLCV infection suggests that these genes play different roles in the complex biotic stress response mechanisms, which could potentially differentiate between sensitive and resistant cultivars. Regardless of the time point difference, the upregulation of these genes indicates their involvement in the plant’s response to TYLCV.
TYLCV and ToCV, which exhibit similar symptoms in plant leaves, are believed to be regulated by leaf senescence processes, although the genes involved in the molecular mechanisms are still largely unknown (Espinoza et al., 2007; Seo et al., 2018). In the same study, it was found that the cell wall invertase 2 gene was significantly upregulated after TYLCV infection, suggesting a general defense response in tomato, and this gene was also upregulated in ToCV-infected plants (Seo et al., 2018). In our study, except for the Csl-H1 gene, the other three CesA/Csl genes were significantly upregulated on the 25th day, particularly in the SH cultivar. For instance, in the SH (SWO) cultivar, Ces-A2 showed a 9-fold increase on the 1st day, while it was upregulated by 69-fold on the 25th day. However, in the MK (MWO) cultivar, no significant difference in upregulation was observed between the 1st and 25th day of ToCV infection.
Studies have shown that drought stress can affect the content of sugars in plants by inhibiting enzymes involved in cellulose synthesis (Foyer et al., 1998). Mutations in cellulose synthase genes, such as AtCesA8 in Arabidopsis, have been found to lead to increased accumulation of soluble sugars and enhanced drought and osmotic tolerance (Chen et al., 2005). This suggests that cellulose synthesis plays an important role in the response of tomato plants to drought and osmotic stress (Khan et al., 2015).
In our study, in the drought-stressed, ToCV and TYLCV-infected SH cultivar, the Csl-D3,1 and Ces-A2 genes showed significant upregulation on the 25th day. However, the upregulation level of the Csl-H1 gene in the same cultivar was lower on the 25th day compared to the 1st day. Similarly, in rice cultivars subjected to drought stress, an increase in the CesA10 gene correlated with improved drought tolerance (Narciso et al., 2010). On the other hand, in the drought-stressed MK cultivar infected with ToCV and TYLCV, no significant upregulation of the 4 genes was observed on the 25th day. This suggests that the MK cultivar, despite being infected with the viruses, did not show a significant correlation with increased drought stress.
In a study on susceptible/resistant maize cultivars subjected to drought stress, significant upregulation of 3 CesA genes was observed in the resistant line (Waititu et al., 2021). The comparison of the 1st and 25th days in our study, particularly in terms of the Csl-D3,1 and Ces-A2 genes, suggests the possibility of the SH cultivar exhibiting higher resistance compared to the MK cultivar. These genes may play a role in distinguishing between resistant and sensitive cultivars under drought stress.
Physiological analysis of TYLCV and ToCV-infected tomatoes
Drought is a significant inhibitor of crop production, including tomatoes, as it affects numerous plant processes and results in a reduction in plant growth. In our current experimental system, tomato plants exposed to water withholding exhibited a dramatic reduction in growth after 10 days. However, grafting of tomato cultivars (MK and SH) with TYLCV and ToCV viruses provided critical protection to the plants from these effects. After 25 days, while the virus-free tomatoes completely wilted and collapsed, the VD plants continued to grow.
Harsh environmental stresses, particularly water deficit, influence plant biomass and its distribution on leaflets. When measuring the dry weights of MK and SH cultivars, we observed significant losses in the leaves of healthy-drought plants compared to HW plants. However, the VD tissues lost much less biomass compared to VW plants, both in the case of TYLCV and ToCV inoculated tomato plants. This suggests that virus infection enhances plant endurance under drought stress, and the allocation of leaflet biomass in TYLCV and ToCV-inoculated tomato plants plays an important role in drought adaptation. Physiological water balance parameters measured in healthy and virus-grafted tomato plants also revealed significant differences in response to drought stress (Mishra et al., 2022).
Similar results were obtained for RWC, where VD-inoculated variants developed drought tolerance over time, enabling the plants to cope with the stress, while mock-inoculated variants gradually withered. Thus, the combined effect of virus and drought in plants activates the phyto-immune system and promotes plant adaptation to adverse climatic conditions. Similar trends were observed for leaf area in plants exposed to dual stress and mock-inoculated samples, whereas these parameters decreased in VW variants. This confirms that the combined effect of biotic and abiotic stresses triggers plant adaptation mechanisms. Specific leaf weight, which is considered an essential physiological indicator of plants, was found to decrease in VW-affected plants compared to HW plants. On the other hand, the increase in specific leaf weight in VD samples indicates the activation of self-resistance mechanisms in plants, approaching the levels observed in HW plants.
The RWC is an indicator of water status in plant cells and is related to drought stress tolerance. In our study, we found that RWC content decreases in HD variants. This is consistent with findings by Sakya et al. (2018), who reported that RWC also decreases during drought stress with an average decrease ranging from 8% to 23%. Additionally, the leaf area was significantly lower under drought stress compared to the control variant, in line with the observations of Zhou et al. (2017), who suggested that the leaf fresh weight of plants significantly decreases under drought and combined stress compared to control plants. Furthermore, the dry biomass of both cultivars showed a significant decrease under drought and combined heat stress compared to the control variant. This is in accordance with findings by Zhou et al. (2017), where the dry biomass of plants was considerably smaller under drought and combined heat stress compared to control plants. The RWC of all cultivars significantly decreased under drought and heat stress compared to control plants (Zhou et al., 2017). The decrease in RWC of tomato plants subjected to drought stress, as shown in Fig. 6, is consistent with the observations of Khan et al. (2015), who reported a decrease in RWC to 89.28% in drought-treated plants compared to control variants. Moreover, RWC values above 75% indicate a normal plant water status even under exposure to drought stress (Patanè et al., 2022).
In the study by Tahi et al. (2007), it was found that RWC decreased by about 88% during the experiment, while plant dry biomass and leaf dry mass decreased significantly by about 30%. In our study, we also observed a sharp decrease in leaf area in both MK and SH cultivars under HD conditions. Similarly, leaf area decreased by about 28% in the experimental plants compared to the control. However, in response to water deficit, there was a significant increase in plant growth in both cultivars.
We further investigated RWC in TYLCV and ToCV-infected tomato cultivars (MK, SH) and found that RWC reduced sharply in VW variants. Hosseini et al. (2018) suggested that the combined effect of drought stress and Cucumber mosaic virus (CMV) led to the lowest RWC in the experimental plants compared to mock-inoculated variants. Additionally, dry mass production was enhanced by 20–25% in drought-treated plants (Sand et al., 2018). In TYLCV-infected plants, we observed a significant decrease in leaf area. Sofy et al. (2021) reported a sharp reduction in shoot and root length (40.00% and 35.82%, respectively), as well as in fresh and dry weight biomass in shoots (38.96% and 61.54%, respectively) in ToMV-infected plants compared to water-treated plants. Plants infected with potato spindle tuber viroid (PSTVd) also exhibited smaller leaf area and 30% reduction in dry weight compared to control variants (Mackie et al., 2019). Bergès et al. (2021) found that the vegetative growth of A. thaliana is reduced when infected with cauliflower mosaic virus (CaMV), but a combination of viral infection and drought stress induces plant growth.
Plant cell walls, composed of polyphenolic structures such as lignin and polysaccharides (e.g., cellulose, pectin, hemicellulose), play a crucial role in plant biomass construction and protection against environmental stresses. They provide structural growth support, define plant shape and cell size, and contribute to various industrial fields (Hu et al., 2018; Somerville, 2006; Wang et al., 2012). The present study highlights the correlation between drought and virus stresses. Virus quantity decreased in infected plants exposed to drought (Corrales-Gutierrez et al., 2020). Tomato plants infected by viruliferous whiteflies, as they naturally occur in the field, showed enhanced resistance to drought. Gargallo-Garriga et al. (2014) suggested that enhanced tolerance is due to the reallocation of various basic metabolites from shoots to roots. The interplay between plant viruses and their hosts can evolve from pathogenic to mutualistic interaction under stress conditions (González et al., 2021; Prasch and Sonnewald, 2013). Studies on various viruses and their effects on drought tolerance in host plants, such as Nicotiana benthamiana, have shown delayed drought symptoms in virus-infected plants compared to mock-inoculated plants (Xu et al., 2008).
Re-watering of virus-infected beet and rice plants resulted in their recovery from drought stress, indicating the possibility of beneficial interactions. Plants in natural and crop field environments are exposed to multiple environmental stresses, and the responses to simultaneous abiotic and biotic stresses are complex (Garrett et al., 2006). In some cases, abiotic and biotic stresses have synergistic effects. For example, common bean and sorghum plants infected with the fungal pathogen Macrophomina phaseolina exhibit more severe charcoal rot symptoms under drought, with increased drought stress effects in the infected plants (Mayek-Pérez et al., 2002). Similar phenomena have been observed in grape plants infected by the bacterial pathogen Xyllela fastidiosa (McElrone et al., 2001). Field studies have also shown that viral infection and drought stress can have dire consequences for crop loss, with varying additive effects influenced by other environmental variables in the field (McLaughlin and Windham, 1996).
Overall, the outcomes of the present study indicate that plants infected with several RNA and DNA genome viruses exhibit better tolerance and survival under drought stress. These plant-virus interactions may be part of the complex mechanisms that plants employ to cope with environmental changes. Virus infection can reduce the size of plant hosts, which can reduce their water requirements and improve survival under extreme drought stress. Virus infection can also lead to physiological modifications in plant tissues, including water content alteration and metabolite synthesis and translocation. For example, CMV infection increased water quantity in above-ground tissues, and infected plants retained water better than mock-inoculated plants (Hull, 2002).
The study aimed to investigate the self-regulation ability of tomato plants in tolerating various stresses such as TYLCV and ToCV infections, as well as water deficit and drought. Physiological indicators were examined, and pathogen-related transcriptional changes were analyzed alongside the recovery process. Ultimately, the research aimed to understand the mechanisms underlying drought stress tolerance in tomato plants to prevent yield loss and address the potential impact of environmental changes.
The results of this study suggest that the combined effects of biotic and abiotic stresses play a crucial role in mitigating the negative consequences of drought and viral infections caused by TYLCV and ToCV. Furthermore, these combined stressors were observed to expedite the recovery time of tomato plants after rehydration. This finding is significant because it implies that by understanding the underlying mechanisms supporting drought stress tolerance, it may be possible to prevent yield losses in tomato crops, which hold immense economic and commercial value. In practical terms, several tomato cultivars or hybrids carrying resistance genes to TYLCV-causing viruses are currently being cultivated in extensive areas to minimize the impact of yield losses. This cultivar selection approach aligns with the findings of this study, reinforcing the validity of studying the self-regulation ability of tomato plants in response to multiple stressors. Given the economic importance of tomato crops and the imminent environmental changes, understanding the mechanisms behind stress tolerance in tomato plants is essential. This knowledge will enable researchers and farmers to implement effective strategies for preserving crop yield under challenging environmental conditions. By embracing this research, the agricultural industry can strive for sustainable practices and secure the future of tomato production.
The commercialization of tomato fruits heavily relies on cultivars resistant to viral infection. However, it is important to note that these cultivars do not develop immunity against viral infection; rather, they only exhibit systemic infection with mild symptomatology. This crucial information was highlighted by Marchant et al. (2020), emphasizing that although resistant cultivars may still succumb to viral infections, the symptoms are much milder than in susceptible cultivars. To supplement this knowledge, Czosnek (2021) provided insights into the pathophysiological behavior of TYLCV-resistant tomato lines under water shortage conditions. Their research shed light on how these resistant plants respond to abiotic stress, particularly water scarcity, while still maintaining their resistance against the viral infection.
Expanding on this research, it would be beneficial to explore the resistance mechanisms of other host plants against different epidemic DNA and RNA genome viruses under various abiotic stresses. This deeper understanding of the perception and interaction between biotic and abiotic stress factors can significantly contribute to global knowledge on plant defense mechanisms. By elucidating how plants respond to both types of stress simultaneously, future genetic agronomic applications can be developed to enhance crop resilience in challenging environments.
In summary, the cultivation of resistant tomato cultivars against viral infection has allowed for the commercialization of tomato fruits. However, these cultivars do not develop immunity, but rather exhibit mild symptomatology. Further research on the resistance mechanisms of host plants to various viral infections under different abiotic stresses will advance our understanding of the combined effects of biotic and abiotic stressors and provide valuable insights for future genetic agronomic applications.
TYLCV and ToCV are two common pathogens that significantly impact tomato production. In this study, we utilized the susceptible MK and the Shalala cultivar (susceptibility or resistance status unknown) to conduct single-leaflet grafting with TYLCV and ToCV. By subjecting the plants to virus infection and drought stress, we aimed to investigate the potential effects on water parameters in the leaves and assess their recovery compared to healthy plants. To gain insight into the plant’s response to the combined stress, we employed qRT-PCR to detect antivirus-related genes or pathways. Specifically, we focused on measuring the expression levels of cellulose synthase and cellulose synthase-like genes, as their biosynthesis is known to play a crucial role in the defense mechanisms of tomatoes against TYLCV and ToCV.
According to our experiments, we have observed certain physiological parameters and gene expressions that lead us to hypothesize that the plant recovery process in both cultivars, MK and SH, cannot be disregarded when considering the effect of combined stress. These findings contribute to a wider understanding of the interplay between host plants and viruses during periods of drought stress, shedding light on the molecular-genetic mechanisms that govern plant adaptation to the simultaneous impacts of biotic and abiotic environmental stresses.
The implications of our research extend beyond the realm of academia, particularly within the current context of climate change. As water scarcity becomes a pressing concern in many regions, it is imperative to develop agricultural practices that can sustain crop growth despite limited water availability. By delving into the intricate relationship between plants, viruses, and drought stress, our work provides valuable insights that can aid in the formulation of new agronomic strategies customized for water-scarce areas. Moreover, our study paves the way for future investigations that aim to explore the dual effects of viruses and additional abiotic stressors on plants. By unraveling the complex interactions between multiple stress factors, we can gain a deeper understanding of plant resilience and uncover novel approaches to mitigate the detrimental effects of environmental challenges on crop productivity.
In conclusion, our research contributes to the expanding knowledge base surrounding plant-virus interactions during drought stress. By elucidating the molecular-genetic mechanisms of plant adaptation to combined stressors, our findings offer valuable insights for combating water scarcity in agriculture. Ultimately, this work serves as a catalyst for further advancements in the field, inviting future research that explores the multifaceted challenges posed by viruses and abiotic stressors on plant health and productivity.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This research was financially supported by a grant from the Islamic Development Bank (ISDB) “Postdoc” Scholarship programme, Scholarship No.: 600047690 (2022). This research was conducted under the supervision of Professor Dr. Ali Ergul at Ankara University, Institute of Biotechnology, located in Ankara, Turkey. We extend our sincere gratitude to Professor Dr. Ali Ergul for his guidance and support throughout the study. Additionally, we would like to express our appreciation for the provision of laboratory space and the necessary chemical reagents, which greatly contributed to the successful completion of this work.