 |
 |
| Plant Pathol J > Volume 39(6); 2023 > Article |
|
Abstract
Active plant immune response involving programmed cell death called the hypersensitive response (HR) is elicited by microbial effectors delivered through the type III secretion system (T3SS). The marine bacterium Hahella chejuensis contains two T3SSs that are similar to those of animal pathogens, but it was able to elicit HR-like cell death in the land plant Nicotiana benthamiana. The cell death was comparable with the transcriptional patterns of H. chejuensis T3SS-1 genes, was mediated by SGT1, a general regulator of plant resistance, and was suppressed by AvrPto1, a type III-secreted effector of a plant pathogen that inhibits HR. Thus, type III-secreted effectors of a marine bacterium are capable of inducing the nonhost HR in a land plant it has never encountered before. This suggests that plants may have evolved to cope with a potential threat posed by alien pathogen effectors. Our work documents an exceptional case of nonhost HR and provides an expanded perspective for studying plant nonhost resistance.
The most prominent strategy of plants to counteract pathogens is rapid and localized programmed cell death (PCD), termed the hypersensitive response (HR), which is similar to apoptosis in animal cells (Heath, 2000). HR initiates with recognition of a pathogen effector protein by a corresponding plant resistance protein with a nucleotide-binding leucine-rich repeat (NLR), in a process known as gene-for-gene resistance (Goodman and Vovacky, 1994). Plants also manifest the HR cell death as part of nonhost resistance against a broad spectrum of plant pathogens.
Bacterial effector proteins are delivered into host cells through specialized type III secretion systems (T3SSs) (Kim, 2001). T3SSs are considered effective weapons for pathogenic bacteria to ensure successful infection by subverting the host immune system or by hijacking host metabolism (Grant et al., 2006; Xian et al., 2020). However, it is not known to science that any animal-pathogenic bacterium including Yersinia species is able to elicit HR in plants by injecting a T3SS effector into the cytoplasm. A T3SS has been reported to possess several elements such as the secretion structure consisting of outer/inner rings with an extracellular needle-like extension, the translocation apparatus to penetrate the host cell surface barriers, effector proteins with unique secretion signals, and customized chaperones for stabilizing the effector proteins during the delivery (Kim and Alfano, 2002). T3SS genes frequently occur in pathogenicity islands that can be transferred among species, probably under certain evolutionary pressures (Yoon et al., 2007).
Hahella chejuensis KCTC 2396T, isolated from coastal marine sediment in the Jeju Province, Korea, is a new species belonging to an oceanic Gammaproteobacteria group (Lee et al., 2001). Analysis of the 7.2-Mb single-circular genome of H. chejuensis provided insights into the lifestyle of this bacterium in the marine environment (Jeong et al., 2005). Interestingly, the H. chejuensis genome contains two sets of genes encoding the T3SS that delivers an array of virulence effectors into eukaryotic host cells (Alfano and Collmer, 1997; Chang et al., 2004; Petnicki-Ocwieja et al., 2002). The amino acid sequences of H. chejuensis T3SSs are highly similar to those of the plasmid encoded T3SSs of Yersinia spp. (Galán et al., 2014; Kim, 2001).
To better understand the pathogenic potential of H. chejuensis, for which the hosts should not be terrestrial plants, and to better understand the nonhost resistance of plants, we used H. chejuensis as a model pathogen and Nicotiana benthamiana as a model plant. We first assessed the expression of T3SS genes in H. chejuensis at different growth stages to explore the functionality of H. chejuensis T3SSs. We then infiltrated H. chejuensis cells into N. benthamiana leaves to observe HR-like cell death. We further evaluated whether HR-like cell death elicited by H. chejuensis can be ameliorated when a bacterial effector that suppresses HR is expressed or a key component of R-gene-mediated resistance is silenced.
Amino acid sequences of the inner membrane channel proteins (StcV, EscV, LcrD, HrcV, and SsaV) of T3SSs were downloaded from the UniProtKB database. In total, 509 protein sequences were aligned using MUSCLE software (Edgar, 2004) with the complete gap deletion option, and evolutionary analyses were conducted using MEGA X (version 10.1.5) (Kumar et al., 2018) based on the Maximum Likelihood method and a JTT matrix-based model. In total, 381 amino acid positions were used to infer evolutionary distances. The constructed phylogenetic tree was exported in Newick tree format and was visualized using iTOL (http://itol.embl.de) (Letunic and Bork, 2019).
Nicotiana benthamiana seeds were obtained from Dr. Kiran Mysore (Oklahoma State University, OK, USA). N. benthamiana were grown in the KRIBB greenhouse and growth chamber facility in Daejeon, South Korea. The temperature in greenhouse was controlled at 25-30°C. For mode of action study, N. benthamiana was grown in a plant growth chamber (Sejong Biotech, Seoul, Korea) at 25°C under a photoperiod of 16 h. When they reached the six-leaf stage in 6 weeks, the plants were subjected to HR-like response assays.
Hahella chejuensis was grown in lysogeny broth (LB) or LB containing 3% NaCl medium supplemented with an appropriate antibiotic (50 μg/ml kanamycin) at 30°C. Escherichia coli EPI300 was grown in LB medium (BD Difco, Sparks, MD, USA) at 37°C, and Pseudomonas syringae pv. tomato DC3000 T1a was grown on King’s B medium (10% proteose peptone, 1.5% K2HPO4, 15% glycerol, and 5 mM MgSO4) or in LB broth at 30°C. Agrobacterium tumefaciens GV3101 containing TRV-VIGS vectors were grown in LB at 28°C. To identify the HR-like response elicited by H. chejuensis on N. benthamiana at different stages of growth, fresh cultures were diluted in LB medium containing 3% NaCl and grown at 30°C until harvesting. Bacteria samples were collected every 2 h for inoculation into N. benthamiana. Each sample was centrifuged and resuspended in phosphate buffered saline (PBS) at a cell density of 1 × 108 cfu/ml and infiltrated into N. benthamiana leaves. Infiltrations were performed by pricking leaves with a needle and then pressing the needless syringe against the leaf surface while supporting the leaf with a finger (Chaudhry et al., 1987). The development of a HR-like response was observed at room temperature.
The plasmid pPTE6::AvrPto1 was transformed into H. chejuensis by electroporation and was selected on LB medium containing kanamycin (50 ng/ml) at 30°C. H. chejuensis (AvrPto1) transformants were confirmed through polymerase chain reaction (PCR) using specific AvrPto1 primers in Supplementary Table 1.
For quantitative reverse transcription PCR (RT-PCR), total RNA was extracted from H. chejuensis using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) and was eluted in RNase-free water. Reverse transcription was carried out using 400 ng total RNA followed by quantitative PCR (qPCR) using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). The qPCR conditions were composed of an initial denaturation step at 95ºC for 10 min, 45 cycles at 95ºC for 30 s, 58ºC for 30 s and 72ºC for 30 s. To calculate the relative fold change, the 2−ΔΔCT method was employed, where ΔΔCT = (CT,Target - CT,rpoA)Time x - (CT,Target - CT,rpoA)Time 4h. In this equation, ‘Time x’ is any time point and ‘Time 4h’ represents the 1X expression of the target gene normalized rpoA, which is a housekeeping gene in H. chejuensis. All primers used are listed in Supplementary Table 1.
The leaf disks containing inoculum were excised at 12 h after inoculation and stained with lactophenol-trypan blue (10 ml of lactic acid, 10 ml of glycerol, 10 g of phenol, and 10 mg of trypan blue dissolved in 10 ml of distilled water) (Keogh et al., 1980). Whole leaves were boiled for 1 min in the staining solution and then decolorized in chloral hydrate (2.5 g of chloral hydrate dissolved in 1 ml of distilled water) for at least 30 min. The bleached (stained) leaves were examined under a compound microscope equipped with interference or phase-contrast optics.
Total RNA was extracted from the leaves of N. benthamiana 12 h after inoculation with H. chejuensis and P. syringae pv. tomato DC3000 as a positive control and PBS as a negative control. To isolate total RNA from plant tissue, leaf disks were collected with a 1 cm diameter cork borer, frozen in liquid nitrogen, ground into powder, and extracted RNA using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and suspended in water. Reverse transcriptase reactions were carried out using 1 μg of total RNA. We used 1 μl of the reverse transcription reaction as a template in 20 μl PCR reactions with primer sets listed in Supplementary Table 1. The RT-PCR products were separated by agarose gel electrophoresis.
Agrobacterium GV3101 containing TRV-VIGS vectors either without an insert (TRV::00) or with an N. benthamiana SGT1 insert (TRV::NbSGT) were grown overnight, and cultures were harvested by centrifugation at 4,000 rpm, followed by washing once in 10 mM MES (pH 5.6), and resuspended in Agrobacterium inoculation buffer (10 mM MgCl2, 10 mM MES [pH 5.6], and 200 μM acetosyringone) to a final concentration of 1 × 108 cfu/ml. These mixtures were incubated for 4 h with shaking at 180 rpm at room temperature before infiltration. Agrobacterium cultures containing TRV-VIGS vectors were infiltrated into the leaves of 14-day-old tobacco plants using 1 ml needleless syringes. After two to three weeks of VIGS, the plants were subjected to H. chejuensis-treatments.
The genome of Hahella chejuensis KCTC 2396T, a prodigiosin-producing member of oceanic Gammaproteobacteria (Kwon et al., 2010; Lee et al., 2001), contains two sets of genes encoding T3SS, which are highly similar to those in the virulence plasmids of animal-pathogenic Yersinia spp. (Fig. 1A) (Jeong et al., 2005; Snellings et al., 2001). The export apparatus (SctRSTUV), inner and outer rings of the needle complex (SctJD/C), ATPase (SctN), the cytoplasmic sorting platform (SctKQ), the stator (SctL), the stalk (SctO), and even a controller of the release and translocation of Yop effector proteins (YopN) and its chaperone protein (SycN) (Cheng et al., 2001; Deng et al., 2017; He et al., 2004) were conserved in H. chejuensis (Fig. 1A). To infer the phylogenetic affiliation of H. chejuensis T3SSs, we compiled all SctV sequences and analyzed their evolutionary relationships (Fig. 1B and C). We designated these genes as hct (Hahella chejuensis T3SS). As with YopN and SycN (data not shown), both HctVs formed sister branches to those of Yersinia-type SctVs (Fig. 1C).
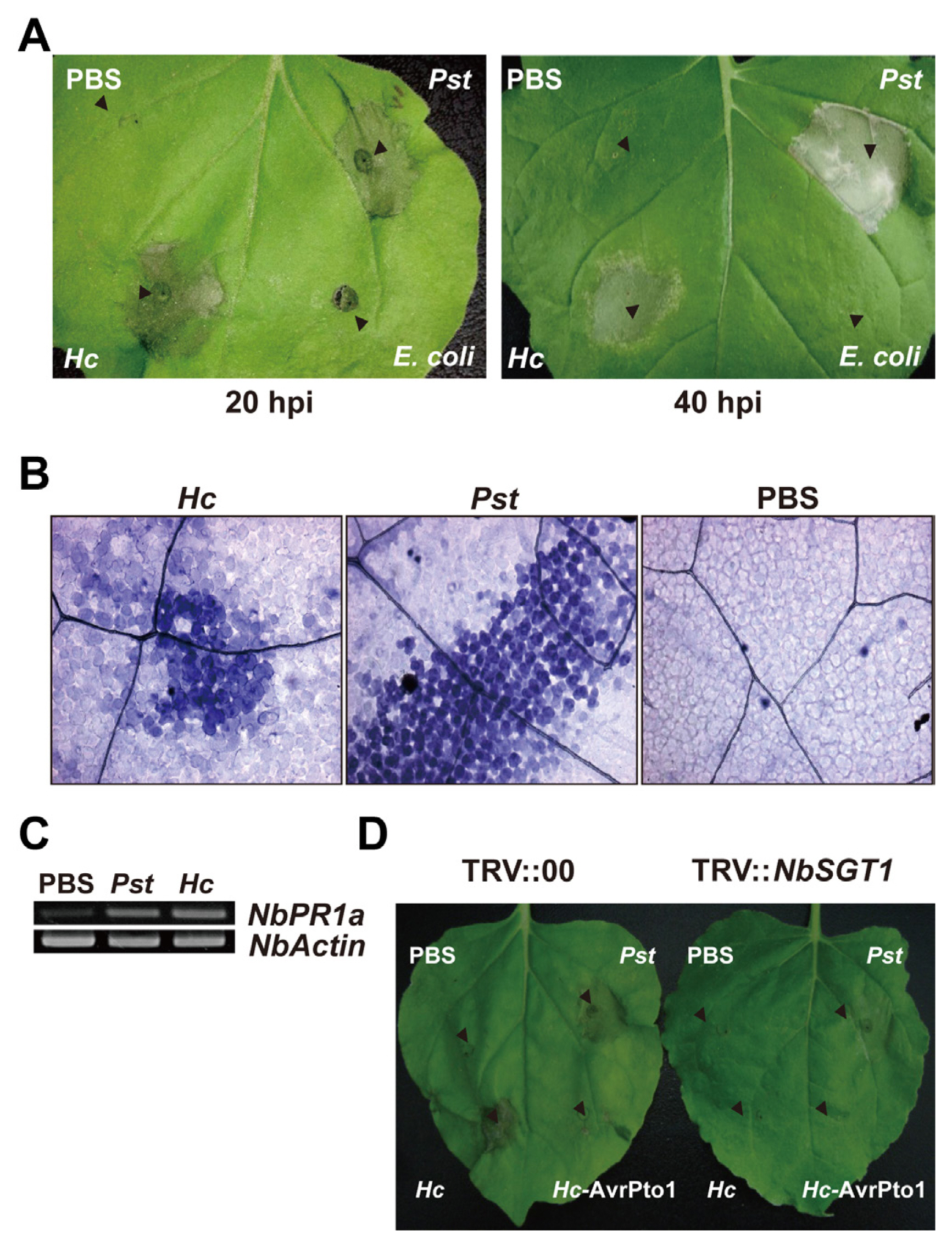
Considering that nonhost resistance of plants is a common form of disease resistance (Senthil-Kumar and Mysore, 2013) and the land plant N. benthamiana obviously is not a host for the T3SS-bearing marine bacterium H. chejuensis, we examined whether N. benthamiana would show HR against H. chejuensis by infiltrating the bacteria at different growth stages into the plant leaves. Surprisingly, necrosis appeared at 20 hours post inoculation (hpi), and the lesion became clear at 40 hpi only in areas where H. chejuensis in the late exponential phase (8 h, OD600nm = 1.3) and in the stationary phase (10 h and 12 h, OD600nm > 1.7) were infiltrated (Fig. 2A and B). Infiltration of Hahella-extracted or synthetic prodigiosin at different concentrations resulted in no visible lesions (data not shown), ruling out that it is involved. In addition, H. chejuensis in the late stationary phase did not cause necrosis, even though the pigment production was higher. This HR-like cell death was comparable with the transcriptional patterns of H. chejuensis T3SS-1 genes, which were increased gradually and showed maximum expression at the early stationary phase (10 h) and then decreased (12 h and 14 h) (Fig. 2C). At the late stationary phase of 14 h when the expression of TTSS-1 genes were low, we could not see the necrosis.
We then tested whether the cell death in N. benthamiana was equivalent to PCD induced by a genuine plant pathogen by comparing the responses of N. benthamiana against H. chejuensis, Pseudomonas syringae pv. tomato DC3000 T1a (a tomato pathogen without avrPto1), Escherichia coli EPI300 (a non-pathogen), or PBS. Upon monitoring the cell death macroscopically (Fig. 3A) and microscopically using dead-cell-staining trypan blue at 20 hpi (Fig. 3B), the infiltration sites of H. chejuensis and P. syringae pv. tomato did not differ. PR-1a gene expression in N. benthamiana (NbPR-1a) for H. chejuensis and P. syringae pv. tomato implicated that the HR-like cell death was a result of active defense (Fig. 3C).
AvrPto1, a T3SS-secreted effector of P. syringae pv. tomato, inhibits HR induced by nonhost-pathogen interactions (Kang et al., 2004). Thus, we examined whether H. chejuensis-induced HR-like cell death in N. benthamiana would be suppressed by H. chejuensis expressing avrPto1. As expected, infiltration of H. chejuensis harboring avrPto1 suppressed HR-like cell death (Fig. 3D), indicating avrPto1 expression in H. chejuensis and AvrPto1 translocaion into the cytoplasm of N. benthamiana through T3SS. To verify that the HR-like cell death follows a typical defense-associated PCD mechanism, we examined the cell death after virus-induced gene silencing of N. benthamiana SGT1 (NbSGT1). SGT1 is a general regulator of plant resistance, and silencing NbSGT1 compromises HR development during R-gene-mediated and nonhost resistance (Peart et al., 2002). The cell death caused by H. chejuensis was completely abolished in NbSGT1-silenced N. benthamiana (Fig. 3D). These observations demonstrate that the PCD of N. benthamiana caused by H. chejuensis is a result of nonhost resistance.
In this study, we provide the first evidence that a land plant, N. benthamiana, can respond to the marine bacterium H. chejuensis with PCD. To date, the natural hosts of H. chejuensis in the marine environment have not been defined yet. Based on the presence of animal-pathogen-type T3SSs and other genes that may play roles in virulence (Jeong et al., 2005), however, we speculate that H. chejuensis may be a pathogen or a symbiont of a marine protist or animal.
Previously, several animal pathogens, such as Pseudomonas aeruginosa, Enterococcus faecalis, and Salmonella enterica, have been reported to infect plants (Prithiviraj et al., 2005; Rahme et al., 1995; Roy et al., 2013). Infection with S. enterica on plants induced a range of basal defense responses, and transient expression of an effector of S. enterica triggered HR-like symptom in N. benthamiana (Roy et al., 2013; Üstün et al., 2012). However, marine bacteria have not been reported to cause HR in terrestrial plants. The analysis of amino acid sequence of H. chejuensis T3SSs revealed those are phylogenetically close to T3SSs of animal-pathogenic Yersinia spp. (Jeong et al., 2005). The major differences between T3SSs of plant- and animal-pathogenic bacteria are the structures of extracellular needle, pilus tip, and translocon pore (Buttner and Bonas, 2003; Kvitko and Collmer, 2023). The length of the needle in plant pathogens can reach up to 2 μm, enabling it to cross the thick plant cell wall. In contrast, the needle length is less than 87 nm in animal pathogens. It is expected that the needle structure of H. chejuensis T3SSs made from HctF might be enough to reach plant cytoplasm and effectively transport effectors. Unlike their animal pathogen counterparts, T3SSs of plant pathogens lack homologs of the conserved pilus tip and translocon (Kvitko and Collmer, 2023). Instead, they encode an array of translocator components, each of which plays a unique role in translocation, with variations in different plant pathogens (Kvitko and Collmer, 2023). Genes encoding the translocon complex (LcrV and YopBD homologs) are missing in H. chejuensis, a feature commonly observed in T3SSs of plant-pathogenic bacteria. Therefore, H. chejuensis is presumed to be a marine animal pathogen, but T3SS-1 of H. chejuensis may exhibit traits similar to those of plant-pathogenic bacteria.
H. chejuensis was isolated from the coastal marine sediment of an island where the annual temperature of water is 12-26°C, and active cell growth and T3SS gene expression of H. chejuensis was observed at 30°C. Among animal pathogenic bacteria including Yersinia spp. and P. aeruginosa, temperature around 37°C is an important factor for inducing T3SS gene expression (Schwiesow et al., 2015; Wurtzel et al., 2012). Although T3SS-1 of H. chejuensis may have a structure similar to those of animal pathogenic bacteria, it might be expressed and form an active structure at low temperatures, allowing effectors to be translocated into the plant cell.
Because of their different habitats, it is unlikely that H. chejuensis and N. benthamiana encounter under natural conditions. Nevertheless, N. benthamiana exhibited AvrPto1-inhibited SGT1-mediated PCD against H. chejuensis. Thus, we hypothesize that N. benthamiana responded either directly to the effectors translocated through T3SS or more likely to the protein modification or metabolic perturbation caused by the activities of translocated effectors, which are also recognized by NLR proteins. It is improbable if not impossible that Hahella effectors absent in plant-pathogenic bacteria or phylogenetically distant are specifically recognized by the plant NLR protein. According to a previous study, NLR- and pattern recognition receptor (PRR)-triggered defense responses contribute to nonhost resistance during increasing phylogenetic divergence time with the increasing effectiveness of PRR-triggered immunity, whereas the contribution of NLR protein-mediated immunity decreases (Schulze-Lefert and Panstruga, 2011). However, in the case of N. benthamiana and H. chejuensis, effectors delivered by T3SS, rather than microbe-associated molecular patterns, may play major roles in inducing the defense response of N. benthamiana, despite the considerable phylogenetic distance between N. benthamiana and the presumed hosts of H. chejuensis. NLR-triggered defense response is often dependent on the recognition of effector-mediated disturbances of target proteins or homeostasis as described in the guard and decoy hypothesis (Mukhtar et al., 2016). A recent study suggests that certain NLRs form complex networks for sensing pathogen invasion and eliciting plant immune responses (Duxbury et al., 2021), and the networks may have functionally redundant nodes that can be targeted by pathogen effectors (Adachi et al., 2019; Derevnina et al., 2021).
To our knowledge, this is the first study to document that a T3SS-harboring marine bacterium elicits PCD in a land plant, which is an extreme example of plant nonhost resistance. Our work that provides novel insights into the inner workings of plant immunity sheds light on the evolution of plant defense systems. Further investigation is warranted to determine the roles of T3SS-translocated effectors in H. chejuensis in subverting the plant homeostasis and eliciting immune responses. Additionally, identification of the plant factors responsible for recognition and response and the underlying mechanism involved are to be elucidated.
Acknowledgments
We thank S. P. Dinesh-Kumar for providing GATEWAY ready TRV-VIGS vectors, Alan Collmer for pCPP3221 plasmid. This work was supported by grants from the National Research Foundation of Korea (2018R1A6A1A03025607 and RS-2023-00211512) and the KRIBB Research Initiative Program.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Type III secretion systems (T3SSs) in the Hahella chejuensis genome and the phylogeny of SctV. (A) Genetic organization of two T3SSs in the H. chejuensis genome with the archetypal T3SS of Yersinia enterocolitica. (B) A topological representation of a prokaryotic type III secretion inner membrane channel protein, SctV. The tree was built using the maximum likelihood method based on 509 protein sequences in 381 conserved amino acid positions. The outmost circle’s color code represents the bacterium’s ecological features, and the inner circle was colored according to the clade information. The branches for H. chejuensis HctVs are shaded in grey. “Chlamy” strands for the clade that includes Chlamydia SctVs; “Desulfo”, Desulfovibrionales, “Hrp1”, Pseudomonas syringae; “Hrp2”, Ralstonia solanacearum; “Myxo”, Myxococcales; “Rhizo”, Rhizobium; “SPI1”, Salmonella enterica Pathogenicity Island 1 and Shigella; SPI2, S. enterica Pathogenicity Island 2; Ysc, Yersinia. Nomenclature for taxonomic clades follows those of (Abby and Rocha, 2012). (C) Detailed maximum likelihood tree of the “Ysc” clade in (B).
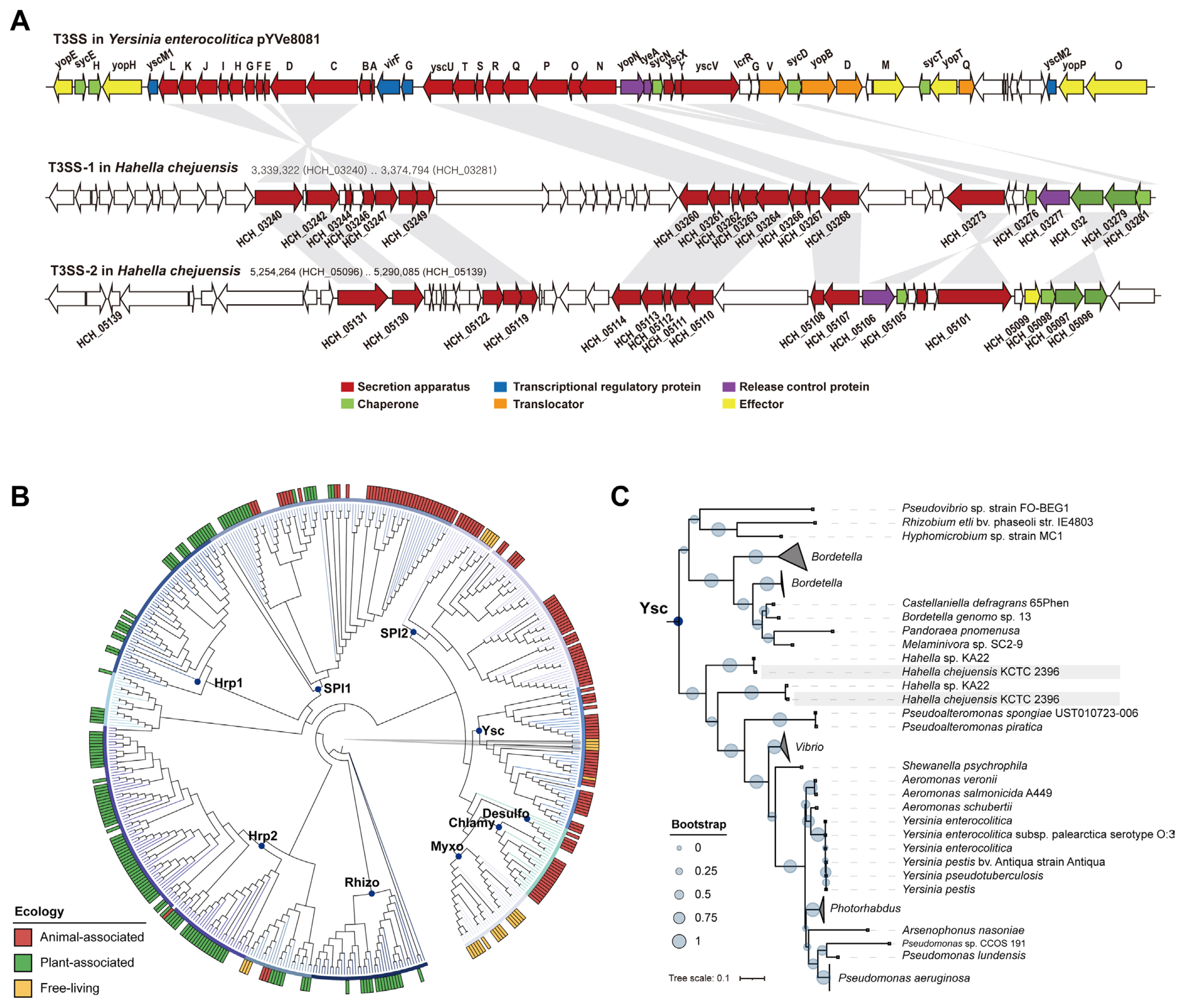
Fig. 2
Hypersensitive response (HR)-like cell death in Nicotiana benthamiana elicited by the marine bacterium Hahella chejuensis and type III secretion systems (T3SSs) gene expression. (A) HR-like cell death in N. benthamiana leaves at 20 (left) and 40 hours post inoculation (hpi) (right) of H. chejuensis. The infiltrated areas are indicated by dashed circles, the red of which indicates the location where HR was observed. (B) Growth patterns of H. chejuensis and red pigment production. (C) Expression of T3SS-1 and T3SS-2 genes in H. chejuensis.

Fig. 3
Programmed cell death in Nicotiana benthamiana induced by the Hahella chejuensis. (A) Hypersensitive response (HR)-like cell death in N. benthamiana infiltrated with bacteria or phosphate buffered saline (PBS) at 20 h and 40 h after infiltration (‘Hc’ stands for H. chejuensis, ‘Pst’ for Pseudomonas syringae pv. tomato DC3000 T1a and ‘Hc-AvrPto1’ for H. chejuensis expressing avrPto1 gene). (B) Microscopic observation of the trypan blue-stained spots. (C) Expression of the NbPR1a gene at 12 hours post inoculation (hpi) of bacteria. (D) Suppression of programmed cell death (PCD) by type III-secreted AvrPto1 or NbSGT1 silencing in N. benthamiana.
References
Abby, S. S. and Rocha, E. P. C. 2012. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 8:e1002983.



Adachi, H., Derevnina, L. and Kamoun, S. 2019. NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 50:121-131.


Alfano, J. R. and Collmer, A. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662.




Büttner, D. and Bonas, U. 2003. Common infection strategies of plant and animal pathogenic bacteria. Curr. Opin. Plant Biol. 6:312-319.


Chang, J. H., Goel, A. K., Grant, S. R. and Dangl, J. L. 2004. Wake of the flood: ascribing functions to the wave of type III effector proteins of phytopathogenic bacteria. Curr. Opin. Microbiol. 7:11-18.


Chaudhry, A. D., Baker, C. J. and Springett, J. A. 1987. Barley seedling establishment by direct drilling in a wet soil. 2. Effects of earthworms, residue and openers. Soil Till. Res. 9:123-133.

Cheng, L. W., Kay, O. and Schneewind, O. 2001. Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J. Bacteriol. 183:5293-5301.




Deng, W., Marshall, N. C., Rowland, J. L., McCoy, J. M., Worrall, L. J., Santos, A. S., Strynadka, N. C. J. and Finlay, B. B. 2017. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15:323-337.



Derevnina, L., Contreras, M. P., Adachi, H., Upson, J., Cruces, A. V., Xie, R., Skłenar, J., Menke, F. L. H., Mugford, S. T., MacLean, D., Ma, W., Hogenhout, S. A., Goverse, A., Maqboo, A., Wu, C.-H. and Kamoun, S. 2021. Plant pathogens convergently evolved to counteract redundant nodes of an NLR immune receptor network. PLoS Biol. 19:e3001136.



Duxbury, Z., Wu, C.-H. and Ding, P. 2021. A comparative overview of the intracellular guardians of plants and animals: NLRs in innate immunity and beyond. Annu. Rev. Plant Biol. 72:155-184.


Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797.



Galán, J. E., Lara-Tejero, M., Marlovits, T. C. and Wagner, S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68:415-438.



Goodman, R. N. and Vovacky, A. J. 1994. The hypersensitive reaction in plants to pathogens. APS Press, St. Paul, MN, USA. pp. 256.
Grant, S. R., Fisher, E. J., Chang, J. H., Mole, B. M. and Dangl, J. L. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60:425-449.


He, S. Y., Nomura, K. and Whittam, T. S. 2004. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim. Biophys. Acta 1694:181-206.


Jeong, H., Yim, J. H., Lee, C., Choi, S.-H., Park, Y. K., Yoon, S. H., Hur, C.-G., Kang, H.-Y., Kim, D., Lee, H. H., Park, K. H., Park, S.-H., Park, H.-S., Lee, H. K., Oh, T. K. and Kim, J. F. 2005. Genomic blueprint of Hahella chejuensis, a marine microbe producing an algicidal agent. Nucleic Acids Res. 33:7066-7073.



Kang, L., Tang, X. and Mysore, K. S. 2004.
Pseudomonas type III effector AvrPto suppresses the programmed cell death induced by two nonhost pathogens in Nicotiana benthamiana and tomato. Mol. Plant-Microbe Interact 17:1328-1336.


Keogh, R. C., Deverall, B. J. and McLeod, S. 1980. Comparison of histological and physiological responses to Phakopsora pachyrhizi in resistant and susceptible soybean. Trans. Br. Mycol. Soc. 74:329-333.

Kim, J. F. 2001. Revisiting the chlamydial type III protein secretion system: clues to the origin of type III protein secretion. Trends Genet. 17:65-69.


Kim, J. F. and Alfano, J. R. 2002. Pathogenicity islands and virulence plasmids of bacterial plant pathogens. Curr. Top. Microbiol. Immunol 264:127-147.

Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol 35:1547-1549.



Kvitko, B. H. and Collmer, A. 2023. Discovery of the Hrp type III secretion system in phytopathogenic bacteria: how investigation of hypersensitive cell death in plants led to a novel protein injector system and a world of inter-organismal molecular interactions within plant cells. Phytopathology 113:626-636.


Kwon, S.-K., Park, Y.-K. and Kim, J. F. 2010. Genome-wide screening and identification of factors affecting the biosynthesis of prodigiosin by Hahella chejuensis, using Escherichia coli as a surrogate host. Appl. Environ. Microbiol. 76:1661-1668.




Lee, H. K., Chun, J., Moon, E. Y., Ko, S. H., Lee, D. S., Lee, H. S. and Bae, K. S. 2001.
Hahella chejuensis gen. nov., sp. nov., an extracellular-polysaccharide-producing marine bacterium. Int. J. Syst. Evol. Microbiol. 51:661-666.


Letunic, I. and Bork, P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47:W256-W259.



Mukhtar, M. S., McCormack, M. E., Argueso, C. T. and Pajerowska-Mukhtar, K. M. 2016. Pathogen tactics to manipulate plant cell death. Curr. Biol. 26:R608-R619.


Peart, J. R., Lu, R., Sadanandom, A., Malcuit, I., Moffett, P., Brice, D. C., Schauser, L., Jaggard, D. A. W., Xiao, S., Coleman, M. J., Dow, M., Jones, J. D. G., Shirasu, K. and Baulcombe, D. C. 2002. Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. U. S. A. 99:10865-10869.



Petnicki-Ocwieja, T., Schneider, D. J., Tam, V. C., Chancey, S. T., Shan, L., Jamir, Y., Schechter, L. M., Janes, M. D., Buell, C. R., Tang, X., Collmer, A. and Alfano, J. R. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 99:7652-7657.



Prithiviraj, B., Weir, T., Bais, H. P., Schweizer, H. P. and Vivanco, J. M. 2005. Plant models for animal pathogenesis. Cell. Microbiol. 7:315-324.


Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G. and Ausubel, F. M. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902.


Roy, D., Panchal, S., Rosa, B. A. and Melotto, M. 2013.
Escherichia coli O157:H7 induces stronger plant immunity than Salmonella enterica Typhimurium SL1344. Phytopathology 103:326-332.



Schulze-Lefert, P. and Panstruga, R. 2011. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 16:117-125.


Schwiesow, L., Lam, H., Dersch, P. and Auerbuch, V. 2015.
Yersinia type III secretion system master regulator LcrF. J. Bacteriol. 198:604-614.




Senthil-Kumar, M. and Mysore, K. S. 2013. Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu. Rev. Phytopathol. 51:407-427.


Snellings, N. J., Popek, M. and Lindler, L. E. 2001. Complete DNA sequence of Yersinia enterocolitica serotype 0:8 low-calcium-response plasmid reveals a new virulence plasmid-associated replicon. Infect. Immun 69:4627-4638.




Üstün, S., Müller, P., Palmisano, R., Hensel, M. and Börnke, F. 2012. SseF, a type III effector protein from the mammalian pathogen Salmonella enterica, requires resistance-gene-mediated signalling to activate cell death in the model plant Nicotiana benthamiana. New Phytol. 194:1046-1060.



Wurtzel, O., Yoder-Himes, D. R., Han, K., Dandekar, A. A., Edelheit, S., Greenberg, E. P., Sorek, R. and Lory, S. 2012. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog 8:e1002945.



- TOOLS



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




