Development of a Selective Medium for Surveillance of Fusarium Head Blight Disease
Article information
Abstract
Fusarium head blight (FHB), predominantly caused by Fusarium graminearum and F. asiaticum, is a significant fungal disease impacting small-grain cereals. The absence of highly resistant cultivars underscores the need for vigilant FHB surveillance to mitigate its detrimental effects. In 2023, a notable FHB outbreak occurred in the southern region of Korea. We assessed FHB disease severity by quantifying infected spikelets and grains. Isolating fungal pathogens from infected samples often encounters interference from various microorganisms. We developed a cost-effective, selective medium, named BGT (Burkholderia glumae Toxoflavin) medium, utilizing B. glumae, which is primarily known for causing bacterial panicle blight in rice. This medium exhibited selective growth properties, predominantly supporting Fusarium spp., while substantially inhibiting the growth of other fungi. Using the BGT medium, we isolated F. graminearum and F. asiaticum from infected wheat and barley samples across Korea. To further streamline the process, we used a direct PCR approach to amplify the translation elongation factor 1-α (TEF-1α) region without a separate genomic DNA extraction step. Phylogenetic analysis of the TEF-1α region revealed that the majority of the isolates were identified as F. asiaticum. Our results demonstrate that BGT medium is an effective tool for FHB diagnosis and Fusarium strain isolation.
Fusarium head blight (FHB), a devastating disease affecting small-grain cereal crops like wheat, barley, and maize, is predominantly caused by the Fusarium graminearum and F. asiaticum species (Dean et al., 2012; Goswami and Kistler, 2004; Xu and Nicholson, 2009). These fungi, part of the Fusarium graminearum species complex (FGSC), which includes at least 16 distinct species, are notorious for their ability to cause significant yield losses during severe epidemics and produce harmful mycotoxins such as deoxynivalenol, nivalenol, and zearalenone, posing risks to human and animal health (Pestka and Smolinski, 2005). Geographically, FGSC species, particularly F. graminearum and F. asiaticum, have a pronounced presence in Asia (O’Donnell et al., 2000, 2004). In Korea, F. asiaticum has been identified as the dominant species causing FHB outbreaks (Ahn et al., 2022; Choi et al., 2023; Lee et al., 2009). However, we still lack effective ways to reduce crop losses caused by FHB, mainly because there are few cultivars that resist this disease. This has necessitated extensive investigations into FHB to develop effective management strategies. Changes in climate and agricultural practices have intensified both the frequency and severity of FHB outbreaks (Zhang et al., 2014). In 2021, an FHB outbreak was recorded in the southern region of South Korea (Choi et al., 2023). This trend continued, and in 2023, another significant outbreak in the same region prompted our in-depth evaluation of the disease’s severity.
Isolation of fungal pathogens, such as Fusarium spp., from infected plants or environmental samples is crucial for various research areas, including population genetics, disease diagnosis, and disease forecasting. Over the years, many researchers have concentrated on developing selective media for Fusarium spp. The peptone-pentachloronitrobenzene (PCNB) medium is among the best-known and most widely used for this purpose (Papavizas, 1967). Variants of this medium include the Nash and Snyder medium, which also contains PCNB, and other formulations like Komada’s medium and Rose Bengal-glycerine-urea medium (Komada, 1975; Leslie and Summerell, 2006; Nash and Snyder, 1962). However, due to safety concerns associated with PCNB, alternative selective inhibitors have been explored. These include dichloran, a combination of iprodione and dichloran, and malachite green (Ashiq et al., 2023; Leslie and Summerell, 2006). A noteworthy development in selective media for Fusarium, specifically F. graminearum, involved using toxoflavin, a bacterial toxin produced by the rice pathogen Burkholderia glumae (Jung et al., 2013; Shahjahan et al., 2000). This toxin effectively inhibits a wide range of fungal genera, including Aspergillus, Colletotrichum, and Penicillium, while Fusarium species, particularly F. graminearum, exhibit high resistance (Jung et al., 2013). Yet, the high cost of toxoflavin has been a limiting factor for producing this medium in large quantities. To overcome this, we developed a novel, cost-effective selective medium using the B. glumae strain, which we have designated as the BGT (Burkholderia glumae Toxoflavin) medium. This medium facilitates efficient isolation of Fusarium spp. suppressing the growth of other microorganisms.
In this study, we utilized the BGT medium to isolate FGSC strains from wheat and barley samples collected during the 2023 FHB outbreak in Korea. To better understand the geographical distribution of the causal agents, we isolated 60 FGSC strains from 115 cereal crop samples collected from 18 sites in this region. Phylogenetic analysis of the translation elongation factor-1α (TEF-1α) region successfully identified the species, revealing the distribution pattern of FGSC strains across Korea. Our findings suggest that this newly developed medium could significantly enhance the efficiency and cost-effectiveness of Fusarium strain isolation, contributing to better management of FHB.
Materials and Methods
Evaluation of FHB disease severity in wheat and barley fields
We assessed the severity of FHB in wheat and barley across various cities and towns in the southern region of Korea. The study areas included Iksan, Gimje, Buan, and Gunsan in Jeollabuk-do; Yeongam, Haenam, Gangjin, Boseong, and Jangheung in Jeollanam-do; Sacheon, Goseong, Jinju, Uiryeong, Hapcheon, and Miryang in Gyeongsangnam-do; as well as Gyeongju, Yeongdeok, and Daegu in Gyeongsangbuk-do. In each location, we selected six to nine fields to assess FHB severity and collect samples. Within each field, we randomly chose five points using a circular frame of 30 cm in diameter. At these points, we counted the total number of wheat and barley spikes within the frame. The severity of FHB was determined by quantifying the affected spikes and grains in both crops. We analyzed the collected data according to the Agricultural Science and Technology Research Analysis Criteria provided by the Rural Development Administration in 2012 (Rural Development Administration, 2012). Further details on the severity of FHB disease, including raw data, are presented in Supplementary Table 1.
Preparation of BGT medium
We used the BGT medium for the specific isolation of Fusarium strains. This medium was prepared by inoculating the B. glumae BGR1 strain, which was generously provided by Dr. Eunhye Goo at Seoul National University, into 5 ml of Luria-Bertani (LB) broth in a test tube (Jeong et al., 2003). We incubated this culture at 37°C overnight with shaking to promote growth. Next, 150 μl of the bacterial culture was spread on a 90 mm diameter LB agar plate (20 ml in volume). This plate was then incubated at 37°C for 24 h to allow for abundant toxoflavin production. Finally, we overlaid the agar plate with 15 ml of potato dextrose agar (PDA) supplemented with 0.1% lactic acid and stored the prepared medium at 4°C, using it within one week.
Selectivity test of the BGT medium
We performed a selectivity test of the BGT medium against a range of fungal plant pathogens, including F. graminearum, F. oxysporum, F. venenatum, Botrytis cinerea, Alternaria alternata, Magnaporthe oryzae, and Colletotrichum scovillei. The PDA supplemented with 0.1% lactic acid (acidified PDA, abbreviated as APDA) was used as a control medium. Plant pathogenic fungi were inoculated on both BGT and APDA plates, then incubated at 25°C in darkness for four days. Further details about the fungal strains are provided in Supplementary Table 2.
Fungal isolation from wheat and barley seeds
In 2023, we isolated fungal strains from barley and wheat seeds, which were harvested from various fields in the southern region of Korea. From each of the 115 wheat and barley fields sampled, we randomly selected five seeds for analysis. The seeds underwent surface sterilization in 1% sodium hypochlorite for one minute, followed by a rinse in sterile distilled water for another minute. Excess water was then gently blotted off using autoclaved paper towels. The sterilized seeds were placed on either BGT plates or APDA and incubated at 25°C for three days. Following incubation, the fungal colony that grew on the surface was transferred onto a fresh APDA plate. Subsequently, the fungal isolate was subcultured onto a fresh PDA plate using the hyphal-tip transfer method (Leslie and Summerell, 2006). Fungal strains were stored in a 20% glycerol solution at −80°C. To assess the prevalence of Fusarium strains, we focused on 20 samples from fields with high infected grains of FHB (as detailed in Supplementary Table 1). Twenty seeds from each of these selected samples were surface sterilized and then placed on BGT plates (5 seeds per plate). After incubating at 25°C for three days, we counted the Fusarium colonies that had grown on the BGT plates, as illustrated in Supplementary Fig. 1.
PCR amplification and sequencing
We subjected the fungal isolates to PCR amplification, targeting the TEF-1α and the internal transcribed spacer (ITS) regions (O’Donnell et al., 1998; Schoch et al., 2012). The oligonucleotide primer pairs EF1T/EF2T for TEF-1α and ITS5/ITS4 for ITS were synthesized by Bioneer (Daejeon, Korea). We employed a direct PCR approach, which allows the amplification of targeted regions without the need for separate fungal DNA extraction (Jeon et al., 2023a). For the amplification reaction, we prepared a total volume of 20 μl using AccuPower PCR Premix (Bioneer), which included 19 μl of distilled water and 0.5 μl of each primer (20 μM). A small sample of mycelium from each fungal isolate grown on PDA was added directly to the reaction mixture using a sterilized pipette tip (Jeon et al., 2023b). The amplification process involved a heat shock treatment to lyse the fungal cells, followed by rapid cooling on ice with vigorous agitation. Post heat shock, the samples were placed in a thermal cycler for the PCR reaction. The amplified PCR products, along with 5 μl of Lambda DNA/HindIII Marker (Thermo Scientific, Waltham, MA, USA), were analyzed on a 1.5% (w/v) agarose gel using Tris-acetate-EDTA buffer. Following electrophoresis, the PCR products were purified using the α+ Solution GEL/PCR Purification Kit (Alphagen, Changzhi, Taiwan). The purified products were then sent to Bioneer for sequencing, using the same oligonucleotides as in the PCR amplification.
Data analysis
The DNA sequences obtained from the fungal isolates were first processed using SeqMan software (DNASTAR Inc., Madison, WI, USA) for trimming. These sequences were then compared against the NCBI GenBank database through a BLAST search to identify the species. For accurate species identification, especially for F. graminearum and F. asiaticum, we performed phylogenetic analysis using the TEF-1α sequences. Reference sequences for F. graminearum (NRRL 5883 and NRRL 6394), F. asiaticum (NRRL 6101 and NRRL 13818), F. boothii (NRRL 29011 and NRRL 29105), and F. pseudograminearum (NRRL 28062) were obtained from NCBI GenBank (O’Donnell et al., 2000). We aligned the TEF-1α sequences using MEGA11 software and constructed a phylogenetic tree through maximum likelihood analysis. This analysis utilized the general time-reversible model with invariant sites and gamma distribution (GTR + GAMMA + I) with 1,000 bootstrap replicates, using RAxML software (Edler et al., 2021). The sequence data presented in the study are deposited in the NCBI repository, accession numbers from OR961135 to OR961194.
In addition to molecular analysis, we quantified the impact of FHB on wheat and barley crops. The FHB index was calculated using Microsoft Excel (Microsoft, Redmond, CA, USA) by analyzing the number of grains and spikelets affected by FHB. The conversion index was determined by multiplying the percentage of infected spikelets and grains per spike, and then dividing by 100. To illustrate the correlation between the FHB index and the isolation frequencies of Fusarium strains, we generated a trend line graph using Microsoft Excel and GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA). Furthermore, a scientific illustration to represent our findings was created with BioRender (https://www.biorender.com).
Results
Assessment of FHB disease severity and isolation of FGSC strains using a BGT medium in southern region of Korea
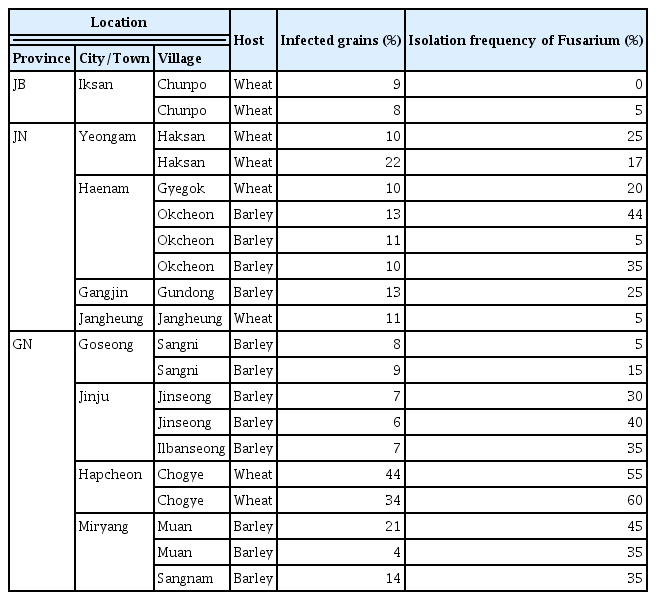
We evaluated FHB disease severity and isolated FGSC strains from 115 fields in the southern region of Korea, spanning 18 cities and towns across four provinces (see Fig. 1A, Supplementary Table 1). Of these fields, 42 were wheat cultivation fields, and the remainder were used for barley. A breakdown by province showed that Jeollabuk-do (JB), Jeollanam-do (JN), and Gyeongsangnam-do (GN) had both barley and wheat fields, while Gyeongsangbuk-do (GB) had only barley fields (Fig. 1B). We measured the number of spikes and grains affected by FHB in each field to calculate the FHB indices (Fig. 1C). The FHB indices showed significant impact in wheat and barley fields in GN (Supplementary Table 1). In particular, the percentage of infected barley spikes in Muan-myeon, Miryang-si, GN was 30%, which was much higher than the average of 5% (Table 1). Three wheat fields in Chogye-myeon, Hapcheon-gun, GN showed the highest percentage of infected grains, with over 20% (Table 1).
Distribution and severity of Fusarium head blight (FHB) disease in southern Korea. (A) A map displaying the southern region of Korea, marking 18 collection sites within four provinces: Jeollabuk-do (JB), Jeollanam-do (JN), Gyeongsangbuk-do (GB), and Gyeongsangnam-do (GN). (B) Pie chart showing the distribution of hosts. Adjacent bar graphs represent the relative abundance of hosts in each province, with numerical values indicating the actual number of the strains. (C) Box plots showing the severity of FHB disease, measured by the percentage of infected spikes and grains in various provinces. The percentage of infected spikes was calculated by averaging the results from five random points within each site. The percentage of infected grains was calculated by averaging the results from five infected spikes. Statistical significance is denoted by asterisks, with ‘ns’ indicating non-significant differences. **P < 0.01, ***P < 0.001.
Selectivity test against fungal representatives using a BGT medium
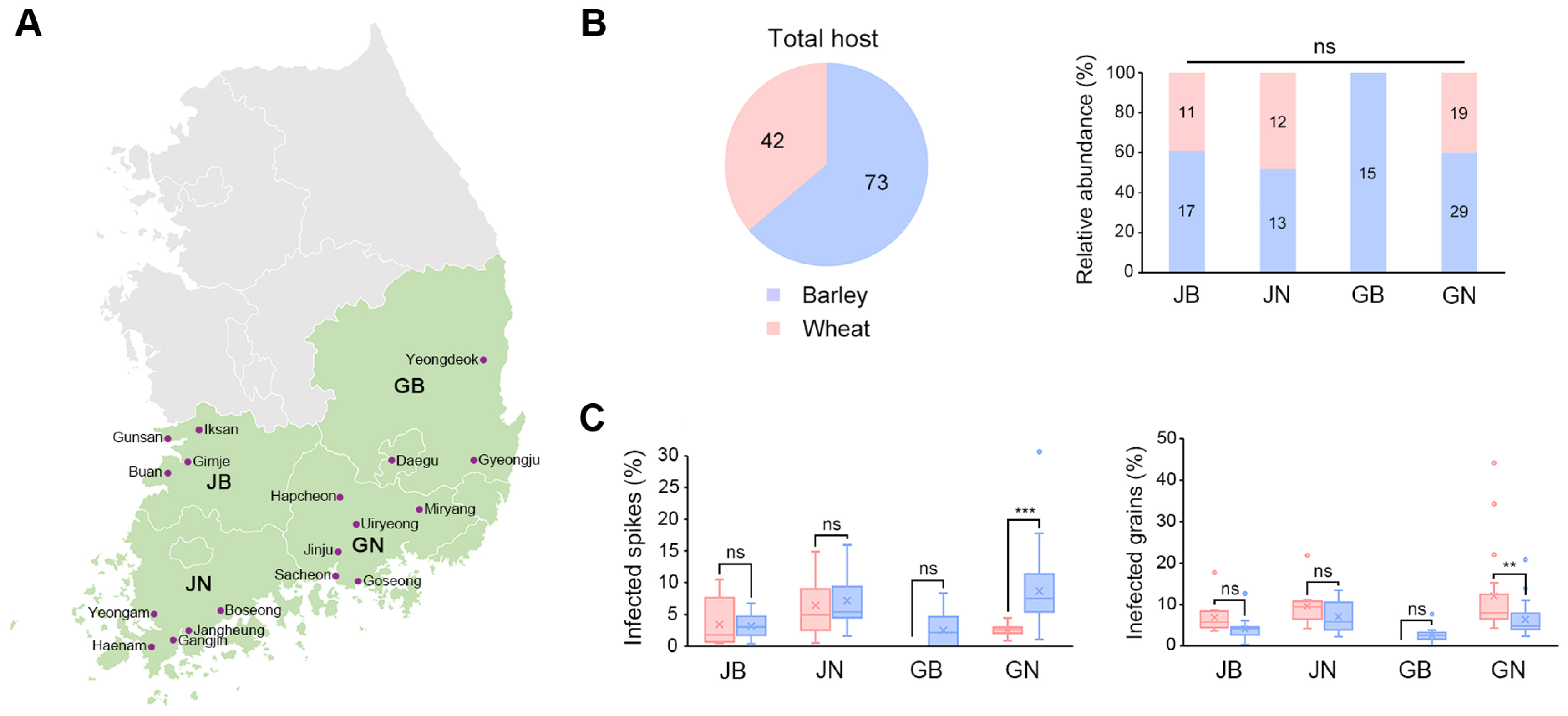
Previous studies developed a selective medium for Fusarium spp. that utilized the resistance of these species to toxoflavin produced by B. glumae, while other fungi were susceptible (Jung et al., 2013, 2018). Despite its efficiency in isolating Fusarium spp., the practical use of this medium was limited by the high cost of toxoflavin. To address this, we developed a new BGT medium using the B. glumae BGR1 strain to produce toxoflavin (experimental scheme in Fig. 2A). We conducted a selectivity test on the BGT medium against various fungal strains, including F. graminearum, F. oxysporum, F. venenatum, B. cinerea, A. alternata, M. oryzae, and C. scovillei. The results showed that Fusarium species grew well on the BGT medium, while other fungal strains exhibited no or significantly reduced growth compared to their growth on control APDA medium (Fig. 2B). Fig. 2C highlights the effectiveness of the BGT medium in selectively isolating Fusarium spp. from surface-sterilized seeds, in contrast to results on APDA.
Validation of the BGT (Burkholderia glumae Toxoflavin) medium for selectivity and efficacy in Fusarium isolation. (A) Diagram illustrating the preparation of BGT medium, starting from spreading Burkholderia glumae cultures on Luria-Bertani (LB) agar and overlaying with APDA. (B) Results of the test on various fungal species using BGT medium, demonstrating selective inhibition. Species tested include F. graminearum (Fg), F. oxysporum (Fo), F. venenatum (Fv), Botrytis cinerea (Bc), Alternaria alternata (Aa), Magnaporthe oryzae (Mo), and Colletotrichum scovillei (Cs). (C) Efficacy of BGT medium in isolating Fusarium spp. from surface-sterilized seeds as compared to APDA. The images showcase the differential growth patterns observed after a 3-day incubation at 25°C.
Phylogenetic analysis of FGSC isolates with BGT media and direct PCR approach
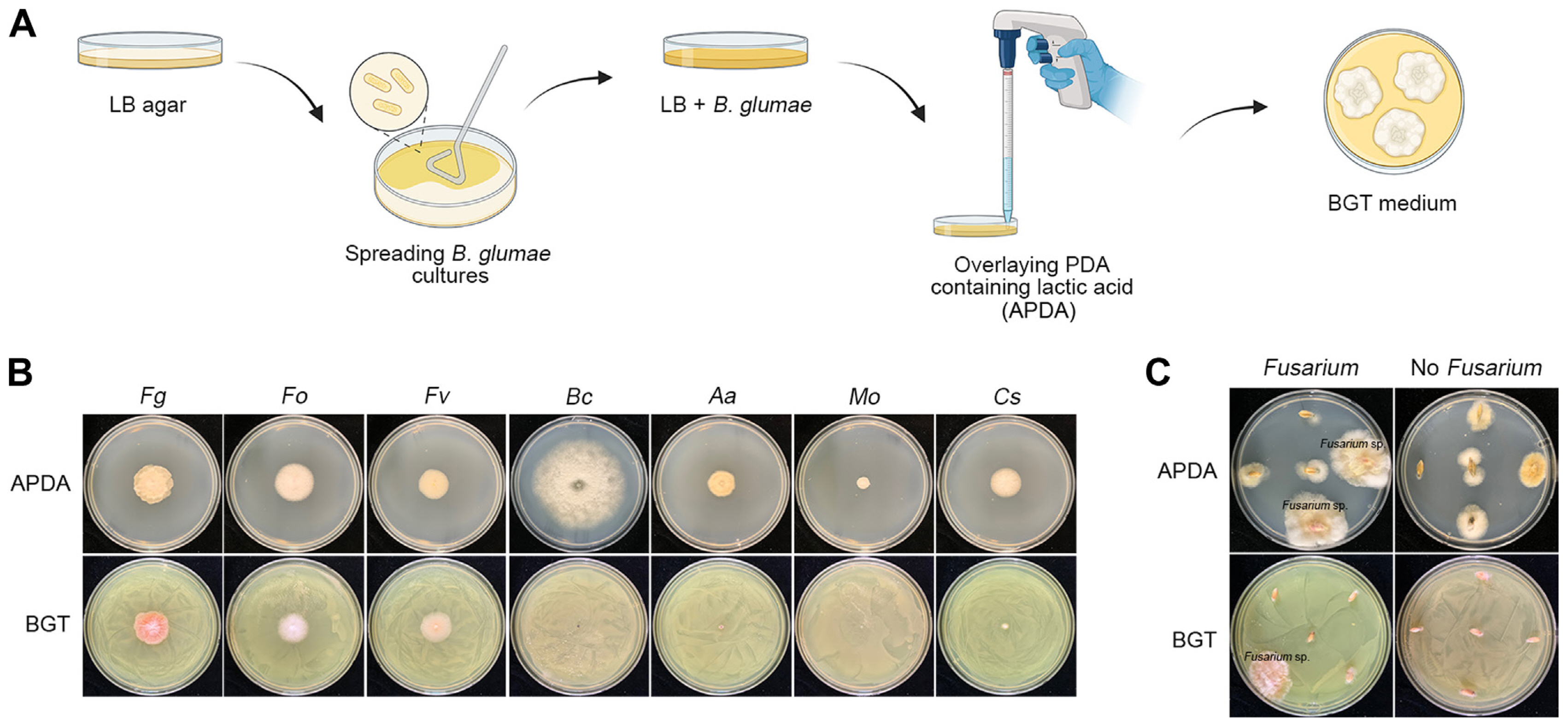
We used the BGT medium for the rapid and selective isolation of FGSC strains from 115 wheat and barley samples, successfully isolating 60 fungal strains. DNA sequences of the TEF-1α region were obtained using direct PCR amplification, bypassing the genomic DNA extraction process (Jeon et al., 2023a). Using BLAST analysis, 51 strains (85%) were identified as F. asiaticum, and the remaining nine strains (15%) as F. graminearum (Fig. 3A). Interestingly, eight F. graminearum strains were isolated from GN and one from GB; however, no F. graminearum strains were isolated in JB and JN. We conducted a phylogenetic analysis of these 60 FGSC strains based on their TEF-1α gene sequences (Fig. 3B). A phylogenetic tree was constructed with reference strains (O’Donnell et al., 2000) using maximum likelihood analysis to validate species identifications. The phylogenetic analysis divided the isolates into two distinct subclades corresponding to F. graminearum and F. asiaticum. Specifically, nine isolates (HJ3, HJ18, HJ25, HJ26, HJ29, HJ35, HJ38, HJ40, and HJ57) were categorized into the F. graminearum subclade, while the rest aligned with F. asiaticum reference strains. This distinction confirmed that the phylogenetic relationships of all isolates were consistent with the species identification obtained through the BLAST search.
Phylogenetic distribution and relative abundance of Fusarium graminearum species complex (FGSC) isolates. (A) The pie chart illustrates the distribution of FGSC isolates, showing that 85% are Fusarium asiaticum, while the remaining 15% are F. graminearum. Bar charts display the relative abundance of these FGSC strains from each host type (wheat and barley) and province (JB, Jeollabuk-do; JN, Jeollanam-do; GB, Gyeongsangbuk-do; and GN, Gyeongsangnam-do), with ‘ns’ indicating non-significant differences in abundance. (B) A phylogenetic tree representing FGSC isolates based on the translation elongation factor-1α (TEF-1α) gene sequence, as determined by maximum likelihood analysis using the GTR + I + G model. The analysis includes bootstrap validation with 1,000 replicates. Isolates are categorized into two main groups: F. graminearum and F. asiaticum, with the color-coding corresponding to each phylogenetic group. TEF-1α sequences of reference strains from GenBank are included for comparison.
Estimation of correlation between FHB index and incidence of Fusarium isolates
We utilized the BGT medium to examine the isolation frequencies of Fusarium strains from samples with high FHB disease indices (Table 1). Twenty such samples were collected from various locations including Iksan in JB; Yeongam, Haenam, Gangjin, and Jangheung in JN; and Goseong, Jinju, Hapcheon, and Miryang in GN. To evaluate the incidence of Fusarium isolates from these samples, we placed 20 surface-sterilized seeds from each sample onto BGT media. We then recorded the number of Fusarium strains that emerged after three days of incubation (details in Table 1, Supplementary Fig. 1). Notably, two samples from Hapcheon exhibited the highest values in both the FHB index of infected grains and the rate of Fusarium incidence, suggesting a potential correlation between FHB severity and Fusarium isolation frequency. To investigate this possibility further, we performed a correlation test between the FHB index and Fusarium isolation rates. Interestingly, the dot plot displayed an upward-sloping diagonal trend line, indicating that the BGT medium could be a valuable tool for exploring the dynamics of FHB disease (Fig. 4).

Correlation between Fusarium head blight (FHB) index and isolation frequency of Fusarium strains. This scatter plot depicts the correlation between the FHB index, determined by the percentage of infected grains in symptomatic spikelets, and the isolation frequencies of Fusarium strains from those grains. The isolation frequencies were calculated after placing infected seeds on a BGT (Burkholderia glumae Toxoflavin) medium and counting the Fusarium isolates that grew after a 3-day incubation period. The trend line indicates a positive correlation, with an R-value of 0.57, suggesting a moderate association between the extent of grain infection and the success rate of isolating Fusarium.
Discussion
The development of selective media for fungi focuses on promoting the growth of specific fungal groups, restricting the microbial contamination, and facilitating identification. Key considerations for these media include the stability and toxicity of the inhibitors used. Historically, PCNB has been effective in inhibiting a wide range of fungi, including zygomycetes, while allowing slow growth of Fusarium (Komada, 1975; Nash and Snyder, 1962; Papavizas, 1967). This has made it a popular choice for Fusarium isolation. However, a significant drawback is that most Fusarium species do not form distinctive colonies on PCNB media, necessitating further subculturing of all colonies (Leslie and Summerell, 2006). Moreover, concerns over PCNB’s potential carcinogenicity and the cancellation of its registration have spurred the development of alternative media, moving away from reliance on PCNB-based formulations (U.S. Environmental Protection Agency, 2018). An alternative to PCNB, malachite green, has been used in selective media (Vujanovic et al., 2002). However, malachite green has been identified as a multi-organ toxin in experimental animals and is known to lose antifungal activity upon light exposure (Culp et al., 2006; Vujanovic et al., 2002). The toxoflavin-based selective medium developed in the previous study showed a high selectivity for Fusarium species (Jung et al., 2013). However, using pure toxoflavin to make large quantities of medium is too expensive. Additionally, due to its acute toxicity (Nagamatsu, 2002), preparing media with toxoflavin poses safety concerns.
To address the challenges associated with traditional Fusarium-selective media, we have developed an innovative solution: the BGT medium. This medium leverages the ability of B. glumae to produce toxoflavin on agar plates (Jeong et al., 2003). After the initial growth of B. glumae, the plates are overlaid with PDA supplemented with lactic acid. The inclusion of lactic acid serves a dual purpose: it not only inhibits the growth of B. glumae but also other bacterial contaminants. Simultaneously, toxoflavin selectively inhibits the growth of a broad spectrum of fungi, except for Fusarium species. This unique combination facilitates the effective isolation of Fusarium spp., eliminating the need for potentially harmful chemicals or costly inhibitors. Our study demonstrated a correlation between the frequency of Fusarium isolation and the percentage of infected grains (Fig. 4). This finding underscores the selectivity and sensitivity of the BGT medium in isolating Fusarium species, making it a valuable tool in Fusarium research. A previous study (Jung et al., 2018) suggested that the resistance of F. graminearum to the oxidative stress caused by toxoflavin is mediated by triacylglycerides containing linolenic acid. Interestingly, the same study also found that B. glumae can physically attach to F. graminearum, facilitating the dispersal. To minimize the risk of B. glumae contamination, we implemented a subculturing process. Following the initial isolation on the BGT medium, the fungal isolates were transferred to fresh APDA. We then employed the hyphal-tip transfer method to further culture the isolates on fresh PDA plates. This step is crucial in ensuring the purity of the fungal cultures.
In this study, we successfully isolated FGSC strains from infected wheat and barley samples across Korea, utilizing the BGT medium. To enhance the efficiency of molecular identification, we employed a direct PCR approach. This technique allowed us to amplify the TEF-1α region directly from the fungal cells, bypassing the need for genomic DNA extraction—a step that often adds time and complexity to the process. The phylogenetic analysis of the TEF-1α region indicated the majority of FGSC isolates in Korea is F. asiaticum. This pattern is consistent with the results of our recent FHB study in 2021 (Choi et al., 2023). In the future, the utilization of the BGT medium, coupled with direct PCR amplification, is expected to streamline the diagnosis and population study of other Fusarium species across various fields.
Notes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Acknowledgments
This work was supported by the National Research Foundation of Korea (2022R1I1A1A01065138 and 2021R1C1C1004200) and the project PJ015741022024 of National Institute of Crop Science, Rural Development Administration, Republic of Korea. We thank Eunhye Goo for providing the B. glumae BGR1 strain.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).