 |
 |
| Plant Pathol J > Volume 40(2); 2024 > Article |
|
Abstract
Erwinia amylovora is a plant pathogen that causes fire blight on apples and pears. Bacteriophages, which are viruses that selectively infect specific species of bacteria and are harmless to animal cells, have been considered as biological control agents for the prevention of bacterial pathogens. In this study, we aimed to use bacteriophages that infect E. amylovora as biocontrol agents against fire blight. We isolated bacteriophages Fifi044 and Fifi318 infecting E. amylovora, and characterized their morphology, plaque form, and genetic diversity to use as cocktails for disease control. The stabilities of the two phages were investigated at various temperatures and pH values and under sunlight, and long-term storage experiment was conducted for a year. To evaluate whether the two phages were suitable for use in cocktail form, growth curves of E. amylovora were prepared after treating the bacterial cells with single phages and a phage cocktail. In addition, a disease control test was conducted using immature apples and in vitro cultured apple plantlets to determine the biocontrol effects of the phage cocktail. The two phages were morphologically and genetically different, and highly stable up to 50°C and pH value from 4 to 10. The phages showed synergistic effect when used as a cocktail in the inhibition of host bacterial growth and the disease control. This study demonstrated that the potential of the phage cocktail as a biocontrol agent for commercial use.
Erwinia amylovora, a Gram-negative bacterium belonging to the family Erwiniaceae, affects a wide range of host species within the family Rosaceae and is the causative agent of fire blight disease, which is a major global threat to commercial apple and pear production. The symptoms of fire blight, which are blackened and disintegrated tissues, were first observed in the United States in the 1780s, and the disease continues to spread worldwide (Denning, 1794). In Korea, it was first reported from a pear orchard in Anseong in 2015 (Park et al., 2016). Since 2015, when fire blight was first reported from 47 orchards, the infected areas have expanded, and the number of orchards affected by the disease has constantly increased (Lee et al., 2022). In particular, the disease affected 774 more orchards in 2020 owing to unusual weather conditions, including frequent rainfall and high humidity and temperature, in the spring and the negligence of workers (Lee et al., 2022). It suggests that the disease can recur if the environmental conditions are appropriate. Therefore, development of effective measures to control this disease is necessary.
In the absence of disease-resistant cultivars, antibiotics have become the best option for bacterial disease control in many pathological systems because they provide the most effective means of reducing bacterial populations and limiting disease outbreaks (Sundin and Wang, 2018). Streptomycin has been used extensively in the United States for more than 50 years to control fire blight outbreaks caused by E. amylovora. However, streptomycin-resistant isolates of E. amylovora were identified in pear orchards in California in 1971 (Miller and Schroth, 1972) and in Washington and Oregon in 1972 (Coyier and Covey, 1975). Moreover, reports of streptomycin-resistance have raised concerns about the sustainability of this antibiotic in the management of fire blight (Tancos et al., 2016). In addition, chemical agents such as copper compounds are not suitable for sustainable agriculture because of the environmental pollution caused by metal accumulation in the soil. Therefore, development of alternatives such as biocontrol agents, which can replace antibiotics and chemical agents without causing environmental problems, is necessary.
Bacteriophages are a type of viruses that infect bacteria and are widely present in nature wherever bacteria exist (Dion et al., 2020). As bacteriophages are generally believed to be environmentally safe and harmless to humans, animals, and plants, they are considered promising alternatives to antibiotics (Sillankorva et al., 2012). Clinical trials for bacteriophage therapy were approved by the Food and Drug Administration (FDA) in 2019 (Voelker, 2019). Moreover, various studies have been conducted on the application of phages in bacterial plant disease control (Flaherty et al., 2001; Gill and Abedon, 2003; Gill et al., 2003; Stefani et al., 2021). At OmniLytics in the United States, there are several commercial products named AgriPhage, comprising with phage cocktails that infect pathogens causing diseases in tomatoes, citrus, apples, pears, nuts, and stone fruits (OmniLytics, 2024).
In this study, we aimed to use bacteriophages that infect E. amylovora as biocontrol agents against fire blight. We selected two bacteriophages, Fifi044 and Fifi318, out of nine bacteriophages reported in a previous study based on their characteristics and host range for E. amylovora (Park et al., 2022). Both phages were identified to be members of Myoviridae, although analysis of the phylogenetic tree constructed using whole-genome sequences revealed that the phages were genetically different. Stability tests under diverse conditions and long-term storage tests were conducted to assess the feasibility of using the phages as biocontrol agents. The effectiveness of the phage cocktail was confirmed in vitro by analyzing growth curves of E. amylovora incubated with single phages and phage cocktails with different multiplicities of infection (MOIs). Additionally, a disease control test was conducted with the phage cocktail using immature apples and in vitro cultured apple plantlets.
In this study, E. amylovora strain YKB14808, isolated from a pear orchard in Seoun-myeon, Anseong-si, in 2019, was used to propagate phages and confirm the formation of plaques. The bacteria were stored in tryptic soy broth (TSB; Difco, Franklin Lakes, NJ, USA) media supplemented with 15% glycerol at −80°C. The bacteria were cultured on tryptic soy agar (Difco) medium and incubated at 28°C for 18 h. Subsequently, the bacterial cells from a single colony were cultured in 10 ml of TSB medium in a shaking incubator at 150 rpm and 28°C for 16 h, and the culture was used for further analysis.
The bacterial cells were cultured overnight, and then, the bacterial cells and bacteriophages were co-cultured (MOI, 0.001) in nutrient broth (NB; Difco) by incubating at 28°C for 18 h at 150 rpm. The liquid culture was centrifuged at 10,000 ×g for 10 min at 4°C, and the bacterial cells and other debris were removed by vacuum filtering the supernatant using a 500 ml Corning vacuum filter/storage bottle system (0.22 μm pore size, Corning Incorporated, Corning, NY, USA). To precipitate the phages, 1/5 volume of polyethylene glycol/NaCl solution (20% w/v PEG 8000; 2.5 M NaCl) was added to the filtrate, and the solution was stored at 4°C for 24 h. Subsequently, the filtrate was centrifuged at 10,000 ×g for 30 min, and the phage pellet was resuspended in SM (sodium chloride-magnesium sulfate) buffer (50 mM Tris-HCl, pH 7.5; 10 mM NaCl; 10 mM MgSO4). The phage titer was measured using the double-layer agar method, as previously described (Bae et al., 2012). The phage stock was stored at 4°C, and the SM buffer was replaced with sterile distilled water (SDW) when necessary, by ultrafiltration (Centricon, PL-70, 30,000 Da NMWL, Millipore, Billerica, MA, USA). High-quality pure phage particles were collected using CsCl-based purification. The phage lysates were centrifuged in CsCl buoyant density gradient at 78,500 ×g and 4°C for 2 h using an ultracentrifuge (Sorvall WX100, Sorvall Surespin-630 rotor, Thermo Fisher Scientific, Waltham, MA, USA). The tubes were punctured with a 20-gauge needle, and the white band obtained after ultracentrifugation, which comprised the phage particles, was collected.
The morphology of bacteriophages Fifi044 and Fifi318 was determined by transmission electron microscopy (TEM; LEO 912AB, LEO Electron Microscopy Inc., Thornwood, NY, USA). The phage stock purified using CsCl gradient ultracentrifugation was used for TEM analysis. Five microliters of the phage stock were loaded onto carbon-coated copper grids and negatively stained with 2% aqueous uranyl acetate (pH 4.0). The phages were examined using a TEM system (LEO 912 AB transmission electron microscope, Carl Zeiss, Wetzlar, Germany) at 80-kV accelerating voltage.
Bacteriophage DNA was extracted using a Norgen Phage DNA isolation kit (Norgen Biotek Corp., Ontario, Canada) according to the manufacturer’s instructions. Purified phage DNA was sequenced using a PacBio RS II sequencing system at Macrogen, Seoul, Korea. The complete dataset, including the genome information of the newly isolated phages and previously reported E. amylovora phages, was subjected to state-of-the-art genome-based analysis using the Virus Classification and Tree Building Online Resource (VICTOR; freely available at https://victor.dsmz.de).
To examine the stability of the bacteriophages at different temperatures and pH levels, the phages were treated under the specified conditions for 1 h. SM buffers with pH ranging from 4 to 11 were prepared by adjusting the pH with 1 M HCl and 1 M NaOH. The titer of the phages was measured by dotting serially diluted phage suspensions onto media that were overlaid with the host bacteria.
The stability of the phages in the presence of ultraviolet (UV) light was tested using a large-area illumination source (100 mW/cm2, Oriel Sol1A Class ABB Solar Simulator, Newport, Yongin, Korea) that could emit light whose spectrum was similar to the solar spectrum. The phage solution (2.5 ml) was added to a 24-well cell culture plate. A transparent quartz plate was placed on the 24-well plate, the plate was irradiated with light for 6 h, and samples were collected every hour to confirm phage activity.
Long-term stability tests of the phages were performed at different temperatures (−80, 4, 27, and 37°C). The initial phage titer was adjusted to 108 pfu/ml using SM buffer, and the phage titer was measured approximately every 30 days for 200 days. To prevent physical damage of phage particles because of the crystallization of water, 15% of glycerol was added to the samples stored at −80°C. Phage activity was measured using the double-layer agar method.
The persistence of infection with Fifi044 and Fifi318 was evaluated using E. amylovora strain YKB14808. Bacterial cells cultured overnight in TSB medium were re-inoculated into fresh TSB medium at 1/100th of the volume and incubated at 27°C with shaking at 150 rpm until the optical density at 600 nm (OD600 nm) reached 0.5 (approximately 1 × 108 cfu/ml). The bacterial suspension was centrifuged at 6,000 ×g for 3 min, and the pellet was washed twice with SDW and resuspended in SDW. The bacterial suspension was inoculated into NB medium (Difco) at 1/1,000 of the volume and divided into 180 μl aliquots in a 96-well plate. Serially diluted phage suspensions (20 μl) were added to the 96-well plate to prepare samples with different MOIs (0.01, 1, and 100). The OD600 nm value was monitored for 24 h at 1 h intervals using a microplate reader (Hidex, Turku, Finland).
To confirm the inhibitory effect of the bacteriophages on E. amylovora, a disease control test was conducted using E. amylovora strain YKB14808 that infected immature apples and in vitro cultured apple plantlets. E. amylovora YKB14808 cells were cultured in TSB overnight and then re-inoculated into fresh TSB and incubated at 28°C with shaking at 150 rpm until an OD600 nm value of 0.5 was obtained. The bacterial culture was centrifuged at 6,000 ×g for 3 min, and the pellet was washed with SDW and resuspended in the same volume of SDW. The bacterial suspension was used for the disease control tests.
To prepare the pathogen inoculum for the disease control tests conducted using immature apples, the prepared bacterial suspension was diluted 10 times with SDW (approximately 107 cfu/ml). Immature apples (Malus pumila var. dulcissima) were sterilized with 70% ethanol. The apples were pierced using a cork borer to create small holes of diameter 5 mm. The immature apples were placed on a plant culture dish (100 mm diameter; SPL Life Sciences, Pochen, Korea) that was covered with tissue paper. To maintain humidity, 5 ml SDW was added to the plant culture dishes. Subsequently, 20 μl of the disease control agents, namely, phages (109 pfu/ml), phage cocktail (109 pfu/ml), antibiotic agent (Buramycin, 50 μg/ml; Farm Hannong, Seoul, Korea), and biological agent (AgriPhage-Fire blight, 109 pfu/ml; Kyungnong, Seoul, Korea), were dropped into the holes made in the apples, and then, the apples were inoculated with 10 μl of the pathogen. The inoculated apples were incubated at 27°C for seven days, and disease severity was assessed according to the criteria mentioned in Supplementary Fig. 1. The total numbers of the plants used in each treatment are shown in Table 1.
To prepare the pathogen inoculum for conducting disease control tests using in vitro cultured apple plantlets (M9 cultivar), the prepared bacterial suspension was diluted 1,000-fold using SDW (approximately 105 cfu/ml). Subsequently, 1 ml suspensions of the disease control agents, namely, phages (108 pfu/ml), phage cocktail (108 pfu/ml), antibiotic agent (Buramycin, 50 μg/ml), and AgriPhage-Fire blight (108 pfu/ml), were sprayed on the plantlets; after 2 h, the plantlets were spray-inoculated with 1 ml suspensions of the pathogen. The plantlets were incubated at 25°C for 2 weeks, and disease severity was assessed according to the criteria mentioned in Supplementary Fig. 2. The total numbers of the plants used in each treatment are shown in Table 2.
Since the group size was small, Kruskal-Wallis test was used for statistical analyses using the R software (version 4.1.3, R Foundation for Statistical Computing, Vienna, Austria). Bonferroni-Dunn test was used to determine which groups had statistically significant differences. To compare the disease severity obtained for the water treatment with those obtained for the other treatments, the Wilcoxon rank-sum test was conducted using R. The significance level was set at P < 0.05.
In a previous study, nine E. amylovora-specific phages were isolated from apple and pear orchards where fire blight was observed in 2020 (Park et al., 2022). Of the nine phages, two phages, Fifi044 and Fifi318, were selected to test the synergistic effects of the phage cocktail since the two phages showed different characteristics including plaque morphologies, restriction fragment length polymorphism patterns, and host range (Park et al., 2022). Additionally, the two phages supplemented their infectivity against some E. amylovora strains each other.
To identify the phages, morphological and genomic characteristics was investigated. TEM analysis revealed that both phages were composed of a head and a contractile tail, which suggested that the phages belonged to Myoviridae (Fig. 1). However, morphological differences were observed between these two phages. Phage Fifi044 had a head diameter of 65 nm and a tail length of 134 ± 10 nm. In contrast, phage Fifi318 had a head diameter of 72 nm and a tail length of 116 ± 10 nm. Since the size of the phages remained constant within the TEM images, and since phage Fifi318 exhibited a larger head and phage Fifi044 displayed a longer tail, the two phages were morphologically distinct.
A phylogenetic tree was constructed based on the whole-genome sequences of 41 phages infecting Erwinia, including Fifi044 and Fifi318, to analyze the genetic differences between the phages (Fig. 2). Fifi044 was similar to pEa_SNUABM_27 (accession no. MW349138), which was isolated from the Republic of Korea. Fifi318 was in the same group as phiEa21-4 (accession no. EU710883), vB EamM SunLIRen (accession no. MH426725), and phiEa104 (accession no. FQ482083), which were isolated from the United States and Canada, respectively (Fig. 2). When the similarity of genome sequences was assessed through alignment using BLAST on the National Center for Biotechnology Information platform, the genome of Fifi044 exhibited high similarity to pEa_SNUABM_27 (coverage, 96%; identity, 9.10%). In contrast, it did not align with the genome sequences of Fifi318 and phiEa21-4. The genome of Fifi318 exhibited high similarity to phiEa21-4 (coverage, 100%; identity, 98.49%), vB EamM SunLIRen (coverage, 99%; identity, 98.06%), and phiEa104 (coverage, 100%; identity, 98.14%). These results demonstrated that there are differences in the genetic characteristics between phage Fifi044 and Fifi318. To be used as a phage cocktail, the phages should be able to affect the target pathogen over as wide a range as possible and complement the infectivity of the host through different infection mechanisms (Kering et al., 2019). Therefore, these two phages, which show differences in various characteristics and complement each other in host range, have the potential to be used as a phage cocktail for disease control.
Investigating several stability factors of phages is important in determining the effectiveness of phages as biocontrol agents, such as treatment method, timing, environmental conditions (pH, temperature, and UV light), and accessibility to target bacteria (Hudson et al., 2005; Ly-Chatain, 2014). The stabilities of the two phages at various temperatures, pH levels, and UV light were examined to evaluate their potential applications in the field (Fig. 3). Both phages were treated at four different temperatures (30, 40, 50, and 60°C). The phages were stable at 30, 40, and 50°C, but lost their infective activity at 60°C within an hour (Fig. 3A and B). Although there are some phages that are highly stable in extremely high-temperature environments (Lin et al., 2011; Wagner et al., 2018), this may be the result of co-evolution of phages that infect bacteria that grow in particularly high-temperature environments, and is not a general situation. Not only E. amylovora phages but also most phages that infect other bacteria drastically lost their infective activity at temperature above 60°C (Park et al., 2018; Rahimi-Midani et al., 2018).
To examine the stability of the two phages under different pH conditions, both phages were treated with SM buffers whose pH ranged from 4 to 11 (Fig. 3C and D). Both the phages were stable over a wide pH range (5-10). However, the titers of the two phages slightly decreased at pH 4, especially for phage Fifi044. In contrast, at pH 11, which represents a highly alkaline condition, phage Fifi318 entirely lost its infective activity, whereas the phage Fifi044 tolerated in the condition. Nonetheless, both phages showed a high degree of stability unless extreme conditions were encountered with respect to temperature or pH.
UV irradiation can inactivate phage infectivity and impede the potential application of phages as biocontrol agents in the field (Born et al., 2015; Diston et al., 2012, 2014; Tyrrell, 1979). To confirm the stability of the phages under sunlight, including the UV spectrum, the phages were exposed to light of the same wavelength as that of the solar spectrum for 6 h using a solar simulator (Fig. 3E and F). Upon exposure to UV light for 6 h, the titer of phage Fifi044 gradually decreased from 7.2 to 5.6 log pfu/ml. The titer of phage Fifi318 also gradually decreased by log 1 point (10 times). The phage Fifi318 showed higher stability compared to that of phage Fifi044. Although the titer of the phages under UV light decreased in some degree, the phages maintained their infectivity even after being exposed to UV light for 6 h, which is comparable to the strongest level in the natural environment. Furthermore, upon exposure to environmental stressors such as UV light, phage particles that survive and subsequently encounter their host bacteria exhibit an increased likelihood of survival. Consequently, the phages Fifi044 and Fifi318 may exhibit a prolonged effect independent of the application timing within a day. However, application during periods of reduced sunlight, such as at sunset, is anticipated to enhance efficacy.
Long-term storage tests were performed at different temperatures to evaluate phage stability during distribution (Fig. 4). Both phages showed high stability at −80°C. In addition, the phages were stable at 4°C for 170 days, although the titers of the phages decreased slightly after 200 days (log pfu/ml values for Fifi044 and Fifi318 decreased by approximately 0.2 and 0.3 points, respectively). However, when the phages were stored at 27°C, the titers of both phages gradually decreased by 2 points of the log values after 200 days, which means that only 1% of the phages survived after 200 days. The titer of phage Fifi044 stored at 37°C decreased rapidly (more than 1 point of the log value per month), and the samples could not form plaques after 170 days. Moreover, the titer of phage Fifi318 stored at 37°C also decreased rapidly, but the rate of decrease was slower than that for phage Fifi044. In addition, the Fifi318 was detectable even after 200 days, although only 0.1% of the phages survived. These results suggest that both phages were stable for more than 6 months when stored at less than 4°C without any adjuvant. There is bound to be a certain amount of time between phages being produced and transported to consumers. Therefore, such tests that investigate the stability of phages for a long period of time would be a desirable example in phage research.
To examine the lytic activity and infection persistence of the phages, the growth curves of E. amylovora cells inoculated with the phages at different MOIs (0.01, 1, and 100) were prepared (Fig. 5). In samples inoculated with phage Fifi044, the bacteria began to grow 20 h after the inoculation, regardless of the MOI concentration. In contrast, in samples inoculated with phage Fifi318, the bacteria began to grow 16 h after the inoculation, but the growth was restricted when high concentrations of the phage were inoculated (Fig. 5C). When E. amylovora cells were inoculated with the phage cocktail, bacterial growth was delayed by up to 23 h after the inoculation, and the growth of the bacteria was insignificant until 24 h. Overall, the phage cocktail showed better performance than single phage treatments, and the phage cocktail at an MOI of 0.01 was sufficient for effective disease control. Since the phage concentration of commercial products can be easily adjusted without the purification of base phage stock, the lower MOI of the product set in the product, the more efficient the product productivity will be. In this study, 0.01 was the lowest MOI value tested with the phage cocktail. However, testing with MOI values lower than 0.01 is necessary to determine the optimal and lowest effective MOI concentration.
The phage cocktail showed better suppression of E. amylovora than the single phages in vitro. Therefore, to confirm the biocontrol effect of the phage cocktail in vivo, disease control tests were conducted using immature apples and in vitro cultured apple plantlets (Tables 1 and 2). We conducted preliminary experiments to analyze the appropriate pathogen concentration for the disease control test and determined that 107 and 105 cfu/ml of the pathogen were appropriate for immature apples and plantlets, respectively. Streptomycin, which is used as a control agent for fire blight disease, and a AgriPhage-Fire blight composed of five phages were used as the control of our phage cocktail. In immature apples, treatment with phage Fifi044 alone and the phage cocktail led to a significant level of disease suppression, whereas treatment with phage Fifi318 alone did not suppress the disease (Table 1). Although treatment with phage Fifi318 alone showed no disease control effect, the phage cocktail treatment showed a superior control effect compared with that of treatment with phage Fifi044 alone. This suggests that the two phages exerted complementary effects when used together, as expected. In addition, the disease control effect of the phage cocktail was better than that of the existing AgriPhage-Fire blight.
In in vitro cultured apple plantlets, the phage cocktail showed significant disease suppression effects, whereas single phage treatments did not (Table 2). Interestingly, the phage cocktail treatments showed a greater control effect than the antibiotic treatments, and fewer plants showed severe disease progression (index 4) when treated with the phage cocktail than that with the antibiotic treatments. To our knowledge, this is the first report of a phage cocktail prepared in Korea whose effect has been verified in vitro using immature apples and tissue culture-generated plantlets.
In conclusion, the phage cocktail comprising phages Fifi044 and Fifi318 showed significant disease control effects, and we believe that an effective and stable biocontrol agent can be produced using this cocktail because it showed great stability under diverse conditions. It is interesting to note that the two phages showed a greater effect when applied as a cocktail than when treated phage alone not only in vitro but also in planta, but additional research will be needed to determine why the two phages show a synergistic effect. Additionally, the process of selection of the members of the phage cocktail used in this study serves as a good precedent for future studies.
Acknowledgments
This work was supported by the Rural Development Administration (RDA; grant number RS-2020-RD008879).
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Fig. 1
Transmission electron micrographs of Erwinia phages Fifi044 (A) and Fifi318 (B). The microscope was operated at 80-kV. Scale bars = 100 nm.
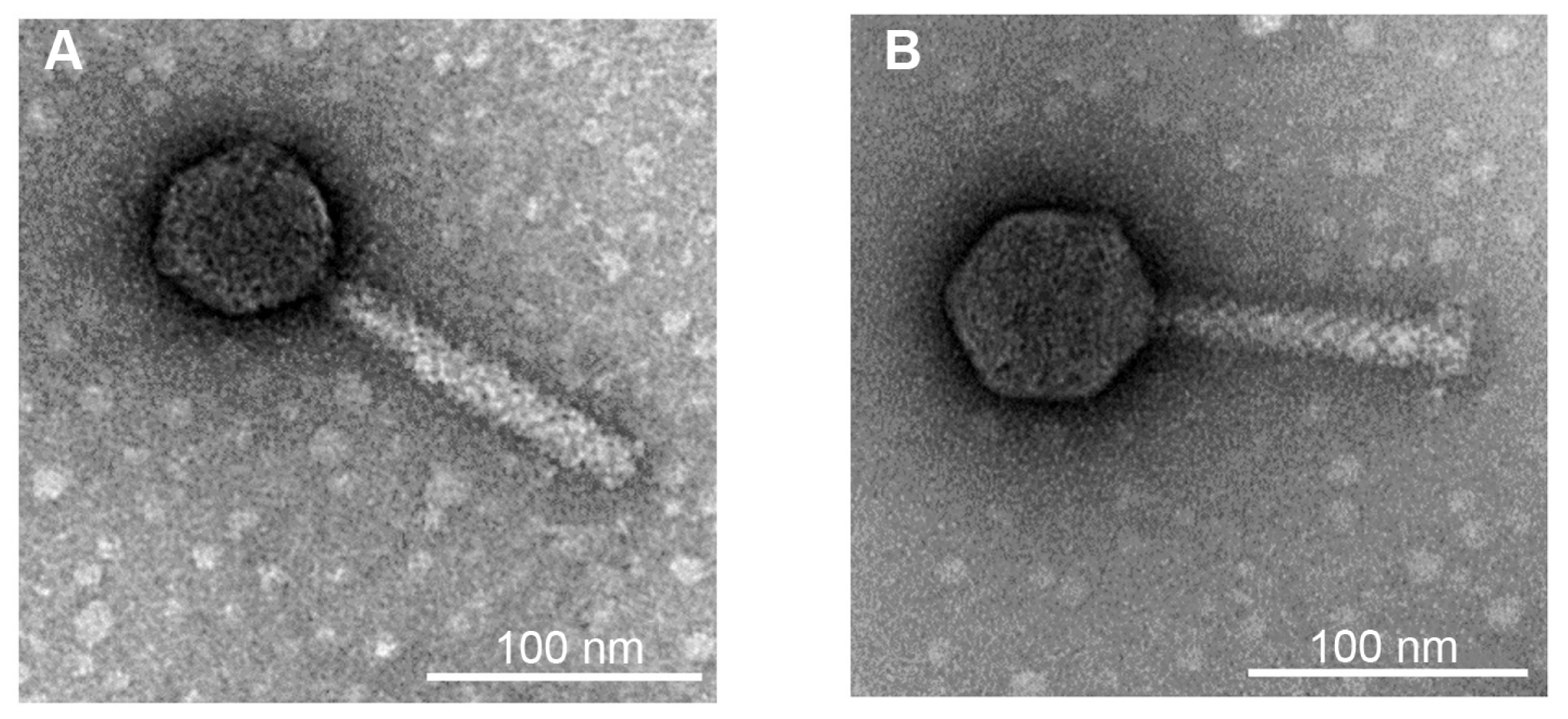
Fig. 2
Whole-genome based phylogenetic analysis of 41 Myoviridae bacteriophages infecting Erwinia amylovora. The genome sequences of 39 phages were obtained from GenBank database, and their names and accession numbers were stated in the figure. The tree was generated using VICTOR software for phages (prokaryotic viruses). The numbers above the branches represent Genome-BLAST Distance Phylogeny (GBDP) pseudo-bootstrap support values based on the conduction of replications. The origin countries of phages were indicated using different colors (United States, navy; Canada, green; Switzerland, yellow; South Korea, red; Belarus, gray; Latvia, black).
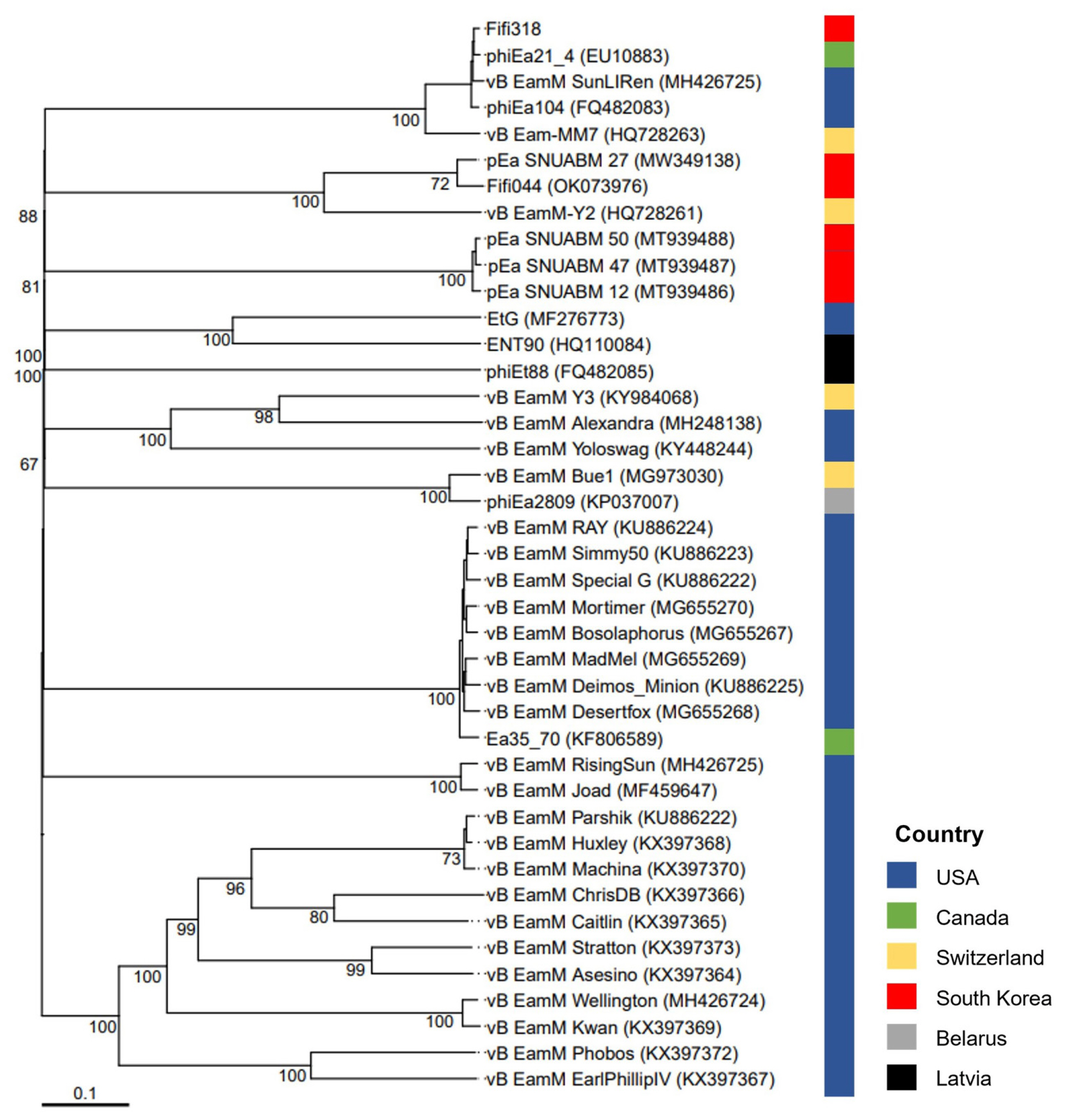
Fig. 3
Stability of Fifi044 and Fifi318 under 30-60°C (A, B), pH4-11 (C, D), and sun light condition (E, F). Different letters (a-b) on the bars indicate that the mean values differ significantly by Kruskal-Wallis test with Bonferroni-Dunn post hoc test at P < 0.05.

Fig. 4
Long-term storage tests of Fifi044 (A) and Fifi318 (B) at different temperatures. Colors ranged from black to white depending on the time when residual phages were measured. Different letters (a-g) on the bars indicate that the mean values differ significantly by Kruskal-Wallis test with Bonferroni-Dunn post hoc test at P < 0.05.
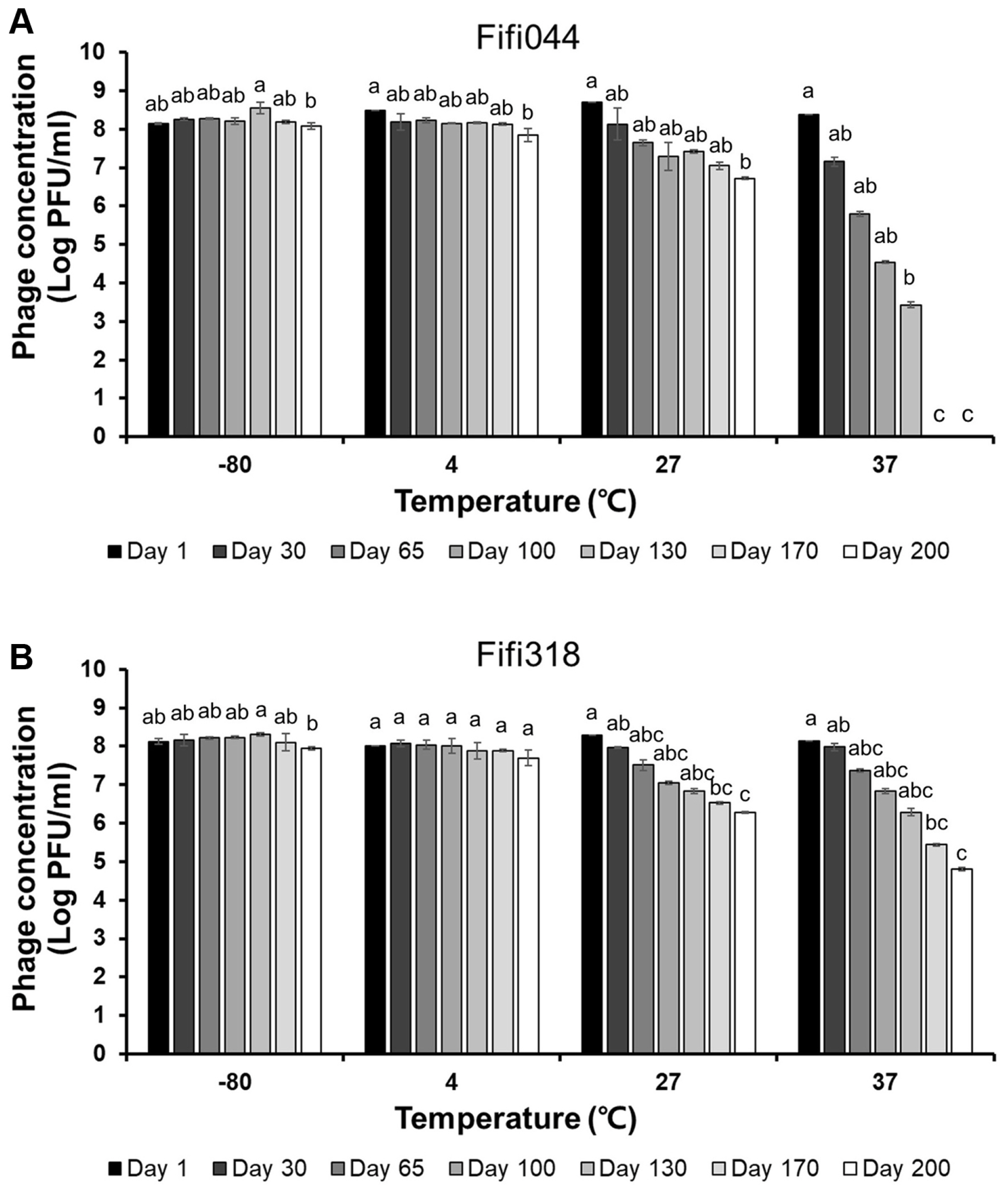
Fig. 5
Lytic activity of two bacteriophages and phage cocktail against Erwinia amylovora YKB14808. The growth curves of E. amylovora mixed with phages in different multiplicities of infection (MOI) were generated. MOI = 0.01 (A), MOI = 1 (B), MOI = 100 (C). Gray circles represent Fifi044, light gray squares represent Fifi318, and white circles represent phage cocktail. E. amylovora cultured without phages (black circle) were used as a control. Each experiment was performed three times with three technical replications. The error bars represent standard deviations.
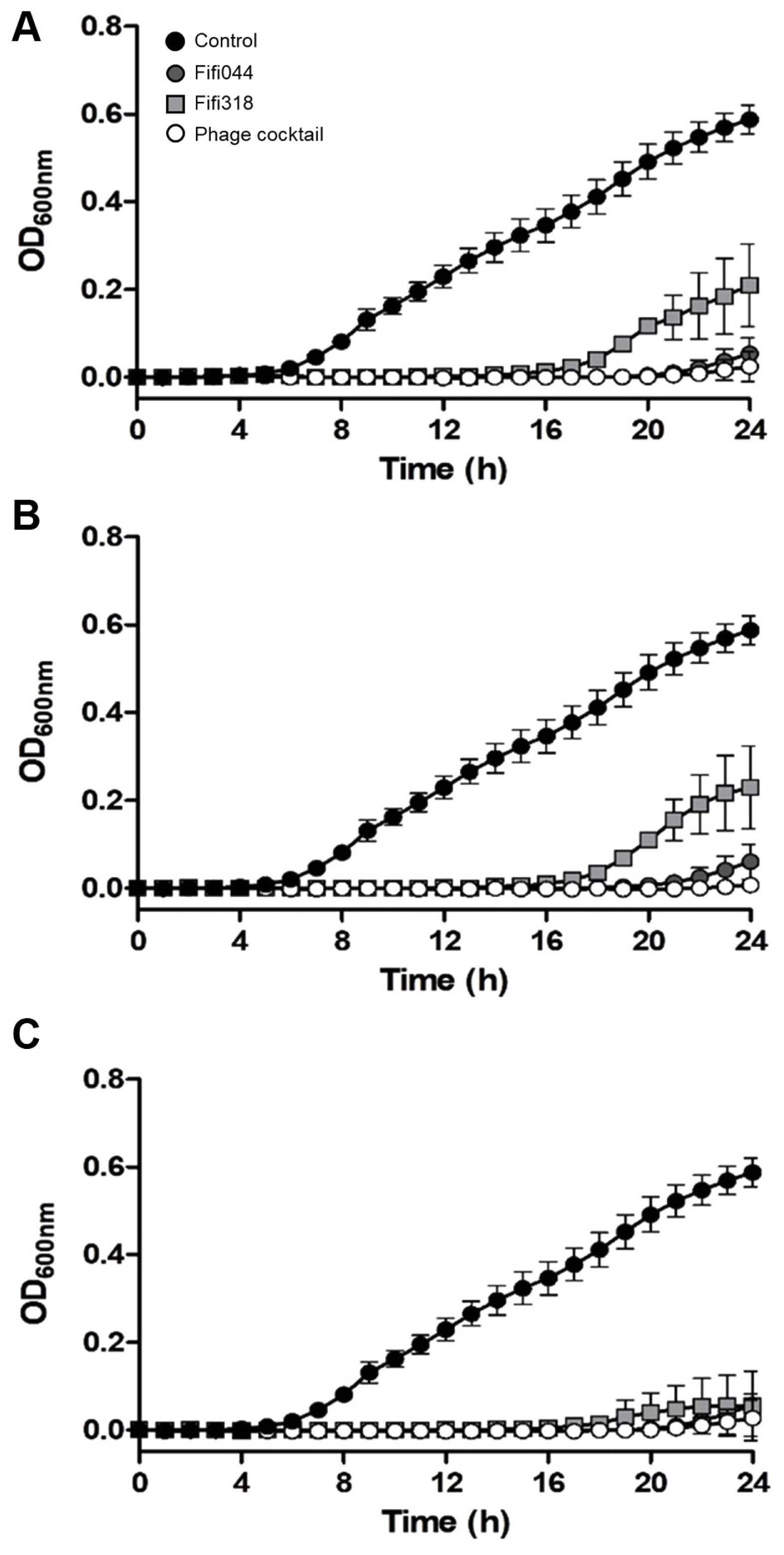
Table 1
The effect of Fifi044, Fifi318, and phage cocktail to control Erwinia amylovora in immature apples
| Symptom index | Percentage of tested plant tissue culture according to the severity of symptoms (No. of apples in each symptom index/Total number of apples in treatment group) | Mean value of pathogenic symptoms | |||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| 0 | 1 | 2 | 3 | 4 | 5 | ||
| Water treatment | 1.32 (1/76) | 15.79 (12/76) | 15.79 (12/76) | 36.84 (28/76) | 26.32 (20/76) | 3.96 (3/76) | 2.83 |
| Antibiotic agent (Buramycin) | 100.00 (58/58) | 0.00 (0/58) | 0.00 (0/58) | 0.00 (0/58) | 0.00 (0/58) | 0.00 (0/58) | 0.00*** |
| Biological agent (AgriPhage-Fire bligt) | 10.77 (7/65) | 35.38 (23/65) | 23.08 (15/65) | 16.92 (11/65) | 4.62 (3/65) | 9.23 (6/65) | 1.97*** |
| Fifi044 | 24.32 (18/74) | 14.86 (11/74) | 12.16 (9/74) | 21.62 (16/74) | 12.16 (9/74) | 14.86 (11/74) | 2.27* |
| Fifi318 | 12.33 (9/73) | 12.33 (9/73) | 15.07 (11/73) | 17.81 (13/73) | 27.40 (20/73) | 15.07 (11/73) | 2.81 |
| Phage cocktail | 29.33 (22/75) | 16.00 (12/75) | 13.33 (10/75) | 29.33 (22/75) | 9.33 (7/75) | 2.67 (2/75) | 1.81*** |
Table 2
The effect of Fifi044, Fifi318 and phage cocktail to control Erwinia amylovora in tissue culture plant (apple)
| Symptom index | Percentage of tested plant tissue culture according to the severity of symptoms (No. of plants in each symptom index/Total number of plants in treatment group) | Mean value of pathogenic symptoms | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| 0 | 1 | 2 | 3 | 4 | ||
| Water treatment | 36.17 (17/46) | 13.04 (6/46) | 15.22 (7/46) | 19.57 (9/46) | 27.78 (7/46) | 1.63 |
| Antibiotic agent (Buramycin) | 58.33 (28/48) | 22.92 (11/48) | 8.33 (4/48) | 4.17 (2/48) | 6.25 (3/48) | 0.77** |
| Biological agent (AgriPhage-Fire bligt) | 13.33 (6/45) | 20.00 (9/45) | 33.33 (15/45) | 24.44 (11/45) | 8.89 (4/45) | 1.95 |
| Fifi044 | 49.12 (28/57) | 21.05 (12/57) | 8.77 (5/57) | 19.30 (11/57) | 1.75 (1/57) | 1.04 |
| Fifi318 | 48.15 (26/54) | 5.56 (3/54) | 7.41 (4/54) | 27.78 (15/54) | 11.11 (6/54) | 1.48 |
| Phage cocktail | 64.91 (37/57) | 12.28 (7/57) | 12.28 (7/57) | 8.77 (5/57) | 1.75 (1/57) | 0.70** |
References
Bae, J. Y., Wu, J., Lee, H. J., Jo, E. J., Murugaiyan, S., Chung, E. and Lee, S.-W. 2012. Biocontrol potential of a lytic bacteriophage PE204 against bacterial wilt of tomato. J. Microbiol. Biotechnol. 22:1613-1620.


Born, Y., Bosshard, L., Duffy, B., Loessner, M. J. and Fieseler, L. 2015. Protection of Erwinia amylovora bacteriophage Y2 from UV-induced damage by natural compounds. Bacteriophage 5:e1074330.



Coyier, D. L. and Covey, R. P. 1975. Tolerance of Erwinia amylovora to streptomycin sulfate in Oregon and Washington. Plant Dis. Rep. 59:849-852.
Denning, W. 1794. On the decay of apple trees. Trans. Soc. Promot. Agric. Arts Manuf. 1:219-222.
Dion, M. B., Oechslin, F. and Moineau, S. 2020. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 18:125-138.



Diston, D., Ebdon, J. E. and Taylor, H. D. 2012. The effect of UV-C radiation (254 nm) on candidate microbial source tracking phages infecting a human-specific strain of Bacteroides fragilis (GB-124). J. Water Health 10:262-270.



Diston, D., Ebdon, J. E. and Taylor, H. D. 2014. Inactivation of bacteriophage infecting Bacteroides strain GB124 using UV-B radiation. Photochem. Photobiol. 90:622-627.

Flaherty, J. E., Harbaugh, B. K., Jones, J. B., Somodi, G. C. and Jackson, L. E. 2001. H-mutant bacteriophages as a potential biocontrol of bacterial blight of geranium. HortScience 36:98-100.

Gill, J. and Abedon, S. T. 2003. Bacteriophage ecology and plants. APSnet Feature Advanced online publication. https://doi.org/10.1094/APSnetFeature-2003-1103.

Gill, J. J., Svircev, A. M., Smith, R. and Castle, A. J. 2003. Bacteriophages of Erwinia amylovora. Appl. Environ. Microbiol. 69:2133-2138.




Hudson, J. A., Billington, C., Carey-Smith, G. and Greening, G. 2005. Bacteriophages as biocontrol agents in food. J. Food Prot. 68:426-437.



Kering, K. K., Kibii, B. J. and Wei, H. 2019. Biocontrol of phytobacteria with bacteriophage cocktails. Pest Manag. Sci. 75:1775-1781.



Lee, H. J., Lee, S. W., Suh, S.-J. and Hyun, I.-H. 2022. Recent spread and potenial pathways for fire blight in South Korea. Bull. OEPP 52:135-140.
Lin, L., Han, J., Ji, X., Hong, W., Huang, L. and Wei, Y. 2011. Isolation and characterization of a new bacteriophage MMP17 from Meiothermus. Extremophiles 15:253-258.



Ly-Chatain, M. H. 2014. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 5:51.

Miller, T. D. and Schroth, M. N. 1972. Monitoring epiphytic population of Erwinia amylovora on pear with a selective medium. Phytopathology 62:1175-1182.

OmniLytics 2024 AgriPhage product-info URL https://www.agriphage.com/product-info/ [12 January 2024].
Park, D. H., Yu, J.-G., Oh, E.-J., Han, K.-S., Yea, M. C., Lee, S. J., Myung, I.-S., Shim, H. S. and Oh, C.-S. 2016. First report of fire blight disease on Asian pear caused by Erwinia amylovora in Korea. Plant Dis. 100:1946.

Park, J., Kim, B., Song, S., Lee, Y. W. and Roh, E. 2022. Isolation of nine bacteriophages shown effective against Erwinia amylovora in Korea. Plant Pathol. J. 38:248-253.




Park, J., Lee, G. M., Kim, D., Park, D. H. and Oh, C.-S. 2018. Characterization of the lytic bacteriophage phiEaP-8 effective against both Erwinia amylovora and Erwinia pyrifoliae causing severe diseases in apple and pear. Plant Pathol. J. 34:445-450.




Rahimi-Midani, A., Lee, Y. S., Kang, S.-W., Kim, M.-K. and Choi, T.-J. 2018. First isolation and molecular characterization of bacteriophages infecting Acidovorax citrulli, the causal agent of bacterial fruit blotch. Plant Pathol. J. 34:59-64.




Sillankorva, S. M., Oliveira, H. and Azeredo, J. 2012. Bacteriophages and their role in food safety. Int. J. Microbiol. 2012:863945.




Stefani, E., Obradović, A., Gašić, K., Altin, I., Nagy, I. K. and Kovács, T. 2021. Bacteriopahge-mediated control of phytopathogenic Xanthomonads: a promising green solution for the future. Microorganisms 9:1056.



Sundin, G. W. and Wang, N. 2018. Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 56:161-180.


Tancos, K. A., Villani, S., Kuehne, S., Borejsza-Wysocka, E., Breth, D., Carol, J., Aldwinckle, H. S. and Cox, K. D. 2016. Prevalence of streptomycin-resistant Erwinia amylovora in New York apple orchards. Plant Dis. 100:802-809.


- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 503 View
- 44 Download
- ORCID iDs
-
Eunjung Roh

https://orcid.org/0000-0003-2999-5817 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement
Supplement Print
Print



