The Transcription Cofactor Swi6 of the Fusarium graminearum Is Involved in Fusarium Graminearum Virus 1 Infection-Induced Phenotypic Alterations
Article information
Abstract
The transcription cofactor Swi6 plays important roles in regulating vegetative growth and meiosis in Saccharomyces cerevisiae. Functions of Swi6 ortholog were also characterized in Fusarium graminearum which is one of the devastating plant pathogenic fungi. Here, we report possible role of FgSwi6 in the interaction between F. graminearum and Fusarium graminearum virus 1 (FgV1) strain DK21. FgV1 perturbs biological characteristics of host fungi such as vegetative growth, sporulation, pigmentation, and reduction of the virulence (hypovirulence) of its fungal host. To characterize function(s) of FgSWI6 gene during FgV1 infection, targeted deletion, over-expression, and complementation mutants were generated and further infected successfully with FgV1. Deletion of FgSwi6 led to severe reduction of vegetative growth even aerial mycelia while over-expression did not affect any remarkable alteration of phenotype in virus-free isolates. Virus-infected (VI) FgSWI6 deletion isolate exhibited completely delayed vegetative growth. However, VI FgSWI6 over-expression mutant grew faster than any other VI isolates. To verify whether these different growth patterns in VI isolates, viral RNA quantification was carried out using qRT-PCR. Surprisingly, viral RNA accumulations in VI isolates were similar regardless of introduced mutations. These results provide evidence that FgSWI6 might play important role(s) in FgV1 induced phenotype alteration such as delayed vegetative growth.
Introduction
Over the many years, a number of mycoviruses have been reported in Fusarium graminearum (Chu et al., 2002, 2004; Cho et al., 2013) which is devastating plant-pathogenic fungi and a causal agent of Fusarium head blight (Son et al., 2011). Among them, one of the well characterized mycovirus is Fusarium graminearum virus 1 (currently named as FgV1) strain DK21. FgV1 infection causes reduced virulence (hypovirulence), delayed mycelial growth, increased pigmentation, and reduced mycotoxin production of the host fungus F. graminearum (Chu et al., 2002; 2004; Kwon et al., 2007). To understand mechanisms underlying these alterations by FgV1 infection, it is important to investigate roles of host factors involved in interaction between mycovirus and fungal host. For identifying putative individual genes or gene products involved in this interaction, comparison of gene expression profiling and proteomic analysis between virus-free (VF) and virus-infected (VI) isolates of F. graminearum using microarray, RNA-Seq, and two dimensional electrophoresis mediated protein analyses were conducted previously (Cho et al., 2012; Kwon et al., 2009; Lee et al., 2014). From the genome-wide transcriptional analysis, patterns of differentially expressed genes which might be related to FgV1 infection were identified between VF and VI isolates.
Morphological change of the fungal host by mycovirus infection is a natural result from complicated molecular biological process of the infected host fungus (Lee et al., 2014). Therefore, it is difficult to understand the mechanism involved in phenotypic alteration of infected host fungus. However, a phenome-based functional analysis of transcription factors (TFs) in F. graminearum enabled to estimate the effect of individual TF deletions especially in morphology (Son et al., 2011). Based on combination of these two studies, transcriptional co-factor F. graminearum SWI6 gene (FgSWI6; locus FGSG_04220) was selected to investigate its role(s) during FgV1–F. graminearum interaction especially on morphological changes caused by FgV1 infection.
The functions of SWI6 gene were previously reported in the model organism yeast. In Saccharomyces cerevisiae, SWI6 involves in regulation of meiotic initiation (Purnapatre et al., 2002). Protein product of SWI6 (Swi6) is major component of CLN3/SWI6/CLN2 pathway which is signaling pathway mediating nutrient environmental controls between growth and meiosis. In fission yeast, Saccharomyces pombe, Swi6 protein composes heterochromatin with HP1 protein and Swi6/HP1 can regulate initiation of replication in silenced chromatin (Hayashi et al., 2009). In the filamentous fungus F. graminearum, transcription cofactor FgSwi6 functions in growth as well as in both sexual and asexual developments (Liu et al., 2013; Son et al., 2011). Deletion of SWI6 gene causes growth defect, reduced production of perithecium and conidia. This gene is also required for cellulose utilization, lithium tolerance, and arginine-induced production of mycotoxin deoxynivalenol (DON) (Liu et al., 2013). Collectively, Swi6 plays critical roles in controlling vegetative growth in a variety of eukaryotes.
Similar with plant and animal viruses, mycoviruses require host factors for maintaining their infection cycle in host cell. Functions of various host factors have been characterized from several host fungi (Son et al., 2015). These host factors involved in alterations of fungal host biology, transmission of mycoviruses, accumulation of viral RNAs, and antiviral mechanism. In one of the model system for studying host fungus–mycovirus interaction, Cryphonectria parasitica–Cryphonectria hypovirus 1-EP713 (CHV1), the host gene NAM-1 modulates symptom induction in the fungus in response to CHV1 infection (Faruk et al., 2008). In F. graminearum, two host genes, HEX1 and HAL2, affecting accumulation of FgV1 viral RNA was reported (Son et al., 2013; Yu et al., 2015). Although much of the research concerning host factors has dealt with general functions in cell growth of fungi, our knowledge about host factors and their roles on mycovirus infection cycles are relatively poor.
The current paper concerns about cellular functions of SWI6 gene of the F. graminearum selected from comparative analysis using previous genome-wide transcriptional patterns and phenome based database. Although previous study already demonstrated cellular functions of FgSWI6 involved in vegetative growth, conidia production, and sexual development (Liu et al., 2013), we introduced targeted gene overexpression strain to strengthen our observations. Using these approaches, we confirmed that FgSwi6 affects vegetative growth and conidia production of F. graminearum. In addition, we provided evidence that the changes of FgSwi6 expression level caused by FgV1 infection might cause the colony morphological changes of infected host.
Materials and Methods
Fungal isolates and culture conditions
All fungal isolates used in this study (Table 1) were stored in 25% (v/v) glycerol at −80°C and were reactivated on potato dextrose agar (PDA; Difco; BD, Franklin Lakes, NJ, USA). For nucleic acid manipulation, all isolates of F. graminearum were grown in 50 ml of a liquid complete medium (CM; Son et al., 2013) at 25°C with shaking (150 rpm) for 5 days. Mycelia were harvested by filtration through miracloth (Calbiochem; EMD Millipore, Billerica, MA, USA).
Computational analysis
Nucleotide sequences from the NCBI database were assembled using the Seqman program in DNASTAR (http://www.dnastar.com). Sequence similarity searches of FgSWI6 and SWI6 homologs were conducted with the NCBI BLAST program. The alignment of FgSwi6 and Swi6 ortholog amino acid sequences was performed by the MegAlign program in DNASTAR, using a default setting and GeneDoc programs (http://www.nrbsc.org/gfx/genedoc/).
Genomic DNA extraction, primers, and PCR conditions
For extraction of genomic DNA, myceilia were ground to a fine powder with liquid nitrogen in a mortar and pestle and further used for extraction as previously described (Son et al., 2013). To construct a PCR fragment for deletion, over-expression, and complementation, a slightly modified double-joint PCR strategy was applied for fusion of PCR products (Son et al., 2013). The PCR construct for over-expression of the target gene was generated by the same procedure that was used for the deletion mutants. Geneticin (gen) and elongation factor 1α promoter were amplified from the pSKGEN plasmid (Son et al., 2013). For the complementation of deletion mutants, the hygromycin resistance gene cassette (hph) was amplified from the pBCATPH plasmid. General PCR was performed following the manufacturer’s instructions (Takara Korea Biomedical Inc., Seoul, Korea). The PCR primers used in this study were listed in Supplementary Table 1.
Fungal transformation for construction of targeted gene deletion, over-expression, and complementation mutants
Preparation of protoplasts of the wild type (WT)-VF isolate and detailed procedure of fungal transformation were conducted as previously described (Son et al., 2013) Transformants with resistance to geneticin or hygromycin B were selected on regeneration medium containing 150 μg/ml of geneticin (Duchefa, Harlem, Netherlands) or hygromycin B (Calbiochem; EMD Millipore). Selected transformants were infected by FgV1 using hyphal fusion-mediated virus transmission, and viral infection was confirmed by RT-PCR (described below). For the Southern blot hybridization of all mutants, we used same procedure as previously described (Son et al., 2013) The [32P]-labeled DNA probes were generated following standard techniques as previously described (Son et al., 2013). The hybridization reaction was performed at 65°C for 16 hours. After hybridization, unhybridized DNA probe was removed by washing with low (2× SSC and 0.1% SDS) and high (0.1× SSC and 0.1% SDS) stringency buffers. The blotting image was visualized using a Fuji BAS-2500 Phosphor Imager and corresponding imaging software (Fuji Film, Tokyo, Japan).
Radial growth, conidial production, and virulence test
Radial growth was measured on PDA, CM, and minimal medium (MM; 0.05% KCl, 0.2% NaNO3, 3% Sucrose, 1% KH2PO4, 0.05% MgSO4·7H2O, 0.02% trace element, 2% agar) from day to day as previously described (Son et al., 2013). To test the effect of FgSWI6 deletion and over-expression on the virulence of F. graminearum, wheat head florets at early-mid anthesis were inoculated with conidial suspensions as described (Son et al., 2013) on wheat cv. Jokyoung. The test was repeated three times. Statistical analysis was performed using IBM SPSS Statistics 23.0 software (IBM Co., Armonk, NY, USA).
RNA preparation and RT-PCR
For total RNA preparation, the powdered mycelia were suspended in Isol-RNA lysis reagent (5 PRIME Inc., Gaithersburg, MD, USA). Nucleic acid was extracted by following the manufacturer’s protocol with slight modification. The extracted total RNAs were purified twice with acid phenol:chloroform (1:1), precipitated with isopropanol, suspended in DEPC-treated water, and further treated with TURBO DNA-free™ (Ambion, Foster City, CA, USA) to remove genomic DNA. The cDNAs were synthesized with M-MLV reverse transcriptase (Promega, Madison, WI, USA) and oligo d(T) primer to quantify SWI6 mRNA expression and viral RNA accumulation. Quantitative real-time RT-PCR (qRT-PCR) was performed on a CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA, USA) using the SsoFast™ EvaGreen® Supermix (Bio-Rad) according to manufacturer’s instructions. After initial denaturation at 95°C for 10 minutes, 40 cycles consisted of 5 seconds at 95°C and 5 seconds at 58°C. Two endogenous reference genes, cyclophilin 1 (CYP1, locus FGSG_07439) and elongation factor 1α (EF1α, locus FGSG_08811), were used in each experiment.
Results
Amino acid alignment of F. graminearum Swi6 and its homologs
The deduced amino acid sequence of Fg-SWI6 genomic DNA and mRNA were investigated using oligonucleotide primers specific to FgSWI6 by PCR and RT-PCR. The 2424 bp coding region contains 55 bp and 49 bp introns, which can encode 807 amino acids and molecular weight of about 87.7 kDa. The protein coding region of FgSWI6 was predicted based on nt sequence of FgSWI6 mRNA (data not shown).
Computer-based comparison of the putative amino acid sequence of FgSWI6 showed high levels of sequence identity with Swi6 homologs from F. oxysporum (88%) and F. verticillioides (88%). In other filamentous fungus Neurospora crassa and Saccharomyces cerevisiae, sequence identities were lower than those of Fusarium species, i.e., N. crassa for 57% and S. cerevisiae for 19%, respectively (Fig. 1).
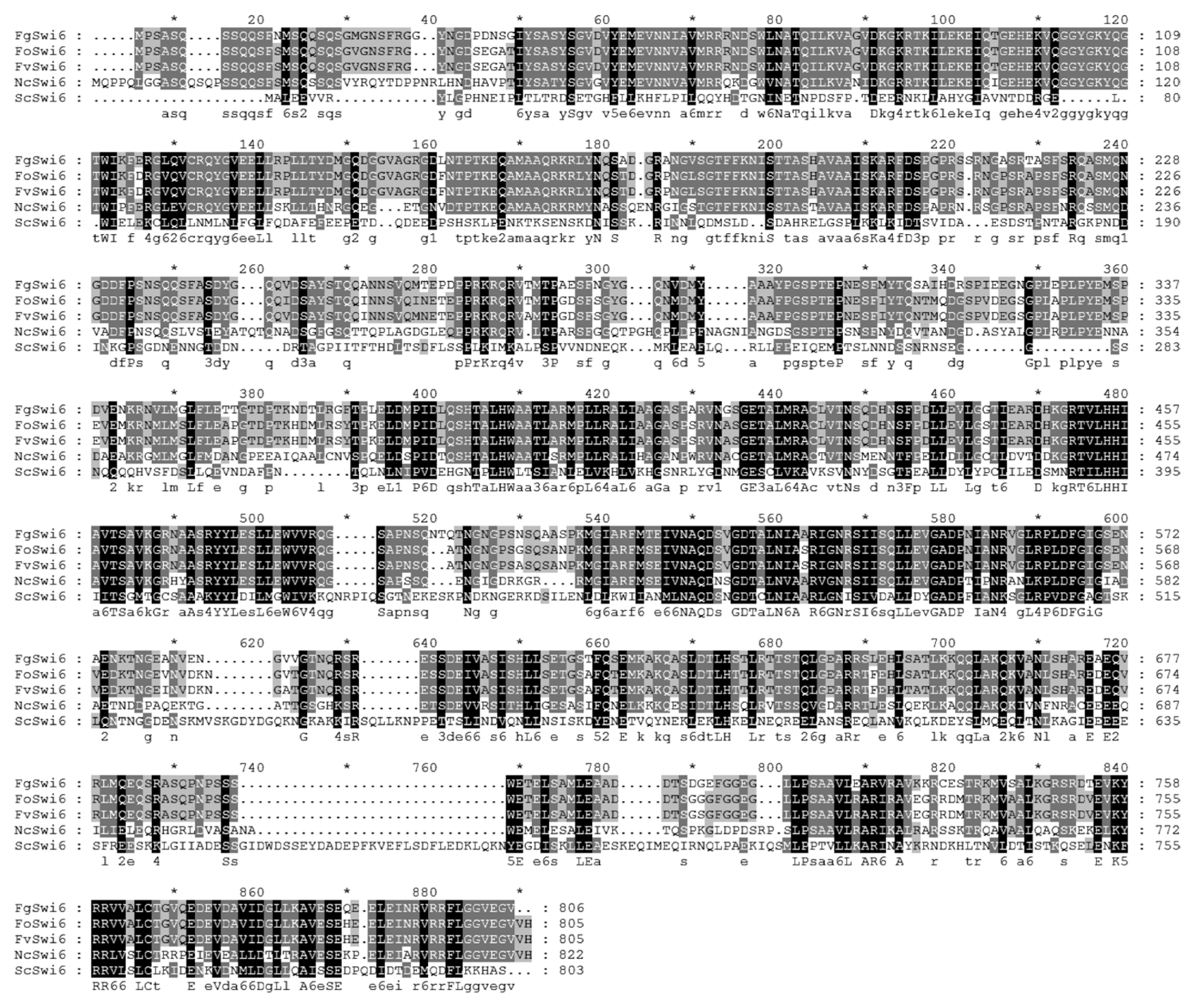
Deduced amino acid sequence alignment FgSWI6 gene with closely related plant-pathogenic fungi. The alignment was conducted using MegAlign in Lasergene using a default setting. Fo, Fv, Nc, and Sc refer Fusarium oxysporum, Fusarium verticillioides, Neurospora crassa, and Saccharomyces cerevisiae, respectively.
Targeted deletion, over-expression, and complementation of the FgSWI6
Targeted gene deletion and over-expression of FgSWI6 mutants were constructed to investigate putative role(s) of the FgSWI6 in F. graminearum biology such as vegetative growth, sporulation, and pathogenicity as well as the effect on the FgV1 viral RNA accumulation. The geneticin resistance gene (gen) was successfully introduced into the locus FgSWI6 by homologous recombination and further complemented with hygromycin registance gene (hph) and designated deletion and complementation mutants as Δ swi6 and Δ swi6::SWI6, respectively (Fig. 2A, B). We also generated FgSWI6 over-expression mutant (SWI6 OE) in which the gene is under the control of the EF1α promoter (PEF1α) from F. verticillioides (Fig. 2C). These FgSWI6 mutants were confirmed by Southern hybridization using 5′ flanking region as a probe (Fig. 2D). Further all mutants were infected with FgV1 by hyphal fusion.
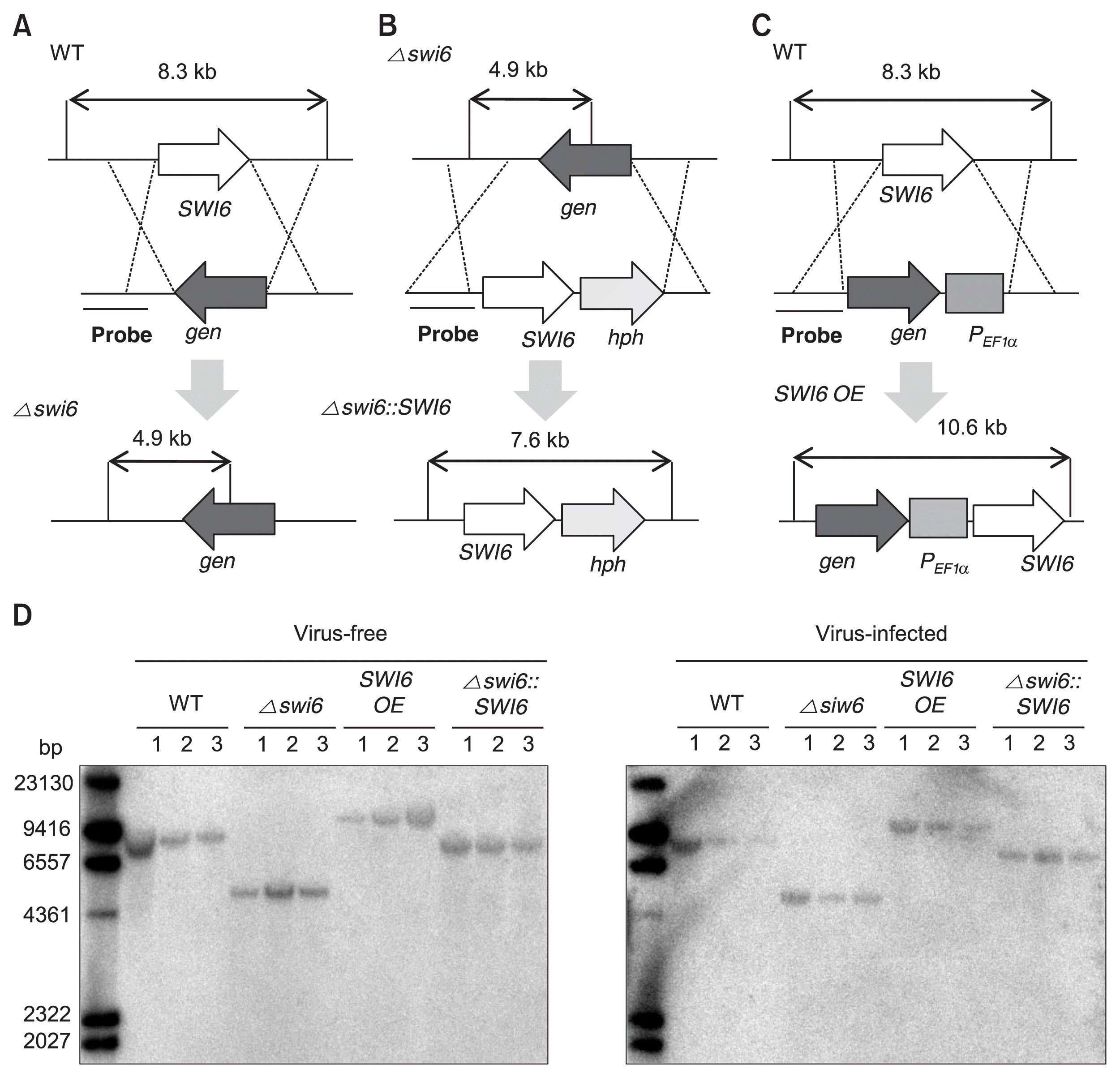
Construction strategy for FgSWI6 gene-deletion, over-expression (OE), and complementation mutants in Fusarium graminearum. (A) The FgSWI6 ORF from the wild type (WT) genome was replaced with the genecitin resistance gene cassette to construct Δ swi6 isolate. (B) The Δ swi6 mutant was complemented by introducing FgSWI6 ORF (Δ swi6::SWI6) with the hygromycin B resistance gene cassette into the Δ swi6 isolate. (C) The geneticin resistance gene cassette and PEF1α fragment were inserted into upstream of FgSWI6 ORF to generate SWI6 OE mutants. (D) These mutants were confirmed by DNA gel blot analysis. When the 5′ flanking region was used as a probe for Southern blot hybridization, the genomic DNA digested with PstI from the four genetically different isolates generated distinguishable hybridized DNA fragments of 8.3-kb for the WT, 4.9-kb for the Δ swi6 mutant, 10.6-kb for the SWI6 OE mutant, and 7.6-kb for the complemented mutant. All isolates were constructed using a PCR-based strategy as described in the Materials and Methods. The numbers above the DNA gel blot image refer three independent biological replicates used in this study.
Colony morphology, vegetative growth, conidial production, and virulence test of FgSWI6 mutants
The colony morphologies of FgSWI6 mutants were investigated on PDA at 5 days post-incubation. The morphologies were very similar among WT-VF, SWI6 OE-VF and Δ swi6::SWI6-VF isolates under standard laboratory conditions (Fig. 3A, top row). However, Δ swi6-VF isolate showed reduced aerial mycelia and growth. All isolates infected with FgV1 showed virus-associated phenotypic changes including reduced growth of aerial mycelia, increased pigmentation, and irregular colony morphology. However, there were significant differences of phenotypic characteristics among VI isolates. The colony diameter was smallest in Δ swi6-VI, biggest for SWI6 OE-VI, and intermediate for WT-VI and complemented isolates (Fig. 3A, bottom row).
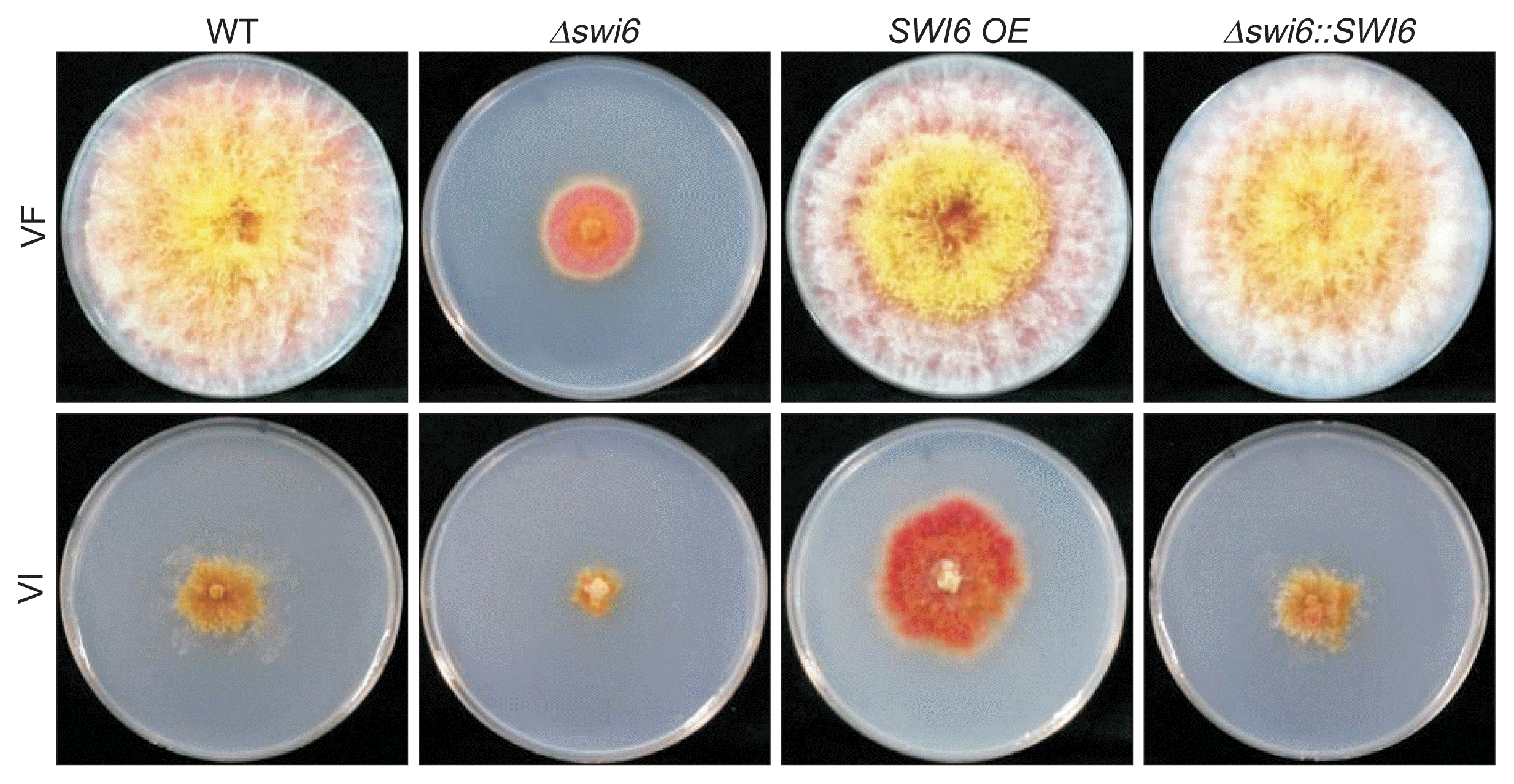
Effect of FgSWI6 gene deletion, over-expression (OE), and complementation on Fusarium graminearum colony morphology. VF and VI indicate virus-free and FgV1-infected isolates of F. graminearum, respectively. WT indicates the wild type. Colonies were photographed after 5 days on potato dextrose agar.
The radial growths on PDA, CM, and MM were investigated to obtain quantitative data concerning vegetative growth. Among VF isolates, radial growth on PDA was significantly affected by FgSWI6 deletion (Fig. 4A). The measured result of radial growth on PDA from day 1 to day 5 showed exactly similar pattern with radial growth data (Supplementary Fig. 1). Among VI isolates, radial growth was greatest for over-expression isolate, smallest for deletion isolate. Results were similar on CM and MM (data not shown).

Effects of FgSWI6 gene deletion, over-expression (OE), and complementation on radial growth and conidia production of Fusarium graminearum. VF and VI indicate virus-free and FgV1-infected isolates of F. graminearum, respectively. WT indicates the wild type. (A) Radial growth after 5 days on potato dextrose agar. (B) Conidial production after 5 days in carboxymethyl cellulose liquid medium. For Fig. 4A and B, error bars indicate the standard deviation, and values with different letters are significantly different at P < 0.05 based on the Tukey test.
Among VF isolates, conidia production form Δ swi6-VF isolate was substantially lowest (Fig. 4B). The conidia production of SWI6 OE-VF isolate was slightly lower than those of WT-VF and complemented isolate. Comparing with conidia production in VF isolates, conidia production was reduced in all four VI isolates. However, general pattern was similar with VF isolates, lowest in deletion isolates and slightly lower in over-expression isolate (Fig. 4B).
The virulence test with wheat plants demonstrated that deletion of the FgSWI6 gene reduced F. graminearum virulence by about 80% among VF isolates (Supplementary Fig. 2). In VI isolates, head blight symptoms were significantly decreased in general. No symptom was observed in Δ swi6-VI isolate inoculated wheat plant that might be caused by severely impaired vegetative growth.
Quantification of FgSWI6 gene expression and FgV1 viral RNA accumulation
The expression levels of FgSWI6 in all isolates were measured using qRT-PCR. The result showed that FgSWI6 expression level was decreased by a fiftieth part approximately in WT-VI compared to that of WT-VF at 5 dpi (Fig. 5A). Relative to expression level in WT-VF, the expression levels were increased by about 10 folds in both VF and VI over-expression isolates (Fig. 5A). The FgSWI6 mRNAs were not detected in both VF and VI deletion (Δ swi6) isolates. Complementation of FgSWI6 gene restored FgSWI6 mRNA expression level to those of WT both in VF and in VI isolates (VF- and VI-Δ swi6::SWI6, respectively; Fig. 5A).

qRT-PCR analysis of expression of FgSWI6 and FgV1 viral RNA accumulation (A) Relative FgSWI6 mRNA expression in the WT-VF, WT-VI, Δ swi6-VF, Δ swi6-VI, SWI6 over-expression (OE)-VF, SWI6 OE-VI, Δ swi6:SWI6-VF, and Δ swi6::SWI6-VI isolates of Fusarium graminearum. Relative transcript levels were normalized using cDNA of elongation factor 1α and cyclophilin 1. (B) Quantification of FgV1 viral RNA accumulation in F. graminearum using qRT-PCR. In Fig. 5A and B, error bars indicate standard deviation. Different letters are significantly different at P < 0.05 based on the Tukey test. VF and VI indicate virus-free and FgV1-infected isolates of F. graminearum, respectively. WT indicates the wild type.
The accumulation levels of FgV1 viral RNA were also quantified in all four VI isolates using qRT-PCR. Relative to WT-VI isolate, the levels were slightly higher in deletion and over-expression isolates, approximately 1.6 folds for deletion and 1.5 folds for over-expression isolates. (Fig. 5B). Complementation of FgSWI6 gene also recovered FgV1 RNA accumulation level similar to that of WT-VI (VI-Δ swi6::SWI6; Fig. 5B).
Discussion
Similar with animal and plant viruses, mycovirus infections also lead to alteration of host fungi in various aspects. Among these alterations induced by mycovirus infections, phenotype changes in vegetative growth, pigmentation, and asexual development are most distinguishable characteristics. However, these easily detectable characteristics are derived from various interactions between viral and host factors. Previous report about FgV1 and host gene expression pattern investigated by microarray and mRNA Seq enabled us to investigate putative individual gene which might play an important role for FgV1-F. graminearum interaction (Cho et al., 2012; Lee et al., 2014). Additionally, phenome-based functional analysis of F. graminearum TFs also provided insight of an individual gene function involved in various phenotypes (Son et al., 2011). Based on these previous studies, we select FgSWI6 gene and generated targeted gene deletion, over-expression, and complementation mutant isolates and report functions of transcription co-factor FgSwi6 which plays a crucial role in FgV1 infection-induced phenotypic alteration.
Although construction of deletion and complementation mutants is generally used method for the functional analysis of the candidate gene as previously conducted for the characterization of FgSwi6 (Liu et al., 2013), we constructed target gene over-expression mutant in addition to the deletion and complementation mutants to support our data more strongly in present study. However, some biological data did not correspond to general expectation. In particular, conidia productions of SWI6 OE-VF and SWI6 OE-VI isolate slightly decreased comparing with WT-VF and WT-VI isolate while those of Δ swi6-VF and Δ swi6-VI also significantly decreased (Fig. 4B). The inconsistency between deletion and over-expression isolate might be explained by the constitutive expression of FgSWI6 gene by EF1α promoter in over-expression isolates. In other words, constitutive over-expression of FgSwi6 might cause defective conidia production. Similarly, inconsistency of biological data between target gene deletion and over-expression isolates was also reported previously in functional analysis of F. graminearum HEX1 gene (FgHEX1; Son et al., 2013). Both deletion and over-expression isolates of FgHEX1 gene showed reduced conidia production and virulence on wheat plant.
Without FgV1 infection, deletion of FgSWI6 gene confers phenotypic alterations such as reduced mycelial growth and aerial mycelia. Interestingly, WT-VI strains also exhibited similar phenotypes, supported by colony morphologies and measurements of radial growth (Fig. 3, 4A). In addition, severely decreased expression level of FgSWI6 was observed in WT-VI strain using qRT-PCR (Fig. 5A). These combined results suggested significantly reduced FgSWI6 expression by FgV1 infection or depletion of FgSWI6 expression by the gene deletion resulted in reduced mycelial growth and aerial mycelia comparing with those of WT-VF isolate.
FgV1 infection confers severe vegetative growth defect on F. graminearum (Fig. 3). Increased accumulation of FgV1 viral RNA caused reduced vegetative growth among genetically different FgV1-infected isolates (Son et al., 2013). However, FgV1-infected FgSWI6 over-expression isolate did not show these general pattern. Although vegetative growth of FgV1 infected FgSWI6 over-expression isolate increased comparing with WT-VI (Fig. 4A), viral RNA accumulation in FgSWI6 over-expression isolate also increased (Fig. 5B). In addition, FgSWI6 deletion isolate showed severe growth defect while viral RNA accumulation level is similar with over-expression isolate. WT-VI isolate also exhibited typical phenotype alteration induced by FgV1 with significantly down-regulated Fg-SWI6 expression level. These combined results indicated that both increased level of FgSWI6 expression and FgV1 accumulation affect vegetative growth of host fungi, but one of the major factor affecting growth seems to be expression level of FgSWI6. Therefore, differentially expressed levels of FgSWI6 might be the major crucial factor for exhibiting typical FgV1 induced phenotypic alteration.
Over the last several years, research on characterization of F. graminearum genes or gene products has greatly increased our understanding of crucial components and their roles in complicated signaling pathways that affect vegetative growth, conidia production, and virulence. For example, mitogen-activated protein (MAP) kinase signaling pathway was already reported as a key player for regulating fungal growth, development, and pathogenicity in response to environmental stimuli. Functional analysis of F. graminearum MAP kinase kinase gene (FgMKK1) revealed that FgMkk1 affects regulation of hyphal growth, pigmentation, conidiation, DON biosynthesis, and virulence of F. graminearum (Yun et al., 2014). The target of rapamycin (TOR) signaling pathway has been known as critical regulator of vegetative growth. Yu et al. (2014) defined a genetic regulatory framework that elucidates how the TOR pathway regulates virulence and vegetative development in F. graminearum by characterizing nine genes encoding components of the TOR pathway followed by investigating biological, genetic, and biochemical functions of each component in F. graminearum. Recently, proteins of the resistance to inhibitors of cholinesterase 8 (Ric8) of F. graminearum (FgRic8) were also reported as component of G-protein signaling pathway (Wu et al., 2015). The FgRic8 also plays important roles in fungal vegetative growth, conidiation, secondary metabolism, and virulence (Wu et al., 2015). Similar with these genes or gene products, FgSwi6 might be component of signaling pathway required for fungal vegetative development. At the initial stage of our study, we tried to identify yeast orthologues, components of CLN3/SWI6/CLN2 pathway in S. cerevisiae, in F. graminearum. Based on sequence homology search tools, some candidate genes were identified. However, their sequence identities were too low and thus could not be determined as orthologues (data not shown). Therefore, we speculated that FgSwi6 might be a component of totally different pathway from S. cerevisiae regulating vegetative development of F. graminearum.
Many host genes interacting with mycoviruses have been identified and characterized from various mycovirus–fungal host systems (Son et al., 2015). These host genes play important roles for the interaction of mycoviruses–fungi. However, only a few genes have been reported for their involvement in mycovirus induced phenotypic alteration (Faruk et al., 2008; Park et al., 2004). Mycoviruses should induce phenotypic alteration of fungal host by affecting various signaling pathways. Therefore, there should be many unknown host genes that affect mycovirus infection-induced phenotypic alteration. Future study should focus on identifying more host genes or on understanding signaling pathways involved in mycovirus-induced host phenotype alteration. Results obtained from these studies will shed light on how individual gene or gene product affects whole signaling pathway during mycovirus-fungus interaction.
Supplemental materials
Acknowledgments
This research was supported in part by grants from the Center for Fungal Pathogenesis (no. 20110000959) funded by the National Research Foundation, the Next-Generation BioGreen 21 Program (no. PJ01101301), Rural Development Administration, and the Vegetable Breeding Research Center (no. 710001-07-05) through the Agriculture Research Center program from the Ministry of Agriculture, Food and Rural Affairs, Republic of Korea. MS was supported by a graduate research fellowship from the Ministry of Education through the Brain Korea 21 Plus Project. We also thank Dr. Son and Professor Lee from the Center for Fungal Pathogenesis at the Seoul National University for the generous gift of pSKGen plasmid and technical helps.
Notes
Articles can be freely viewed online at www.ppjonline.org.
