Mixed Infection of Sugarcane Yellow Leaf Virus and Grassy Shoot Phytoplasma in Yellow Leaf Affected Indian Sugarcane Cultivars
Article information
Abstract
Sugarcane is an important sugar crop contributes more than 80% of world sugar production. Mosaic, leaf fleck, and yellow leaf (YL) are the major viral diseases affecting sugarcane, amongst YL occurrence is widely reported in all the sugarcane growing countries. It is caused by Sugarcane yellow leaf virus (SCYLV) and detailed works were done on complete genome characterization, transmission, and management. However, in countries like Egypt, South Africa, Cuba, Mauritius and Hawaii, the disease was reported to the cause of sugarcane yellow leaf phytoplasma (SCYP) and/or SCYLV as single/combined infections. Hence, we have investigated in detail to identify the exact Candidatus phytoplasma taxon associated in Indian cultivars affected with YL. The sequencing results and the restriction fragment length polymorphism pattern of the PCR products using the universal phytoplasma primers confirmed presence of sugarcane grassy shoot (SCGS) phytoplasma (16SrXI group) in the YL-affected plants. Mixed infection of SCYLV and SCGS phytoplasma was estimated as 32.8% in YL affected plants. Evolutionary genetic relationship between SCYP and SCGS phytoplasma representatively taken from different countries showed that SCYP from South Africa and Cuba were diverged from others and had a highest similarity with SCGS phytoplasma. Although we wanted to identify SCYP from YL affected Indian sugarcane cultivars, the study clearly indicated a clear absence of SCYP in YL affected plants and we found SCYLV as the primary cause for the disease.
Sugarcane is one of the most important commercial crops grown mainly for sugar in many countries and also for bio energy production from its by-products such as, bagasse and molasses. It is being affected by several biotic factors like fungi, bacteria, viruses and phytoplasma causing reduction in production and productivity according to its severity. Among the viral diseases affecting sugarcane, yellow leaf (YL) (showing yellowing of midrib and lamina and in severe cases drying of entire leaf canopy) occurrence is reported in most of the sugarcane growing countries sometimes with up to cent percent incidence (Lehrer et al., 2008; Rassaby et al., 2004; Viswanathan, 2016). In 1960s, it was suspected to occur from East Africa and authentic reports of the disease had come from Hawaii in 1980s (Schenck 1990) followed by South Africa (Cronje et al., 1998) and Cuba (Arocha et al., 1999) and now distributed in all the sugarcane-growing countries causing losses up to 60% in susceptible varieties (Arocha et al., 2000; Comstock et al., 1998). In India, it was first reported during the year 1999 (Viswanathan, 2002; Viswanathan et al., 1999) since then its occurrence and severity continued in all the varieties (Viswanathan et al., 2006). The causative Sugarcane yellow leaf virus (SCYLV) belonging to Polerovirus; family Luteoviridae was reported in Hawaii, Brazil, Florida, and Australia whereas only phytoplasma association was reported from South Africa and Cuba and named as Sugar cane yellow leaf phytoplasma (SCYP) (Arocha et al., 1999; Cronje et al., 1998). However, detection of both SCYLV and SCYP was reported in YL plants from Mauritius (Aljanabi et al., 2001; Parmessur et al., 2002) with suggestion of both were increasing the disease severity.
In India, SCYLV was identified as a causative agent of the disease (Rao et al., 2000; Viswanathan, 2002) through double antibody sandwich enzyme-linked immunosorbent assay (Gaur et al. 2003; Viswanathan and Balamuralikrishnan, 2004), reverse transcription PCR (RT-PCR) (Viswanathan et al., 2008, 2009). Subsequently detailed studies on disease epidemiology, aphid vector transmission, its impact on cane growth, yield and physiological parameters (Chinnaraja and Viswanathan, 2015a, 2015b; Viswanathan et al., 2008, 2014), complete genome characterization of the virus (Chinnaraja et al., 2013), real-time quantitative PCR assays to quantify virus titre in asymptomatic, symptomatic, and tissue culture derived plants (Chinnaraja et al., 2014) and tissue culture nurseries to manage the disease epidemics have been done effectively (Viswanathan, 2016; Viswanathan et al., 2018). Other than SCYLV, the phytoplasma disease, grassy shoot caused by sugarcane grassy shoot (SCGS) phytoplasma belonging to Rice Yellow Dwarf group or 16SrXI is also a major production constraint in sugarcane cultivation and reported to cause 5% to 70% yield reduction in plant crop and up to 100% in ratoon crops (Rao et al., 2012, 2014, 2017; Viswanathan, 2000; Viswanathan and Rao, 2011; Viswanathan et al., 2011). In severe cases, it totally affects the millable cane production and the affected clump produces only chlorotic thin tillers in numerous numbers which gives the typical grassy appearance. By this, the economically important cane stalks are lost, leading to heavy losses in cane yield under field conditions.
The SCGS disease causing phytoplasma is a small, oval to spherical shaped, obligate, endophytic, cell wall-less intracellular parasites resides in the phloem sieve tube and nearby companion cells, and vascular bundles of the host with 0.816-1.603 μm diameter in size (Nithya, unpublished) and have very small genome size of 505.17 kb (Kirdat et al., 2020). They are capable of invading and replicating in both plant and insect hosts. In plants, they inhabit in the nutrient-rich phloem tissues and spread systemically through sieve pores and induce various symptoms such as chlorotic leaves, stunting, and proliferation of axillary shoots, etc. due to abnormal endogenous phytohormone signaling (Bertaccini et al., 2014). In India, sugarcane is widely cultivated in ~5.2 M ha in varying climatic conditions and YL has been distributed throughout the country. Although SCYLV has been found in all the states, association of SCYP is not known except the report of Gaur et al. (2008) and Kumar et al. (2015) from subtropical India. Hence, an attempt was made to investigate the presence of SCYP in YL affected sugarcane cultivars from different geographical locations in India. Our studies clearly proved that SCYLV is the primary cause of YL disease in sugarcane cultivars and no SCYP association was found in any of the YL-affected plants, whereas some of the SCYLV infected plants had SCGS phytoplasma infection.
Materials and Methods
Sample collection
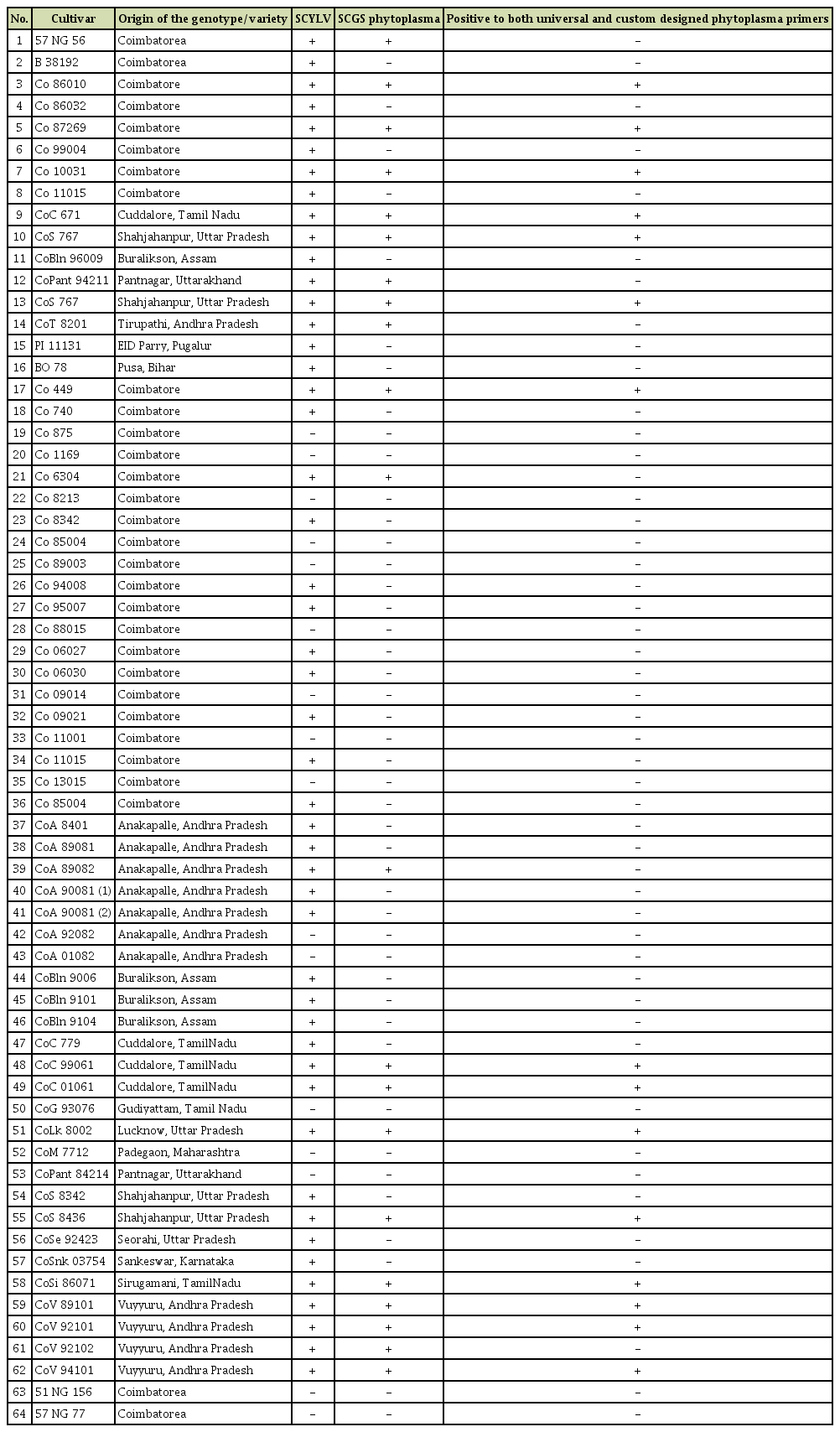
During the year 2016-2018, sugarcane leaves showing YL symptoms with severity grades of 3-4 with intense yellowing of leaf mid rib spread along the lamina region and severe drying of leaf from tip to downwards (Viswanathan et al., 2016) (Fig. 1A and B) were collected from Plant Pathology Farm, ICAR-Sugarcane Breeding Institute (ICAR-SBI) Coimbatore, and at ICARSBI regional Centre, Agali, Kerala and stored at –80°C deep freezer until further processing. Amongst, the cv. Co 10031 in advanced varietal trial was strongly suspected for phytoplasma association as it had YL symptoms with severity grade of 4 along with stunting compared to other YL plants. All the leaves, roots and different internode (3rd, 6th, and 9th) samples were taken for the diagnosis. The sample collection details were given in Table 1.

(A) Severe expression of yellow leaf symptoms on sugarcane. All the leaves in the canopy express yellowing of mid rib and adjoin laminar region. (B) Severe expression of yellow leaf leads to extensive drying of foliage during harvest.
RNA extraction and RT-PCR for SCYLV diagnosis
Total RNA was extracted from the collected leaf samples, ground in liquid nitrogen and resuspended with 1 ml TRI reagent (Sigma, St. Louis, MO, USA) by following the manufacturer’s protocol. The pellet was dissolved in a final volume of 30 ml RNase free water and stored at –80°C. It was treated with DNase I (1 U/ml) (Thermo Fischer Scientific, Waltham, MA, USA) along with 10× reaction buffer with MgCl2 and RNase free water to make up the final volume 10 ml, kept at 37°C for 2 h incubation in water bath and were enzyme de activated by adding 1 ml of 50 mM EDTA and continued incubation at 60-65°C for 10 min. The integrity, concentration and purity of RNA were assessed on 1% EtBr-stained agarose gel visualized under UV light and on a Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific) and stored at –80°C. The SCYLV primer was designed from the consensus coat protein regions through primer 3.0 and sigma oligo evaluator, named as SCYLV-FP (5′-CTGAGAATTCATGGATACGGGCGCTAACCGCTC-3′) and SCYLV-RP (5′-CTGCAAGCTT TTATTTGGGATTCTGGAATA-3′) which can amplify complete ORF 3 of coat protein gene were used in this study. One microgram of total RNA per reaction was reverse transcribed using RevertAid H Minus First Strand cDNA Synthesis Kit (MBI, Fermentas, Waltham, MA, USA), primed with 50 pmol of SCYLV-615R by following the manufacturer’s protocol in a thermal cycler (Eppendorf, Hamburg, Germany). The PCR reaction was performed in a total volume of 25 μl containing 2.5 μl of 10× Taq buffer with 15 mM MgCl2, 0.5 μl of 10 mM dNTP mix, 10 μM of each forward and reverse primers, 1.25 units of Taq polymerase (Origin, Kerala, India), 2 μl of cDNA and sterile MilliQ water to make up the final volume. The PCR programme was performed with initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 4 min, annealing at 65°C for 1 min, extension at 72°C for 1min and final extension at 72°C for 10 min. All the amplified products were run on 1.5% agarose gels stained with ethidium bromide (0.5 μg/ml) and gel images were documented in a gel documentation unit (G-BOX EF, Syngene, Cambridge, UK).
DNA extraction and nested PCR for phytoplasma diagnosis
Total genomic DNA was extracted from 0.5 g leaves, ground in liquid nitrogen by following the cetyl trimethyl ammonium bromide method (Doyle and Doyle, 1990), treated with RNase I (10 U/ml) (Thermo Fischer Scientific) and purified. The integrity, concentration and purity of DNA was assessed on 0.8% EtBr-stained agarose gel visualized under UV light and on a Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific) and stored at –20°C. Based on the genomic DNA concentration it was diluted up to 50 ng/ml for the PCR analysis. PCR was performed using the universal phytoplasma specific primers P1/P7 (Deng and Hiruki, 1991) in the first round PCR and R16F2n 5′-GAAACGACTGCTAAGACTGG-3′ and R16R2 5′-TGACGGGCGGTGTGTACAAACCCCG-3′ primer pairs were used in the nested PCR (Gunderson and Lee, 1996; Lee et al., 1993) for the amplification of both large (23S), small (16S), and 16S-23S ribosomal spacer regions. The PCR reaction was performed in a total volume of 25 μl containing, 2.5 μl of 10× Taq buffer with 15 mM MgCl2, 2 μl of 2.5 mM dNTP mix, 10 μM of each forward and reverse primers, 0.33 units of Taq polymerase (Origin), 1 μl of genomic DNA and sterile MilliQ water to make up the final volume.
The PCR cyclic conditions for the P1/P7 primer was initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 1 min 30 s, extension at 72°C for 1 min 30 s, and a final extension at 72°C for 15 min following that nested PCR was performed using the 1:10 dilution of P1/P7 product with the same cyclic conditions except the annealing temperature at 58°C for 1 min 30 s. Besides, a custom designed primer was synthesized through primer 3.0 and sigma oligo evaluator based on all the available SCYP sequence submissions at GenBank database for the SCYP diagnosis in the collected YL samples. The primers were named as YL FP1 (5′-CAATAGGTATGCTTAGGGAGGAGCTTGCGTCAC-3′) and YL RP 1 (5′-CCTCCACTGTGTTTCTACAGCTTTGCAG-3′), initially standardized with gradient PCR and the cyclic conditions were initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min 30 s, extension at 72°C for 1 min 30 s and a final extension at 72°C for 10 min. All the three PCR amplified products were run on 1.2% agarose gels stained with ethidium bromide (0.5 μg/ml) and gel documented. The positive samples were eluted from the low melting agarose gel using GenElute Gel Extraction Kit (Sigma), purified and was further checked in agarose gel and quantified in Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific). The positive samples were sent for double pass sequencing to commercial sequencing firm Bioserve Technologies, Hyderabad, India.
Restriction fragment length polymorphism (RFLP) analyses
Restriction fragment length polymorphism analysis was performed to the nested PCR product of the YL samples amplified by R16F2n/R16R2n primers. The restriction enzymes Bfa I (FspB I), Msp I (Hpa II), Mse I, Hpa I (KspA I), and Hinf I (10 U/μl) (Thermo Fischer Scientific) were selected based on the in silico virtual RFLP iphyclassifier enzymes list to identify the phytoplasma. The reaction was performed with reaction volume of 20 ml containing 2 ml of 10× restriction digestion buffer with bovine serum albumin (Tango/R/B buffers according to the enzymes), 3 ml of PCR product (500-1,000 ng/ml), 2 ml of restriction enzymes and sterile MilliQ water to make up the final volume kept at 37°C for 2 h incubation in water bath and were run on 1.5% agarose gels and documented. SCGS phytoplasma cv. CoV 92101 was used as a positive control to confirm the restriction pattern of the phytoplasma in YL samples.
Phylogenetic analysis
Quality of all the samples sequences were confirmed based on the q-value (>30). All the sequences were subjected into BLASTn searches at NCBI website (Altschul et al., 1990) and pair wise multiple sequence alignment of the selected sequences were made through Bio edit (Hall, 1999) and maximum likelihood Tamura-Nei model based phylogenetic tree analysis was performed with nearest neighbour interchange tree options (MEGA X v.10.1.6) with 1,000 bootstrap replications to know the evolutionary genetic relatedness (Kumar et al., 2018) of all the samples collected from different varieties and fields.
Results
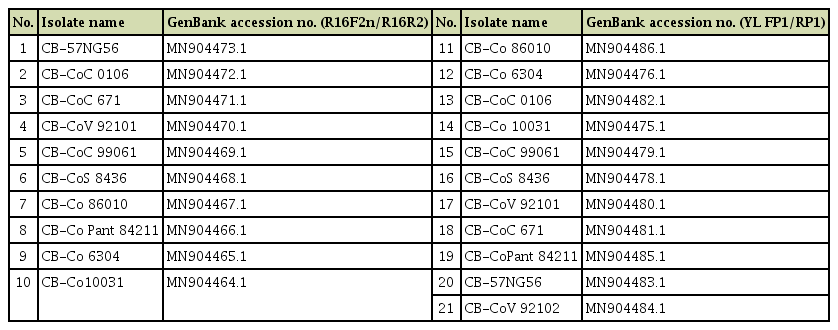
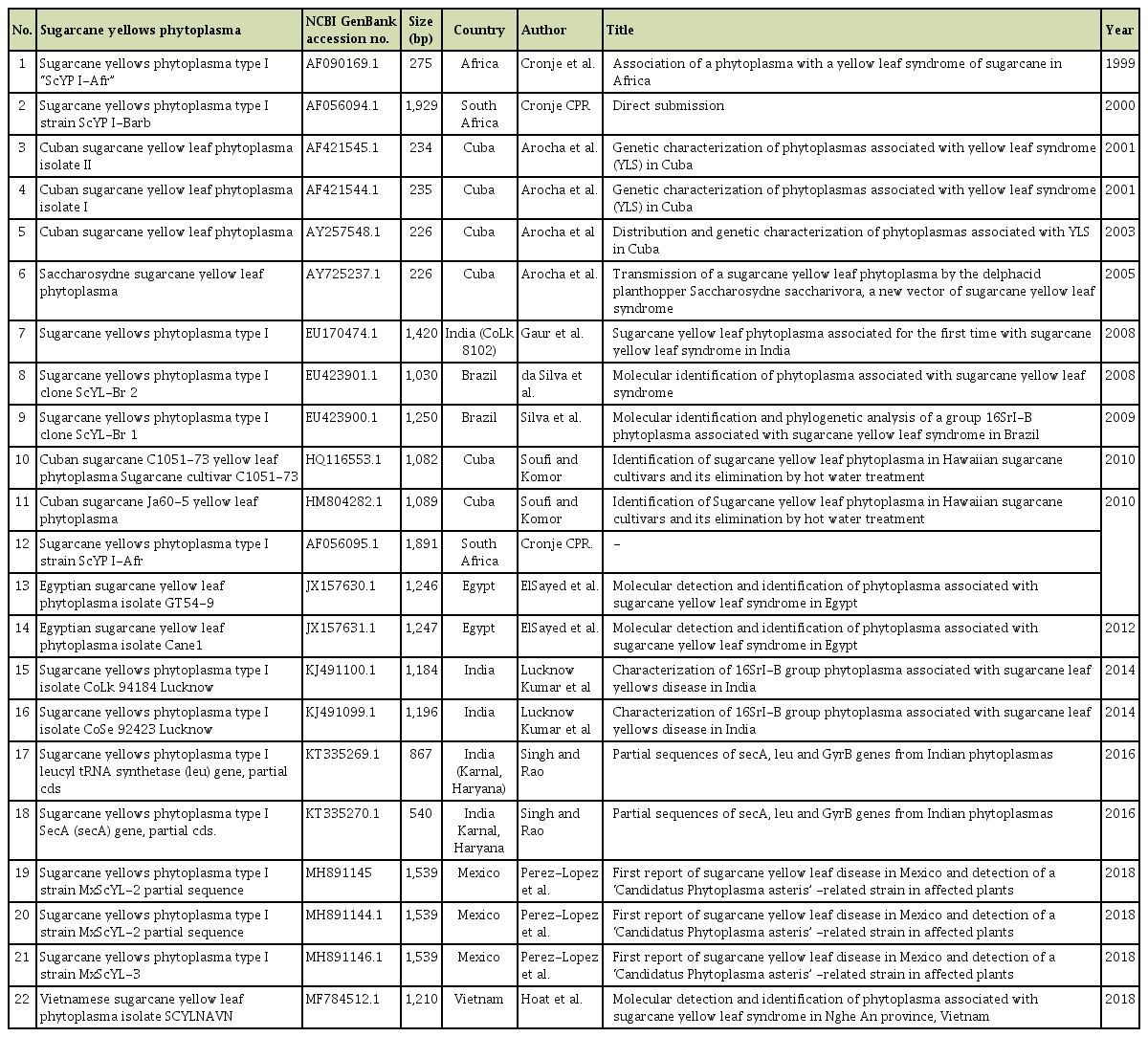
Out of 64 samples tested, SCYLV was detected from 48 samples with the expected amplification of 591 bp in RTPCR assays (Fig. 2). In case of phytoplasma, none of the samples had shown any amplification in the first round of direct PCR using P1/P7 primers except the positive SCGS phytoplasma cv. CoV 92101. Whereas, in nested PCR analysis 14 samples were found positive to both the phytoplasma primers R16F2n/R16R2 and YL FP1/RP1 with the expected amplification of 1,200 bp and 900 bp, respectively (Figs. 3 and 4). The YL affected cv Co 10031 leaf, root and internode (3rd, 6th, and 9th) samples were found positive to YL FP1/RP1 phytoplasma primers with the expected amplification of 900 bp (Fig. 5). The primers YL FP1/RP1 designed from ribosomal RNA regions from SCYP consensus sequences were found better than R16F2n/R16R2 in terms of intense amplification and wide sample coverage. Out of 64 samples tested, YL FP1/RP1 shown positive to 21 samples and R16F2n/R16R2 shown positive to 15 samples and both the primers shown intense amplification to 13 samples viz. 57 NG 56, Co 449, Co 6304, Co 86010, Co 86032, Co 87269, Co 10031, CoC 671, CoC 99061, CoPant 94211, CoS 8436, CoV 92102, and CoV 94101. Mixed infection of SCYLV and SCGS phytoplasma was estimated as 32.8% in YL affected plants. All the genomic sequences analyzed through Blastn had shown 98.92% to 99.82% similarity with 98.25-100% query coverage to other SCGS phytoplasma isolates, amongst maximum similarity was found with sugarcane SCGS phytoplasma sequences submitted from India, Thailand and with white leaf phytoplasma from Sri Lanka (MN174860.2). All the sequences of both the primers have been submitted at GenBank with the accession numbers MN904464.1 to MN904486.1 (Table 2). Since, the SCGS phytoplasma was confirmed in the YL plants, its similarity with the available 22 SCYP sequences as well as from other SCGS and sugarcane white leaf (SCWL) phytoplasma sequences submitted from different countries were analyzed for evolutionary genetic relatedness. But, the SCYP sequences submitted from Cuba during the years 2001 (2 numbers), 2003 and 2005 and one SCYP isolate from Africa (Cronje at al., 1999) were <250 nucleotides in length compared to others hence, were not considered for the analysis (Table 3).

Specific amplification SCYLV-CP gene by RT-PCR assay from different sugarcane cultivars. SCYLV, Sugarcane yellow leaf virus; RT-PCR, reverse transcription PCR.
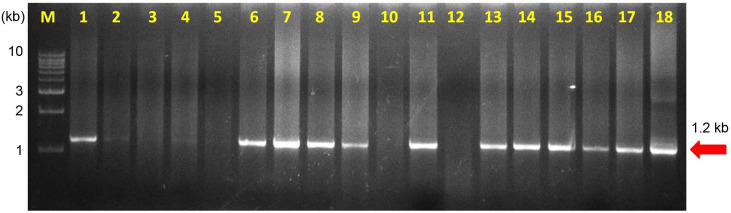
Detection of phytoplasma in yellow leaf affected sugarcane cultivars by nested PCR using R16F2n/R16R2.

Detection of phytoplasma in YL affected sugarcane cultivars by nested PCR using YLD FP1/RP1 primers.
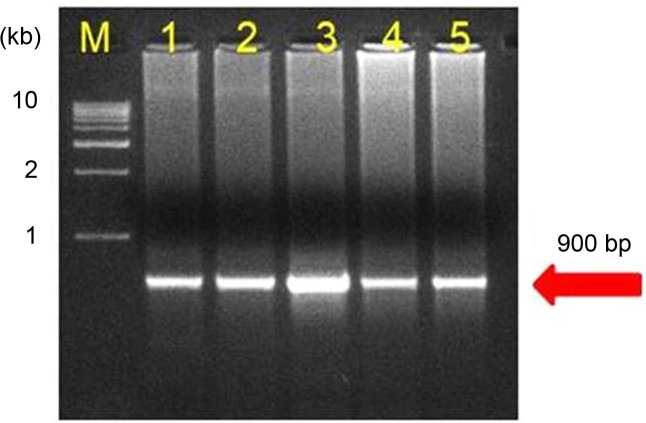
Detection of sugarcane grassy shoot-phytoplasma in different tissues of yellow leaf affected sugarcane cv Co 10031 by nested PCR assay.
The results of the remaining 15 SCYP sequences and other SCGS and SCWL phytoplasma sequences representatively taken from different countries such as India, Thailand, Hawaii, Sri Lanka, China, and Vietnam along with phytoplasma sequences obtained from the YL plants in this study were subjected to phylogenetic analyses. The results of 31 sequences segregated into three major clusters, the first one contained all the SCGS, SCWL phytoplasmas belonging to 16SrXI (Rice yellow dwarf) group and the second one contained all the SCYP belonging to 16SrI (aster yellows; Ca. Phytoplasma asteris) group and the third one contained the SCYP from South Africa and Cuba, both shared the maximum similarity with 16SrXI group. All the SCGS phytoplasma used in this study and SCWL phytoplasma reported from Thailand, Vietnam, and China showed highest similarity of >80% with SCYP India, 2014 followed by SCYP Cuba, 2010 (HM804282.1) and SCYP Brazil, 2009. Acholeplasma sp. was used as out group in the tree (Fig. 6, Supplementary Table 1).
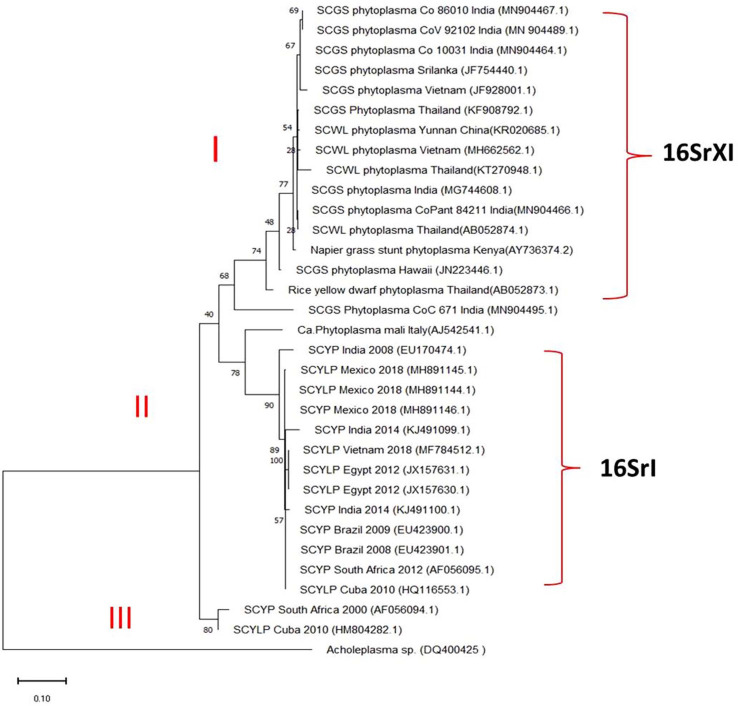
Phylogenetic tree was constructed by maximum likelihood neighbour joining method using MEGA-X v10.1.6 showing the relationship between the sugarcane grassy shoot phytoplasma and sugarcane yellow leaf phytoplasma. Bootstrap values were expressed as percentage of 1,000 replications and branch lengths are proportional to the number of substitutions. Acholeplasma sp. was used to out group the tree.
Of the total 22 SCYP sequences, 20 only were analyzed in pairwise multiple sequence alignment using Clustal W (leaving the two sequences submitted from India during the year 2016 from sec A gene and LeucyltRNA gene partial sequences) in that, the South African sequence submitted by Cronje in the year 2000 (AF056094.1) has the maximum length i.e. 1,929 nucleotides. Hence, all the SCYP sequences were compared by taking that one as a reference in SCGS, sugarcane grassy shoot; YL, yellow leaf.that the Cuban isolate (HM 804282.1) had shown highest similarity with this sequence. The results of RFLP analysis showed that all the YL samples nested PCR product restriction profiles were as similar as to SCGS phytoplasma cv. CoV 92101 in all the enzymes tested and the enzymes Hinf I and Bfa I had shown intense digestion. However, in Hpa I, one sample restriction pattern was different compared to others may be due star activity or over digestion (Fig. 7).
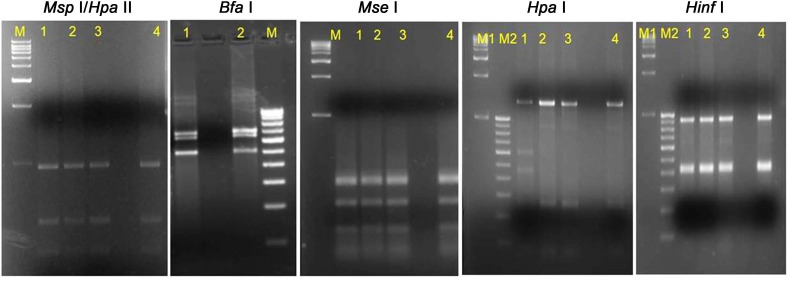
Restriction fragment length polymorphism patterns of phytoplasma from yellow leaf affected sugarcane samples. M1, M2, 1 kb and 100 bp ladder (GeNei StepUp, India); lane 1-3, restriction digestion of nested PCR product of yellow leaf samples DNA amplified by R16F2n/R16R2 primers; lane 4, positive sugarcane grassy shoot (SCGS) sample cv CoV 92101; except 7b) Bfa I where, lane 2 is SCGS sample cv CoV 92101.
Discussion
SCYLV was confirmed as the causative agent of the YL disease in Indian cultivars and its characterization was done up to complete genome level and existence of different genotypes of the virus was reported including the new genotype SCYLV-IND (Chinnaraja et al., 2013, Viswanathan et al., 2008). Since mixed infections of SCYLV and SCYP were reported in some countries, we suspected the possible association of SCYP in YL of sugarcane hence, investigated on their probable mixed infections in Indian sugarcane cultivars, and importantly to develop effective diagnostics and to characterize exact Ca. phytoplasma taxon associated with Indian YL disease. SCYP was first reported in South Africa and Cuba (Arocha et al., 1999; Cronje et al., 1998), and in Mauritius mixed infections of SCYLV and SCYP were reported (Aljanabi et al., 2001; Parmessur et al., 2002). In Africa, two different phytoplasma viz. western Xdisease and SCWL phytoplasma were reported (Aljanabi et al., 2001; Cronje and Bailey, 1999) with 98.8% sequence similarity. In Australia, the single sugarcane plant was reported with multiple infections of two or more distinctly different phytoplasma groups, most of them were infected by the tomato big bud (TBB) and less frequent were sun hemp witches’-broom (SUNHP) of the Faba Bean Phyllody group, stylosanthes little leaf (StLL) of the loofah witches-broom (LfWB) group and Maryland aster yellows (AY1) of the AY group (Tran-Nguyen et al., 2000). Also, the less frequently detected phytoplasmas were mainly from asymptomatic plants while the TBB phytoplasma was detected with equal frequency in both symptomatic and asymptomatic plants (Schneider et al., 1999). Recently, ‘Ca. P. asteris’ subgroup (16Sr I-B) in Brazil and Egypt (Silva et al., 2009; ElSayed and Boulila, 2014); white leaf phytoplasma in Thailand (Soufi et al., 2013); latent infections of 16SrI (aster yellows) and 16SrXI (Rice yellow dwarf) groups in asymptomatic Hawaiian sugarcane cultivars (Soufi and Komor, 2014) and Egyptian sugarcane cultivars based on sequencing and RFLP analyses (ElSayed et al., 2016) were reported from YL affected plants. In India, 16SrXII (Stolbur group) (Gaur et al., 2008) and 16SrI-B (Aster Yellow group) (Kumar et al., 2015) were reported from sub-tropical cvs CoLk 8102, CoLk 94184, and CoSe 92423.
As on date, SCYP has been reported in seven countries viz. South Africa, Cuba, Brazil, Egypt, Mexico, India and recently in Vietnam. Although 22 SCYP partial sequences were submitted in GenBank database, no reports are available on any phytoplasma specific symptom changes in the YL plants other than the characteristic SCYLV symptoms. Also the first reports from South Africa and Cuba have not ruled out absence of SCYLV from the YL affected plants (Arocha et al., 1999; Cronje et al., 1998) hence, all these SCYP reports speculate that there could have been SCYLV infection with additional infection of phytoplasma as reported by different researchers.
Despite the report of different groups of phytoplasma as mixed infections, the white leaf phytoplasma was reported along with that in most of the countries. In sugarcane, grassy shoot disease (GSD) and white leaf disease (WLD) both were named according to its symptom expression on the infected pants. In case of GSD, the infected plant does not produce any millable canes, instead produces only grassy chlorotic tillers hence, named as grassy shoot disease (Viswanathan, 2000) and in case of WLD, the infected clump produces the same symptom with complete white/creamy tillers. In India, two kinds of phenotypes are very common and WLD is a misnomer for Indian scenario (Nasare et al., 2007; Viswanathan et al., 2011). However, both are belonging to the same rice yellow dwarf/16SrXI group. WLD occurrence is widely reported in South East Asian countries viz. Indonesia, Thailand, Sri Lanka, and also in China and GSD occurrence is reported more from India only.
The results obtained in this work clearly evidenced that YL in India is caused only by SCYLV and the phytoplasma associated in the YL plant is SCGS phytoplasma which has been confirmed based on the sequencing results, phylogenetic analysis as well as by RFLP using specific enzymes. In support of our findings, presence of both YL and GSD symptoms in the same clumps were recorded in many popular varieties such as, Co 86032, Co 88028, CoV 92101, CoV 94101, CoV 89101, CoV 09356, etc. in both plant and ratoon crops in the institute fields as well as in farmers’ holdings in that only few millable cane formation used to be there and the remaining tillers show grassy shoot symptoms (Fig. 8) whereas, in some cases, the sett material taken from the severely affected YL plant or the ratoon of the YL affected clump shows the GSD expression even in the tillering phase of the crop due to build of high phytoplasma titre in the previous crop. Although our results were contradictory with SCYP (16Sr-I aster yellow group) association in YL plants of sub-tropical cultivars, only SCGS phytoplasma association was confirmed from both tropical and sub-tropical cultivars in India. Such type of both group of phytoplasma associations (16SrI and 16SrXI) were earlier reported from YL plants in Hawaii (Soufi and Komor, 2014) and, in Egypt, SCYLV as well as 16SrI and 16SrXI groups of phytoplasma were reported on cvs H73-6110, G03-47, G84-47 (ElSayed et al., 2016). All the sugarcane viruses Sugarcane Mosaic Virus (SCMV), Sugarcane Streak Mosaic Virus (SCSMV), and SCYLV were diagnosed even in asymptomatic plants in RT-PCR assays (Viswanathan et al., 2008, 2009; Viswanathan, 2016) due to latent stage or low titre of viruses, the same can be applicable to SCGS phytoplasma. Both, SCYLV and SCGS phytoplasma are phloem limited pathogens and the SCYLV titre might be more than the SCGS phytoplasma in all the collected YL plants in this study. Because of mixed infections of both the pathogens and further inoculum build up in subsequent ratoons, the growth and yield of many high yield and high sugar varieties were reduced. Non-fungal pathogens, especially virus and bacteria are associated with such varietal degeneration in India; pathogen elimination through tissue culture combined with molecular diagnosis has addressed such constraints under field conditions (Rao et al., 2012; Viswanathan, 2016; Viswanathan and Rao, 2011; Viswanathan et al., 2018). In this regard, our study clearly indicates SCYLV only causes the YL disease in sugarcane and SCYP association is not found with the disease.

Different sugarcane varieties showing both yellow leaf and grassy shoot symptoms in the same clump. Top leaf canopy showing the typical yellowing of leaf mid rib with yellow leaf symptoms and the same bottom were with chlorotic leaves and thin grassy tillers of grassy shoot disease symptoms.
Overall, the study has revealed lack of evidences for the occurrences of SCYP in YL affected sugarcane in India, the major sugarcane producer in the world after Brazil. A clarity on the YL associated pathogen has been brought out and incidentally we could detect mixed infections of SCYLV and SCGS-phytoplasma in YL affected sugarcane plants. Further studies are required on the combined infections of these phloem limiting pathogens in sugarcane on YL symptom expression, its severity and their interactions.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
Acknowledgments
The authors thank the Director, ICAR-Sugarcane Breeding Institute, Coimbatore for providing all the facilities.