 |
 |
| Plant Pathol J > Volume 32(4); 2016 > Article |
Abstract
Plants protect themselves from pathogen attacks via several mechanisms, including hypersensitive cell death. Recognition of pathogen attack by the plant resistance gene triggers expression of carboxylesterase genes associated with hypersensitive response. We identified six transcripts of carboxylesterase genes, Vitis flexuosa carboxylesterase 5585 (VfCXE5585), VfCXE12827, VfCXE13132, VfCXE17159, VfCXE18231, and VfCXE47674, which showed different expression patterns upon transcriptome analysis of V. flexuosa inoculated with Elsinoe ampelina. The lengths of genes ranged from 1,098 to 1,629 bp, and their encoded proteins consisted of 309 to 335 amino acids. The predicted amino acid sequences showed hydrolase like domains in all six transcripts and contained two conserved motifs, GXSXG of serine hydrolase characteristics and HGGGF related to the carboxylesterase family. The deduced amino acid sequence also contained a potential catalytic triad consisted of serine, aspartic acid and histidine. Of the six transcripts, VfCXE12827 showed upregulated expression against E. ampelina at all time points. Three genes (VfCXE5585, VfCXE12827, and VfCXE13132) showed upregulation, while others (VfCXE17159, VfCXE18231, and VfCXE47674) were down regulated in grapevines infected with Botrytis cinerea. All transcripts showed upregulated expression against Rhizobium vitis at early and later time points except VfCXE12827, and were downregulated for up to 48 hours post inoculation (hpi) after upregulation at 1 hpi in response to R. vitis infection. All tested genes showed high and differential expression in response to pathogens, indicating that they all may play a role in defense pathways during pathogen infection in grapevines.
Plants have evolved several mechanisms to protect themselves against pathogen attacks. Hypersensitive response (HR) is one of the most common and efficient plant reactions to pathogens, although its precise role is not yet clear (Macro et al., 1990). The characteristics of hypersensitivity include rapid local cell death at the site of infection to limit further spread of invading micro organisms (Hammond-Kosack and Jones, 1997). Hypersensitive cell death occurs via the recognition of pathogen avirulence (avr) genes by plant resistance (R) genes (Liu et al., 2007). Although hypersensitive cell death is triggered by direct R genes, several hypersensitive related genes have been identified and characterized (Baudouin et al., 1997; Bézier et al., 2002; Macro et al., 1990; Pontier et al., 1998b; Tronchet et al., 2001).
Hypersensitivity-related genes, including hsr203J (tobacco), Lehsr203 (tomato), BIG8.1 (Botrytis-induced grapevine), PepEST (pepper), and SOBER1 (Arabidopsis), were characterized as carboxylesterase members of the serine hydrolase family (Baudouin et al.,1997; Bézier et al., 2002; Ko et al., 2005; Tronchet et al., 2001). Serine hydrolases comprise a large number of enzymes, including esterases, lipases, proteases and transferases (Kaschani et al., 2009). The motif ‘GXSXG’ is characteristic of many members of the serine hydrolase family, including lipases, esterases and proteases (Baudouin et al., 1997). Pentapeptide ‘HGGGF’ is observed in many lipase sequences (Baudouin et al., 1997). Carboxylesterases (esterases and lipases) catalyze the hydrolysis of compounds containing an ester bond, while esterases hydrolyze water-soluble compounds such as short acyl chain esters and are inactive against water-insoluble long chain triacylglycerols, which, in turn, are specifically hydrolyzed by lipases (Chahinian and Sarda, 2009). Several esterase and lipase genes have been characterized from various plants such as tobacco, tomato, grapevine, Arabidopsis, and pepper.
Cultivated grapevines (Vitis sp.) are exposed to many pathogenic fungi, such as Elsinoe ampelina and Botrytis cinerea, which cause anthracnose and gray mold, respectively, as well as bacteria such as Rhizobium vitis, which is responsible for crown gall and viruses (Wang et al., 2011). The development of new varieties resistant to diseases can result in the cost saving and convenient management of various diseases in grape production. Several carboxylesterase genes from various plants including hsr203J, Lehsr203, EDS1, PAD4, DAD1, PRLIP, SABP2, and PepEST have been shown to function in different developmental mechanisms and resistant pathways against pathogens. In this study, we isolated six carboxylesterase genes in Vitis flexuosa, analyzed their structural features and homology, evaluated their phylogenetic relationship with other plants, and investigated expression patterns against infection of E. ampelina, B cinerea, and R. vitis.
Plants of V. flexuosa VISKO001 were cultured in a grapevine germplasm collection field of Yeungnam University, Gyeongsan, Korea for leaf production. Leaves were used for gene expression analysis following pathogen inoculation. The pathogens used in this study were virulent strains of E. ampelina (EA-1) and B. cinerea (B1035) isolated from infected grapes by Dr. W.K. Kim, National Academy of Agricultural Science, Rural Development Administration, Korea, as well as R. vitis (strain Cheonan 493) kindly provided by Prof. J.S. Cha, Chungbuk National University, Korea.
Spores of E. ampelina (106 spores/ml) were sprayed onto leaves after scraping off of plates with sterile distilled water according to the method described by Yun et al. (2003). Spore suspensions (106 spores/ml) of B. cinerea in making by 0.24% potato dextrose broth solution were sprayed onto the leaves for inoculation. Additionally, to induce defense responses against bacteria in the leaves, 20 μl cell suspensions (OD600 = 1) of R. vitis grown in YEP medium (yeast extract 10 g, bactopeptone 5 g, NaCl 5 g/l, and pH 7.0) at 28°C in a shaking incubator for 16 to 18 hours were dropped onto the wounded portion of leaves that had been injured slightly with a pencil tip (Choi et al., 2010). Leaves inoculated with spore suspensions were then incubated in a moist box at 22-28°C for 48 hours to induce early hypersentive responses by pathogen infections. Leaves were subsequently harvested at the indicated time points (0, 1, 6, 12, 24, and 48 hours post inoculation [hpi]), immediately frozen in liquid nitrogen and then stored at −80°C for future use.
For RNA isolation, desired leaf samples were ground in liquid nitrogen using a mortar and pestle and total RNA was extracted by the modified pine tree method (Chang et al., 1993). The RNA quality was determined based on the absorbance at 230, 260, and 280 nm, which was measured using a Nano Drop spectrophotometer (ACTGene ASP-3700; ACTGene Inc., Piscataway, NJ, USA). The GoScript™ Reverse Transcription System (Promega, Madison, WI, USA) was used to synthesize first-strand cDNA from the total RNA (500 ng), which was subsequently used as a template for PCR. Real-time PCR was performed on a C1000™ Thermal Cycler (CFX96™ Real-Time System; Bio-Rad, Hercules, CA, USA) using SYBR Premix Ex Taq (TaKaRa Bio Inc., Osaka, Japan) as the fluorescent dye. Amplification was conducted by subjecting the samples to 95°C for 30 seconds, followed by 40 cycles of 95°C for 5 seconds and 60°C for 30 seconds. The standard-curve method was employed to determine the transcript levels. Transcripts were normalized against the grapevine actin gene (AB372563) as an internal control and non-treated leaves (at time zero) as a reference. Melting curves of the amplified products were also recorded. For each gene, the reference sample was defined as the 1 × expression level and the results were expressed as the fold increase in mRNA level over the reference sample. To minimize error, all reactions were replicated three times. The specific primers used in real-time PCR are listed in Table 1.
To verify the presence of the hydrolase like domain, the VfCXE protein sequences were analyzed using the Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1). Nucleotide sequences of genes were transmuted to amino acid sequences using translation software (http://web.expasy.org/translate/). The primary structure analysis of genes was performed using protParam (http://web.expasy.org/protparam/), while the secondary structure was analyzed by the Self-Optimized Prediction Method with Alignment (SOPMA; https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html). The tertiary structure of the protein was predicted by SWISS-MODEL (http://swissmodel.expasy.org/interactive). Multiple alignment of protein sequences was performed using ClustalW (http://www.genome.jp/tools/clustalw/). Homologue protein sequences of the genes were identified by a BLAST (Basic Local Alignment Search Tool) search of the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) by the BLASTp tool using the “nr” database. The serine hydrolase genes were blasted (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against each other to check for gene duplication events. Phylogenetic neighbor-joining analyses of gene sequences were performed using Molecular Evolutionary Genetics Analysis (MEGA) ver. 6.0. The tree branches were evaluated using the bootstrap method.
Six full length transcripts of the carboxylesterase gene were selected from transcriptome analysis (NABIC, NN-0197-000001) by next generation sequencing (NGS) of V. flexuosa VISKO001 inoculated with E. ampelina (Ahn et al., 2014), and verified as serine hydrolase-like genes by detection of the hydrolase like domain using SMART. In this study, these genes were characterized and referred to as V. flexuosa carboxylesterase 5585 (VfCXE5585), VfCXE12827, VfCXE13132, VfCXE17159, VfCXE18231, and VfCXE47674 and deposited in the National Agricultural Biotechnology Information Center (NABIC), Rural Development Administration, Korea under accession numbers NABIC NS-0001-1 to NS-0006-1, respectively. The VfCXE genes were then compared by BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of each other to investigate gene duplication events. The maximum similarity of the predicted amino acid sequences (77%) was observed between VfCXE5585 and VfCXE12827 (Table 2), while the similarity between two amino acid sequences among other genes ranged from 28% to 58%. All six VfCXE genes tested in this study were confirmed to be independent upon gene duplication analysis based on the index proposed by Kong et al. (2013). The primary structure and the characteristics of the six VfCXE genes were analyzed using the bioinformatic tool protParam (http://web.expasy.org/protparam/) (Table 3). The size of the six VfCXE genes extended from 1,098 to 1,629 bp, while the open reading frame varied from 930 to 1,008, encoding 309 to 335 amino acids (34.2 to 37.7 kDa) with predicted isoelectric points ranging from 5.15 to 8.16. The predicted isoelectric points of these genes showed basic characters except VfCXE17159. The VfCXE13132 and VfCXE18231 proteins predicted from nucleotide sequences of the VfCXE transcripts showed an instability index of less than 40 (Table 3), which was considered stable based on the instability index (Guruprasad et al., 1990). The secondary structures of the proteins predicted from the six loci of the VfCXE gene were analyzed by SOPMA (Table 4). The results revealed variations in the proportion of random coils (33.54-41.79%), alpha helices (26.01-31.66%), beta turns (8.06-11.58%), and extended strands (18.27-25.70%) in amino acids from each locus. These results suggest that all six loci of VfCXE genes formed abundant random coils in their amino acid sequences, which are important elements influencing protein flexibility and stabilization of protein folding (Sharmin et al., 2011).
Multiple sequence alignment of the predicted proteins from the six loci of VfCXE genes including Vitis and tobacco serine hydrolase proteins was conducted using ClustalW to analyze the sequence characteristics. Many lipases and esterases contained the consensus motif ‘HGGGF’ in their upstream regions (Baudouin et al., 1997; Bézier et al., 2002) and the motif (GXSXG) is a highly conserved sequence located around the middle of amino acid sequences that contain an active side residue of serine hydrolase family genes, including lipases, esterases and proteases (Baudouin et al., 1997; Bézier et al., 2002; Lee et al., 2010; Reina et al., 2007). The alignment showed that all proteins predicted from VfCXE genes contained two consensus sequence motifs, ‘HGGGF’ related to the lipase/esterase group and a highly conserved serine hydrolase motif (GXSXG) in the middle of the amino acid sequences, which contain an active side residue of serine (Fig. 1). In addition, the catalytic triad, serine (S), situated in the motif ‘GXSXG’, aspartic acid (D), and histidine (H) were also strictly conserved at the C-terminal of all proteins predicted from the six VfCXE genes (Fig. 1). The position of the residue of S among VfCXE ranged from 154-167th, D varied from 253-269th, and H ranged from 283-299th amino acids. The residues of the catalytic triad were dispersed in primary amino acid sequences, but came together in the tertiary structure (Fig. 1, 2), which is essential for activity of hormone sensitive lipase in humans (Osterlund et al., 1996). The integrated results of sequence analyses revealed that the proteins predicted from the CXE genes of V. flexuosa belong to the carboxylesterase family in the serine hydrolase superfamily.
To investigate the genetic relationships of VfCXEs with other Vitis species proteins, homologous protein sequences of VfCXE genes were collected by BLAST searches of the NCBI database. A phylogenetic tree was then constructed from the CXE amino acid sequences of Vitis by the neighbor joining method using MEGA ver. 6.0 (Fig. 3). The CXE proteins predicted from the VfCXE transcripts tested in this study were divided into two groups (group I and II) with strong bootstrap support for the monophyly of each clade. VfCXE18231, VfCXE47674, and VfCXE13132 were clustered into group I, whereas VfCXE17159, VfCXE12827, and VfCXE5585 were clustered into group II. A BLAST search of the NCBI database also indicated that the deduced protein sequences of the six loci of the VfCXE gene showed a high degree of similarity to CXE protein sequences from other plants. All VfCXE proteins were highly homologous to protein sequences that originated from Vitis vinifera, with greater than 92% query coverage values and E values of 0.0, indicating their relatively conserved evolutionary relationship at the protein level (Table 5).
To investigate the expression patterns of VfCXE genes against several pathogen infections, expression analysis was performed by quantitative real-time PCR using gene specific primers based on nucleotide sequence alignment. The expression levels of tested genes in grapevine leaves infected with pathogens were examined as expression relative to uninoculated control samples. The expression levels of six VfCXE genes against E. ampelina pathogens are shown in Fig. 4A. VfCXE12827 was upregulated at all time points, whereas the remaining genes (except for VfCXE5585) were downregulated at all time points during infection of E. ampelina. The highest expression was observed in VfCXE12827 at 48 hpi, which was upregulated 3-fold relative to the control. The responses of VfCXE genes to B. cinerea infection are presented in Fig. 4B. All genes showed active response against B. cinerea, with three genes (VfCXE5585, VfCXE12827, and VfCXE13132) being upregulated and the remaining three (VfCXE17159, VfCXE18231, and VfCXE47674) being downregulated. VfCXE12827 showed the highest expression, which was 3.5 fold greater than the control at 48 hpi with B. cinerea. All transcripts showed active responses against R. vitis inoculation (Fig. 4C), and all genes showed upregulated expression at 1 hpi and abruptly decreased expression at 6 hpi. At 48 hpi with R. vitis, all genes were again upregulated, except for VfCXE12827, which was downregulated. The highest expression was observed at 24 hpi in VfCXE12827 by R. vitis inoculation, which was downregulated by 2-fold relative to the control.
Generally, hypersensitive cell death through the recognition of pathogen avr genes by plant R genes occurs at early stage of resistant response against pathogen infections (Liu et al., 2007). Bézier et al. (2002) reported that hsr203J (tobacco) showed highest response at 48 hpi. It also reported that Ralstonia solanacearum induced the development of HR response on tobacco leaves within 18-24 hpi (Pontier et al., 1998a). Therfore, we were to analyze expression pattern of HR related genes up to 48 hpi of pathogens in the grapevine leaves.
Carboxylesterases belong to the serine hydrolase superfamily, which catalyzes the hydrolysis of compounds containing an ester linkage (Baudouin et al., 1997). Carboxylesterase proteins share a conserved motif of pentapeptide (GXSXG). The main feature of carboxylesterases is the conserved catalytic triad composed of an active site serine surrounded by the conserved consensus sequence, GXSXG, an aspartic acid or a glutamic acid and a histidine (Marshall et al., 2003). In the present study, the molecular structure was analyzed to detect common characteristic features in six VfCXE transcripts based on their predicted amino acid sequences in V. flexuosa. Hydrolase like domains in all six VfCXE proteins were detected by the SMART program in this study. All VfCXE proteins tested in this study contained two conserved motifs (GXSXG and HGGGF), as well as a catalytic triad of serine, aspartic acid and histidine (Fig. 1, 2), which are characteristic features of carboxylesterase in the serine hydrolase superfamily.
Resistance to plant diseases is characterized by either partial or complete suppression of pathogen growth at the site of infection. To protect themselves, plants employ several defense mechanisms against invading pathogens. One typical feature of disease resistance is death of plant cells at the site of infection to confine the growth of pathogens, known as the HR (Greenberg, 1996). Generally, HR occurs in response to interaction between plant R genes and corresponding pathogen avr genes (Ade et al., 2007; Hammond-Kosack and Jones, 1996). In addition to R genes, some genes belonging to the carboxylesterase family are reportedly expressed during HR (Baudouin et al., 1997; Bézier et al., 2002; Marshall et al., 2003; Tronchet et al., 2001). Carboxylesterase genes such as EDS1, PAD4, HSR203J, PRLIP1, and PepEST have been isolated from plant-microbe interactions (Ko et al., 2005). HSR203J is reportedly a tobacco carboxylesterase gene that is rapidly activated in a highly localized and specific manner to enable incompatible interactions between tobacco and the bacterial pathogens R. solanacerum, Pseudomonas syringae pv. pisi, and Erwinia amylovora (Pontier et al., 1998a). Pontier et al. (1998a) also reported that Lehsr203, a tomato carboxylesterase gene, was rapidly and transiently induced in leaves of the tomato containing Cf-9 disease resistance gene following Avr9 product infiltration, but not in the absence of Cf-9. A pepper carboxylesterase gene PepEST showed induced expression in fruit following infection with Colletotrichum gloeosporioides fungus (Ko et al., 2005).
HSR203J was considered to be a HR-specific marker because its expression was strongly correlated with programmed cell death in response to different HR inducing pathogens and elicitors, as well as in response to various cell death triggering extra cellular agents such as heavy metals, but not following exposure to virulent pathogens and elicitors that did not induce cell death (Pontier et al., 1994, 1998b). In Arabidopsis, EDS1 is a signaling component of disease resistance pathways activated by the TIR-NBS-LRR class of R genes (Aarts et al., 1998; Parker et al., 1996). The PAD4 gene was found to be required for expression of multiple defense genes against pathogens (Jirage et al., 1999).
Ir was reported that HR activation in plants is mediated by different rapid changes, such as the production of reactive oxygen species (ROS) such as H2O2 (Levine et al., 1994), ionic fluxes (Mittler et al., 1995; Nürnberger et al., 1994), and protein phosphorylation (Dunigan and Madlener, 1995). In contrast, Zurbriggen et al. (2010) reported that it was not clear if ROS participate in triggering localized cell death, in the induction of pathogenesis-related genes, or in both pathways and that ROS-independent processes also contributed to the HR. Tada et al. (2004) reported that ROS are not essential mediators for the initiation of hypersensitive cell death. Pontier et al. (1998b) suggested that a pathway not requiring H2O2 (ROS) is involved in the hsr203J gene activation or that ROS is insufficient as a cell death activator under these conditions. Like hsr203J gene, hypersensitivity related genes studied in this experiment, may have no strong relations with ROS. Although HR responses including ROS accumulation in plants were not analyzed resistnt response in plants, expression genes related with HR in initial stages of resistant responses suggested that HR was induced by patyhogen infections in grapevines.
Thomma et al. (1998) reported that over expression of PepEST in transgenic Arabidopsis plants showed restriction of Alternaria brassicicola colonization by inhibiting spore production. Ko et al. (2005) reported that PepEST accumulation was localized in epidermal and cortical cell layers in infected ripe fruit during immunochemical examination. Lee et al. (2010) also reported that some Brassica oleracea chlorophyllase isozymes which are serine hydrolase family enzymes degraded chlorophyll. Specifically, it has been reported that a detoxifying esterase from the bacterium Pantoea dispersa abolished the function of albicidin phytotoxin, which is a potent pathogenicity factor produced during the infection of sugarcane by Xanthomonas albilineans that causes a devastating disease known as leaf scald (Zhang and Birch, 1997).
It has been reported that a grape carboxylesterase gene, BIG8.1, was induced in grape leaves during the infection of B. cinerea (Bézier et al., 2002). Additionally, it was reported that a conserved carboxylesterase inhibit resistant phenotypes triggered by AvrBsT, a type III effector from Xanthomonas campestris pv. vesicatoria that is translocated into plant cells during infection in Arabidopsis (Cunnac et al., 2007). Tronchet et al. (2001) reported that antisense suppression of a tobacco gene HSR203J showed accelerated HR against R. solanacearum and increased resistance against P. syringae pv. Pisi and Phytophthora parasitica, suggesting that HSR203J may be involved in the suppression of HR cell death. It suggests that carboxylesterases are involved in HR in plants infected with pathogens, and have roles in synthesis or degradation of a molecule in signal transduction pathways for disease resistance responses in plants. The exact functional mechanisms of CXEs in plants is not well understood, even though several studies have characterized carboxylesterase activity (Gershater and Edwards, 2007).
We attempted to investigate the response of six VfCXE genes against infection with fungi and bacteria in grapevines. In the expression study, all VfCXE genes showed changes in their expression in response to infection by all pathogens. VfCXE5585 and VfCXE12827 commonly showed upregulation and VfCXE17159, VfCXE18231, and VfCXE47674 were commonly downregulated, whereas VfCXE13132 showed reverse expression by fungal pathogens tested in this study (Fig. 4A, B). In the case of R. vitis infection, all transcripts showed upregulation at early and later time points, except VfCXE12827, which decreased rapidly after showing upregulation at 1 hpi and downregulation for up to 48 hpi (Fig. 4C). Among all genes, VfCXE12827 exhibited strong and interesting expression against fungal and bacterial pathogens as indicated by upregulationin response to E. ampelina and B. cinerea at all time points and downregulation after 1 hpi against R. vitis. Although it is well known that pathogen responsive carboxylesterase genes are upregulated against pathogen attacks, we observed both up and down regulated response of VFCXE genes in response to plant pathogens. It has also been reported that V. vinifera HSR1 transcripts were undetectable at 18 to 48 hpi and were upregulated at later time points after inoculation of Plasmopara viticola (Chong et al., 2008), which is similar to the expression of VfCXE transcripts against R. vitis. Islam et al. (2015a, 2015b) also reported that several receptor-like protein kinases and CC-NBS-LRR genes, which are considered potential plant resistance genes, showed up-and down-regulated expression against plant pathogens. Therefore, the change in expression level of all tested VfCXE genes after pathogen infection indicated that VfCXE might play a role in signal transduction pathways for disease resistance or a correlated response, similar to other characterized carboxylesterase genes.
In conclusion, six transcripts of the VfCXE gene were identified and characterized in this study that showed various expression levels in the transcriptome of grapevines inoculated with E. ampelina. The molecular structure was analyzed for characteristic features in the six VfCXE genes based on their predicted amino acid sequences in V. flexuosa, and the expression pattern of VfCXE transcripts was investigated by real-time PCR using gene specific primers in response to pathogen infection in grapevines. Six non-duplicated VfCXE genes were identified from transcriptome analysis by NGS of V. flexuosa inoculated with E. ampelina. Structural analysis and a comparison study confirmed that the tested genes are members of the carboxylesterase gene of the serine hydrolase superfamily. All of these genes also showed a high degree of homology with other carboxylesterase genes. Expression analysis revealed that all tested genes showed responses to differential expression against all tested pathogens, indicating that these genes may be related to plant resistance responses against different pathogens in grapevines. Among tested genes, VfCXE12827 showed upregulated expression at 1 hpi against all tested pathogens and showed upregulation against E. ampelina and B. cinerea at all time points. VfCXE5585 and VfCXE13132 showed upregulation in grapevines infected with B. cinerea. All transcripts showed upregulated expression at early time points in response to R. vitis infection. Therefore, it suggests that VfCXE12827 was commonly upregulated gene against fungal and bacterial pathogens, VfCXE5585 and VfCXE13132 were upregulated specifically in response to B. cinerea, and all tested VfCXE genes may involve in signaling role at early stage in resistant resposes against R. vitis infection. Taken together, these results provide valuable information for elucidating the complex molecular mechanisms of responses resistant to diseases in grapevines.
Acknowledgments
This work was supported by a grant (Grant No. PJ011631) from Agricultural R&D Project, Rural Development Administration, Republic of Korea. We thank Dr. Cha for donating pathogenic isolate of Rhizobium vitis used in this study.
Fig. 1
Multiple alignment of six predicted proteins from the VfCXE gene and two other CXE proteins. The two motifs characteristic of carboxylesterase are shown by bars under the motifs and asterisks indicate the positions of the catalytic triad inferred from HSR203J. Data from the article of Baudouin et al. (1997) (Eur. J. Biochem. 248:700- 706). Vf1, VfCXE5585; Vf2, VfCXE12827; Vf3, VfCXE13132; Vf4, VfCXE17159; Vf5, VfCXE18231; Vf6, VfCXE47674; Vv1, BIG8.1; Nt1, HSR203J. The position of the residue of S among VfCXE ranged from 154-167th, D varied from 253-269th, and H ranged from 283-299th amino acids (asterisks).

Fig. 2
Representative tertiary structure of VfCXE protein using VfCXE5585 amino acid sequence created by PyMOL (Schrodinger, New York, NY, USA). Arrows indicate conserved catalytic triad.
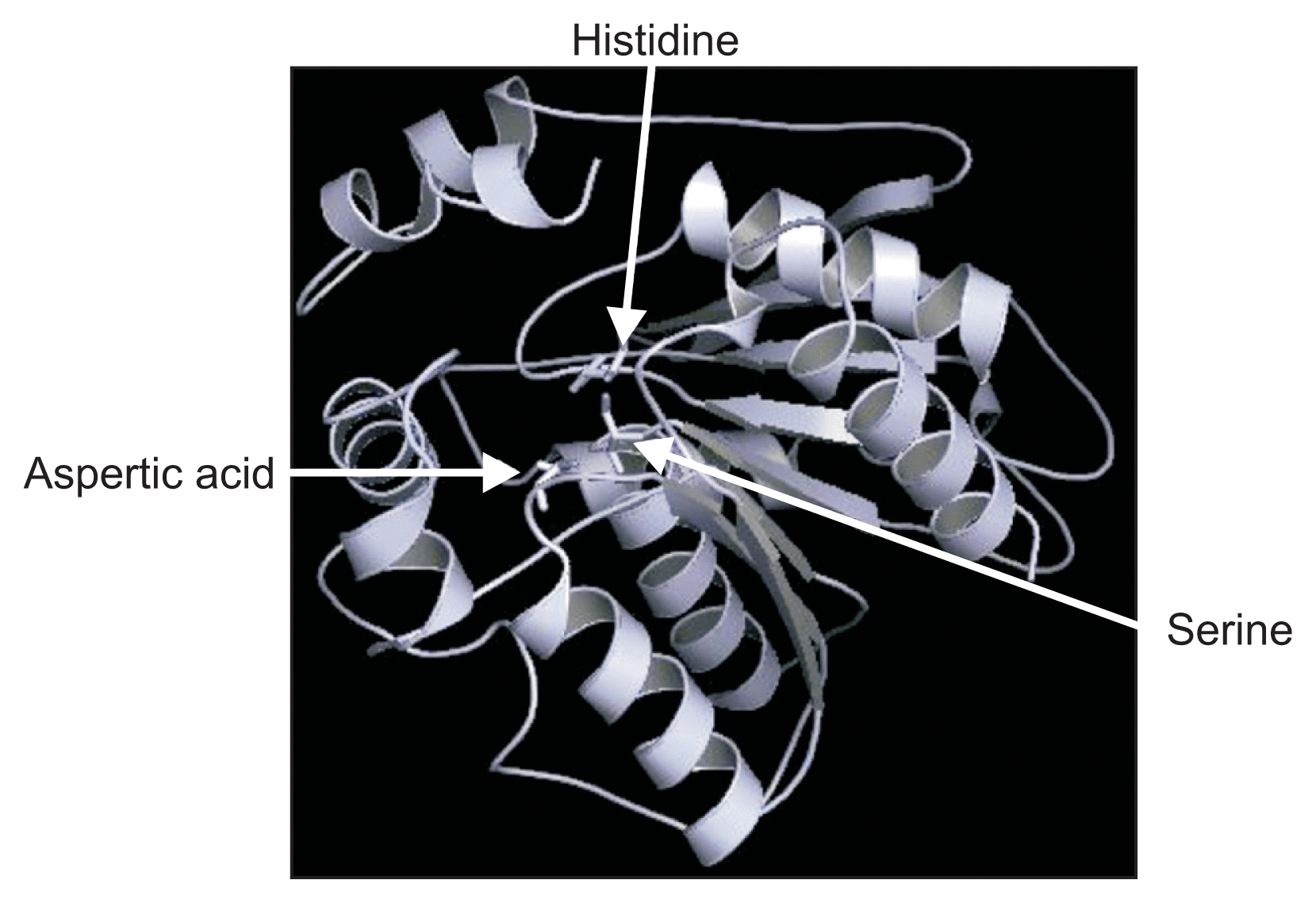
Fig. 3
Phylogenetic tree of predicted proteins from the VfCXE genes with other CXE proteins. The unrooted tree was generated using the MEGA ver. 6.0 software and the neighbor-joining method. Bootstrap values (above 70%) from 1,000 replicates are indicated at each node.
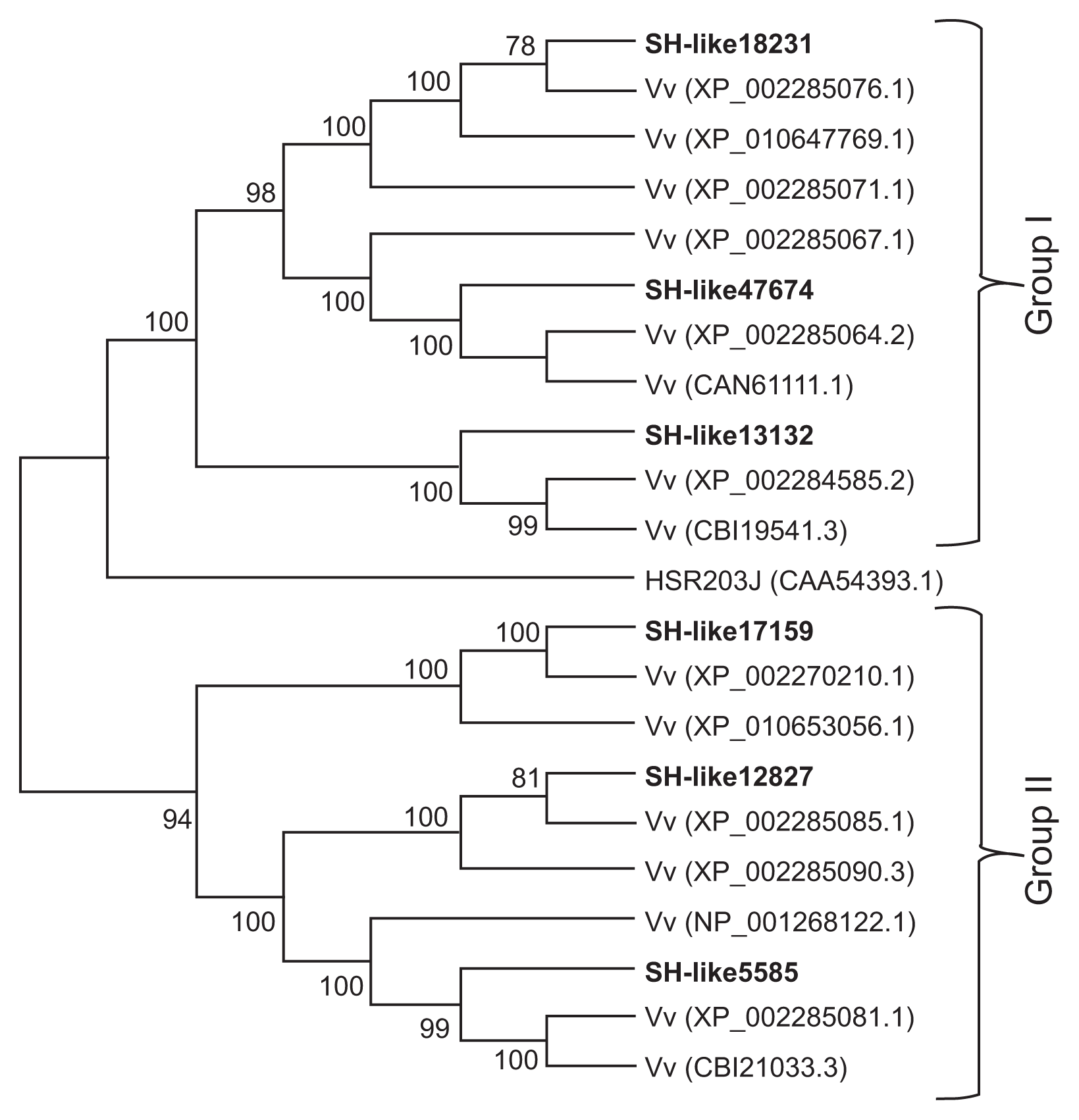
Fig. 4
Expression pattern of six CXE genes of Vitis flexuosa against Elsinoe ampelina (A), Botrytis cinerea (B), and Rhizobium vitis (C). The error bars represent the standard error of the means of three independent replicates.
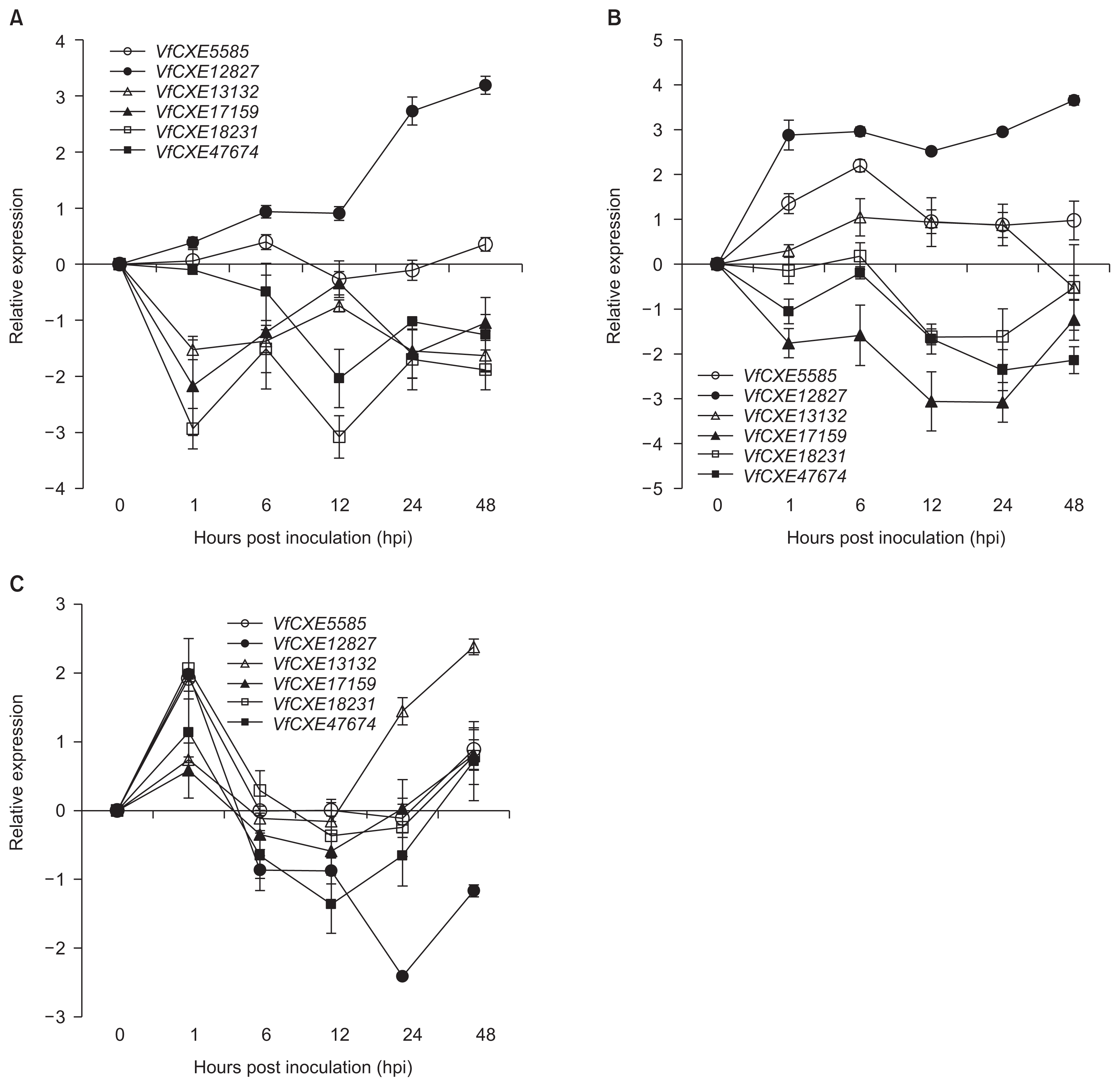
Table 1
Specific primers based on alignment of six VfCXE genes used for real-time PCR
Table 2
Percent identities of six predicted proteins from the VfCXE gene
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| 1 | - | 77 | 35 | 36 | 39 | 35 |
| 2 | 77 | - | 37 | 37 | 39 | 38 |
| 3 | 35 | 37 | - | 33 | 53 | 54 |
| 4 | 36 | 37 | 33 | - | 28 | 30 |
| 5 | 39 | 39 | 53 | 28 | - | 58 |
| 6 | 35 | 38 | 54 | 30 | 58 | - |
Table 3
Predicted primary structure of six CXE genes from Vitis flexuosa
Table 4
Predicted secondary structures of six proteins from the VfCXE gene
Table 5
Homology analysis of six predicted proteins from the VfCXE gene of Vitis flexuosa
References
Aarts, N, Metz, M, Holub, E, Staskawicz, BJ, Daniels, MJ and Parker, JE 1998. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci U S A. 95:10306-10311.



Ade, J, DeYoung, BJ, Golstein, C and Innes, RW 2007. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A. 104:2531-2536.



Ahn, SY, Kim, SA, Jo, SH and Yun, HK 2014. De novo transcriptome assembly of Vitis flexuosa grapevine inoculated with Elsinoe ampelina. Plant Genetic Resour. 12:S130-S133.
Baudouin, E, Charpenteau, M, Roby, D, Marco, Y, Ranjeva, R and Ranty, B 1997. Functional expression of a tobacco gene related to the serine hydrolase family: esterase activity towards short-chain dinitrophenyl acylesters. Eur J Biochem. 248:700-706.


Bézier, A, Lambert, B and Baillieul, F 2002. Cloning of a grapevine Botrytis-responsive gene that has homology to the tobacco hypersensitivity-related hsr203J. J Exp Bot. 53:2279-2280.


Chahinian, H and Sarda, L 2009. Distinction between esterases and lipases: comparative biochemical properties of sequence-related carboxylesterases. Protein Pept Lett. 16:1149-1161.


Chang, S, Puryear, J and Cairney, J 1993. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol. 11:113-116.


Choi, YJ, Yun, HK, Park, KS, Noh, JH, Heo, YY, Kim, SH, Kim, DW and Lee, HJ 2010. Transcriptional profiling of ESTs responsive to Rhizobium vitis from ‘Tamnara’ grapevines (Vitis sp.). J Plant Physiol. 167:1084-1092.


Chong, J, Le Henanff, G, Bertsch, C and Walter, B 2008. Identification, expression analysis and characterization of defense and signaling genes in Vitis vinifera. Plant Physiol Biochem. 46:469-481.


Cunnac, S, Wilson, A, Nuwer, J, Kirik, A, Baranage, G and Mudgett, MB 2007. A conserved carboxylesterase is a supressor of AvrBst-elicited resistance in Arabidopsis. Plant Cell. 19:688-705.


Dunigan, DD and Madlener, JC 1995. Serine/threonine protein phosphatase is required for tobacco mosaic virus-mediated programmed cell death. Virology. 207:460-6.


Gershater, MC and Edwards, R 2007. Regulating biological activity in plants with carboxylesterase. Plant Sci. 173:579-588.
Greenberg, JT 1996. Programmed cell death: a way of life for plants. Proc Natl Acad Sci U S A. 93:12094-12097.



Guruprasad, K, Reddy, BV and Pandit, MW 1990. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 4:155-161.


Hammond-Kosack, KE and Jones, JD 1996. Resistance gene-dependent plant defense responses. Plant Cell. 8:1773-1791.



Hammond-Kosack, KE and Jones, JD 1997. Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol. 48:575-607.


Islam, MZ, Ahn, SY and Yun, HK 2015a. Identification of six transcripts encoding putative receptor-likeprotein kinase (RLK) and their expression profiles in Vitis flexuosa infected with pathogens. Sci Hortic. 192:108-116.

Islam, MZ, Ahn, SY and Yun, HK 2015b. Analysis of structure and differential expression of Pseudomonas syringae 5-like (RPS5-like) genes in pathogen-infected Vitis flexuosa. Turk J Biol. 39:775-789.

Jirage, D, Tootle, TL, Reubert, TL, Frost, LN, Feyes, BJ, Parker, JE, Ausubel, FM and Glazebrook, J 1999. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci U S A. 96:13583-13588.



Kaschani, F, Gu, C, Niessen, S, Hoover, H, Cravatt, BF and van der Hoorn, RA 2009. Diversity of serine hydrolase activities of unchallenged and Botrytis-infected Arabidopsis thaliana. Mol Cell Proteomics. 8:1082-1093.



Ko, MK, Jeon, WB, Kim, KS, Lee, HH, Seo, HH, Kim, YS and Oh, BJ 2005. A Colletotrichum gloeosporioides-induced esterase gene of nonclimacteric pepper (Capsicum annuum) fruit during ripening plays a role in resistance against fungal infection. Plant Mol Biol. 58:529-541.



Kong, X, Lv, W, Jiang, S, Zhang, D, Cai, G, Pan, J and Li, D 2013. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics. 14:433




Lee, GC, Chepyshko, H, Chen, HH, Chu, CC, Chou, YF, Akoh, CC and Shaw, JF 2010. Genes and biochemical characterization of three novel chlorophyllase isozymes from Brassica oleracea. J Agric Food Chem. 58:8651-8657.


Levine, A, Tenhaken, R, Dixon, R and Lamb, C 1994. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 79:583-593.


Liu, J, Liu, X, Dai, L and Wang, G 2007. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genomics. 34:765-776.


Macro, YJ, Ragheh, F, Godiard, L and Froissard, D 1990. Transcriptional activation of 2 classes of genes during the hypersensitive reaction of tobacco leaves infiltrated with an incompatible isolate of the phytopathogenic bacterium Pseudomonas solanacearum. Plant Mol Biol. 15:145-154.



Marshall, SD, Putterill, JJ, Plummer, KM and Newcomb, RD 2003. The carboxylesterase gene family from Arabidopsis thaliana. J Mol Evol. 57:487-500.



Mittler, R, Shulaev, V and Lam, E 1995. Coordinated activation of programmed cell death and defense mechanisms in transgenic tobacco plants expressing a bacterial proton pump. Plant Cell. 7:29-42.



Nürnberger, T, Nennstiel, D, Jabs, D, Sacks, T, Hahlbrock, K and Scheel, D 1994. High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell. 78:449-460.


Osterlund, T, Danielsson, B, Degerman, E, Contreras, JE, Edgren, G, Davis, RC, Schotz, MC and Holm, C 1996. Domain-structure analysis of recombinant rat hormone-sensitive lipase. Biochem J. 319:411-420.




Parker, JE, Holub, EB, Frost, LN, Falk, A, Gunn, ND and Daniel, MJ 1996. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 8:2033-2046.



Pontier, D, Balague, C and Roby, D 1998a. The hypersensitive response. A programmed cell death associated with plant resistance. C R Acad Sci III. 321:721-734.


Pontier, D, Godiard, L, Marco, Y and Roby, D 1994. hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J. 5:507-521.


Pontier, D, Tronchet, M, Rogowsky, P, Lam, E and Roby, D 1998b. Activation of hsr203, a plant gene expressed during incompatible plant-pathogen interactions, is correlated with programmed cell death. Mol Plant-Microbe Interact. 11:544-554.


Reina, JJ, Guerrero, C and Heredia, A 2007. Isolation, characterization, and localization of AgaSGNH cDNA: a new SGNH-motif plant hydrolase specific to Agave americana L. leaf epidermis. J Exp Bot. 258:2717-2731.

Sharmin, S, Moosa, MM, Islam, MS, Kabir, I, Akter, A and Khan, H 2011. Identification of a novel dehydration responsive transcript from tossa jute (Corchorus olitirius L.). J Cell Mol Biol. 9:21-29.
Tada, Y, Mori, T, Shinogi, T, Yao, N, Takahashi, S, Betsuyaku, S, Sakamoto, M, Park, P, Nakayashiki, H, Tosa, Y and Mayama, S 2004. Nitric oxide and reactive oxygen species do not elicit hypersensitive cell death but induce apoptosis in the adjacent cells during the defense response of oat. Mol Plant-Microbe Interact. 17:245-253.


Thomma, BP, Eggermont, K, Penninckx, IA, Mauch-Mani, B, Vogelsang, R, Cammue, BP and Broekaert, WF 1998. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci U S A. 95:15107-15111.



Tronchet, M, Ranty, B, Macro, Y and Roby, D 2001. HSR203 antisense suppression in tobacco accelerates development of hypersensitive cell death. Plant J. 27:115-127.


Wang, Q, Zhang, Y, Gao, M, Jiao, C and Wang, X 2011. Identification and expression analysis of a pathogen responsive PR-1 gene from Chinese wild Vitis quinquangularis. Afr J Biotechnol. 10:17062-17069.
Yun, HK, Park, KS, Rho, JH, Kwon, BO and Jeong, SB 2003. Development of an efficient screening system for anthracnose resistance in grapes. J Korean Soc Hortic Sci. 44:809-812.



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




