Xanthomonas euvesicatoria Causes Bacterial Spot Disease on Pepper Plant in Korea
Article information
Abstract
In 2004, bacterial spot-causing xanthomonads (BSX) were reclassified into 4 species—Xanthomonas euvesicatoria, X. vesicatoria, X. perforans, and X. gardneri. Bacterial spot disease on pepper plant in Korea is known to be caused by both X. axonopodis pv. vesicatoria and X. vesicatoria. Here, we reidentified the pathogen causing bacterial spots on pepper plant based on the new classification. Accordingly, 72 pathogenic isolates were obtained from the lesions on pepper plants at 42 different locations. All isolates were negative for pectolytic activity. Five isolates were positive for amylolytic activity. All of the Korean pepper isolates had a 32 kDa-protein unique to X. euvesicatoria and had the same band pattern of the rpoB gene as that of X. euvesicatoria and X. perforans as indicated by PCR-restriction fragment length polymorphism analysis. A phylogenetic tree of 16S rDNA sequences showed that all of the Korean pepper plant isolates fit into the same group as did all the reference strains of X. euvesicatoria and X. perforans. A phylogenetic tree of the nucleotide sequences of 3 housekeeping genes—gapA, gyrB, and lepA showed that all of the Korean pepper plant isolates fit into the same group as did all of the references strains of X. euvesicatoria. Based on the phenotypic and genotypic characteristics, we identified the pathogen as X. euvesicatoria. Neither X. vesicatoria, the known pathogen of pepper bacterial spot, nor X. perforans, the known pathogen of tomato plant, was isolated. Thus, we suggest that the pathogen causing bacterial spot disease of pepper plants in Korea is X. euvesicatoria.
Introduction
Bacterial spot disease occurs on pepper (Capsicum annum L.) and tomato (Solanum lycopersicum L.) in warm, humid areas worldwide and causes lesions on the leaves, stems, and fruits (Jones et al., 2000; Stall et al., 1994). Yellow haloes appear around the lesions; smaller lesions coalesce into larger ones. Leaf infection results in blight, necrosis, and early leaf fall. These cause a reduction in photosynthesis and fruit infection, resulting in direct economic loss (Jones et al., 1991; Obradovic et al., 2004; Stall et al., 1994). Contaminated seeds and plant debris are common inoculum sources, and the disease is also transmitted by rain splash (Jones et al., 1991).
The pathogens causing bacterial spot disease were originally identified as Bacterium vesicatoria (Doidge, 1921) and B. exitiosum (Gardner and Kendrick, 1921). The 2 bacteria were later classified as Xanthomonas vesicatoria and then as X. campestris pv. vesicatoria by Young et al. (1978). Based on DNA homology by Vauterin et al. (1995), X. campestris pv. vesicatoria was separated into 2 species—X. vesicatoria and X. axonopodis pv. vesicatoria. Pseudomonas gardneri was first reported as the pathogen causing bacterial spot on tomato (Šutic, 1957) but was later reclassified as X. gardneri (De Ley, 1978; Dye, 1966). Jones et al. (2004) reported that all of the bacterial spot-causing xanthomonads (BSX) were reclassified as 4 species—X. euvesicatoria, X. vesicatoria, X. perforans, and X. gardneri. Among them, X. euvesicatoria and X. vesicatoria cause diseases on both pepper and tomato, while X. perforans and X. gardneri are known to infect only tomato. Recently, however, X. perforans was isolated from the pepper plant (Potnis et al., 2015).
X. vesicatoria and X. perforans have strong amylolytic and pectolytic activity, but X. euvesicatoria and X. gardneri do not (Bouzar et al., 1994; Jones et al., 2000, 2004). X. euvesicatoria has a unique 32 kDa protein, while the other BSX have a 27 kDa protein (Bouzar et al., 1994; Jones et al., 2004). In addition, there are differences in carbon source utilization among BSX species (Jones et al., 2004; Stoyanova et al., 2014; Vauterin et al., 1995). RpoB based restriction fragment length polymorphism (RFLP) (Ferreira-Tonin et al., 2012), amplified fragment length polymorphism (AFLP) (Hamza et al., 2012) and multilocus sequence analysis (Almeida et al., 2010; Hamza et al., 2012; Kebede et al., 2014; Timilsina et al., 2015) were used to differentiate 4 species of BSX.
Bacterial spot is a common disease on pepper plants in Korea (Kim, 2004; Lee and Cho, 1996; Lee et al., 1999; Myung et al., 2005, 2006), and X. axonopodis pv. vesicatoria and X. vesicatoria are listed as the causative pathogens (Yoo, 2009). X. perforans was reported as the causal agent of bacterial spot on tomato for the first time from a nursery farm in Korea (Myung et al., 2009). It is not clear as to which pathogens cause bacterial spot disease of pepper in Korea since X. axonopodis pv. vesicatoria is no longer included in the list of BSX, and since X. perforans, which was known to cause the disease on pepper plant, has been isolated only from tomato. The correct identification of the bacterial spot pathogen on pepper is important for plant quarantine, disease management, and breeding for resistance. In this study, the pathogen causing bacterial spot disease of pepper was reidentified by the isolation and identification of bacterial spot disease pathogens throughout Korea. To ensure correct identification, several phenotypic and genotypic characteristics were used, including amylolytic activity, pectolytic activity, unique protein band on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), rpoB based RFLP, phylogenetic analysis with 16S rDNA sequences, and sequences of 3 housekeeping genes.
Materials and Methods
Isolation and pathogenicity test
Pepper leaves showing typical bacterial spot lesions were collected throughout Korea during 2013–2015. Small pieces of leaf lesions were macerated in sterile water, and the resulting suspension was streaked on nutrient agar (NA) (DifcoTM; BD, Sparks, MD, USA). After incubation at 27°C for 3–5 days, distinct single colonies were purified by subculturing. Isolates were stored in a deep freezer. Bacterial suspensions, optical density measured at a wavelength of 600 nm (OD600) = 0.1 (ca. 1.0 × 108 cfu/ml) were prepared on NA in sterile water using 3-day-old cell cultures, and the suspensions were sprayed on pepper and tomato seedlings. The inoculated plants were saturated and maintained in a humid environment for 48 h and then in the greenhouse. Bacterial spot symptoms were observed 3 weeks post-inoculation.
Reference strains
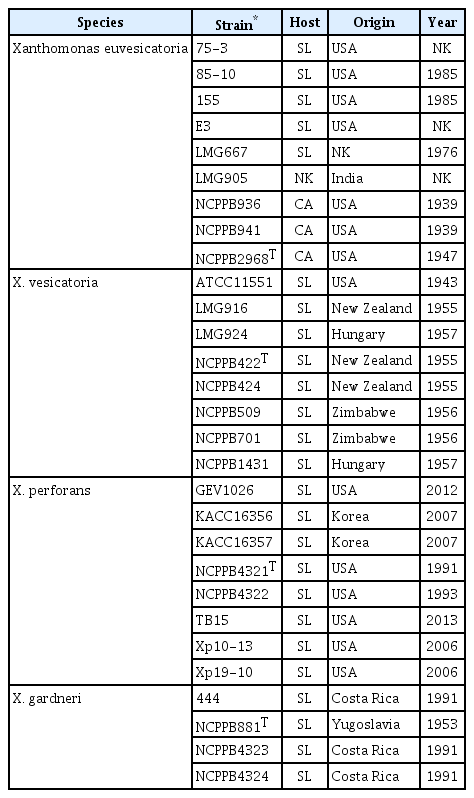
Twenty-nine different strains from 4 BSX species were used as reference strains in this study (Table 1).
Amylolytic and pectolytic assays
Amylolytic and pectolytic assays were carried out according to the method of Bouzar et al. (1994). Bacteria were streaked on brilliant cresyl blue-starch (BS) agar and incubated at 27°C for 2 days. Haloes around the colonies indicated that the strain was positive for amylolytic activity. For pectolytic assay, bacterial cells were spotted on crystal violet pectate (CVP) agar and incubated at 27°C for 2 days. Dents around the colonies indicated that the strain was positive for pectolytic activity.
Observation of unique proteins by SDS-PAGE
SDS-PAGE for the observation of proteins unique to BSX species was carried out according to the method of Bouzar et al. (1994). Bacteria were cultured in 3 ml NA (BD DifcoTM) at 27°C for 18 h. Two milliliters of bacterial culture were harvested by centrifugation (> 13,000 × g) for 10 min, and the bacterial cells were washed twice in sterile water. The cell pellet was resuspended in 180 μl of 10% sorbitol and the bacterial suspension was mixed with an equal volume of 2 × sampling buffer (125 mM Tris-HCl, pH 6.8, 20% glycerol, 2% β-mercaptoethanol, 0.04% bromophenol blue, and 4% SDS). After heating at 100°C for 10 min, 10 μl of suspension were electrophoresed in 12% resolving gel. The gel was stained with Coomassie R250 staining solution (0.1% Coomassie Blue R250 in 10% acetic acid, 45% methanol, 45% H2O) for more than 1 h and destained for more than 2 h.
rpoB gene based RFLP
The rpoB based RFLP was carried out according to the method of Ferreira-Tonin et al. (2012). The rpoB gene was amplified with rpoB2F (5′-TCA AGG AGC GTC TGT CGA T-3′) and rpoB3R (5′-TCT GCC TCG TTG ACC TTG A-3′) primers. PCR amplification was performed in PCR reaction mixture (25 μl) of Takara Ex Taq PCR kit (Takara Co., Shiga, Japan) containing 1 μl of each primer (10 pmol/μl) and 10 μl of genomic DNA (20 ng/μl). The PCR conditions were as follows: an initial denaturation at 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR product was purified and 300 ng of purified PCR product was restricted with HaeIII (FastDigest-Thermo Fisher Scientific Inc., Waltham, MA, USA). The resulting DNA bands were observed after electrophoresis on a 4% agarose gel.
Phylogenetic tree with 16S rDNA sequences
The 16S rDNA was amplified with 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′). The amplicons were sequenced Macrogen Co. (Seoul, Korea). Phylogenetic analysis was carried out using the MEGA 6.0 program with neighbor-joining tree, Kimura 2-parameter model, and 3,000 bootstrap value.
Phylogenetic tree with multilocus sequences
Multilocus sequence analysis (MLSA) was carried out using 3 housekeeping genes—gapA, gyrB, and lepA. The PCR were carried out according to the method of Almeida et al. (2010). PCR primers were gap-1-F (5′-GGC AAT CAA GGT TGG YAT CAA CG-3′) and gap-1-R (5′-ATC TCC AGG CAC TTG TTS GAR TAG-3′) for gapA, gyrB-F (5′-AAG TTC GAC GAC AAC AGC TAC AA-3′) and gyrB-R (5′-GAM AGC ACY GCG ATC ATG CCT TC-3′) for gyrB, and lepA-F (5′-AAG CSC AGG TGC TCG ACT CCA AC-3′) and lepA-R (5′-CGT TCC TGC ACG ATT TCC ATG TG-3′). PCR reactions were performed in reaction mixture (25 μl) of Takara Ex Taq PCR kit containing 1 μl of each primer (10 pmol/μl) and 10 μl of genomic DNA (10 ng/μl). The PCR conditions were as follows: an initial denaturation at 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 7 min. The amplicons were sequenced at Macrogen Co. The concatenated sequence was 444 bp of gapA, 411 bp of gyrB, and 390 of lepA. Phylogenetic analysis was carried out using the MEGA 6.0 program with neighbor-joining tree, Kimura 2-parameter model, and 3,000 bootstrap value.
Results
Pathogen isolation
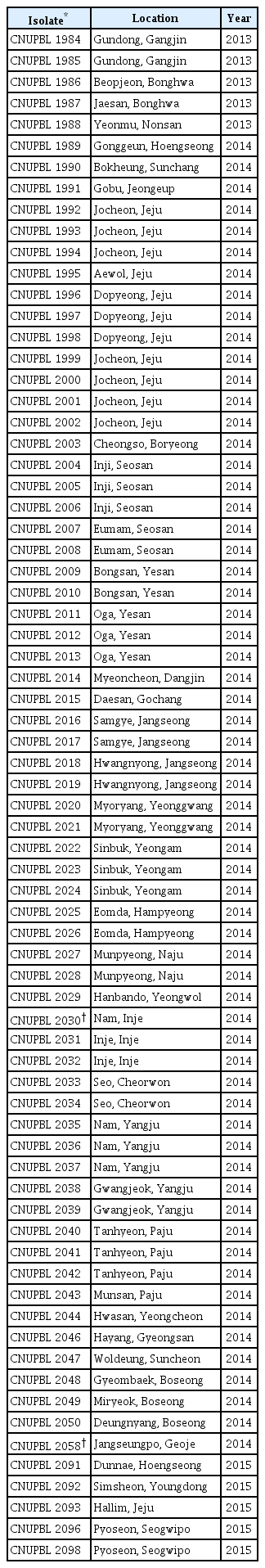
All isolates from the bacterial spot lesions of pepper plants were tested for pathogenicity on both pepper and tomato plants. About 5–10 days after inoculation, water soaked spots started to appear on the lower epidermis of leaves. Circular dark brown and black spots appeared, followed by yellow haloes around some of the spots (Fig. 1). A total of 72 isolates caused typical bacterial spot symptoms on both pepper and tomato plants. Our tests indicated that all of the isolates are pathogenic to both pepper and tomato plants despite the fact that all of them were isolated from only pepper plants. The pathogens were isolated from isolates collected from 42 different locations that cover all provinces of Korea, including Jeju Island (Table 2). The 2 pathogenic isolates CNUPBL 2030 and CNUPBL 2058 were deposited in Korean Agricultural Culture Collection as KACC18722 and KACC18723.
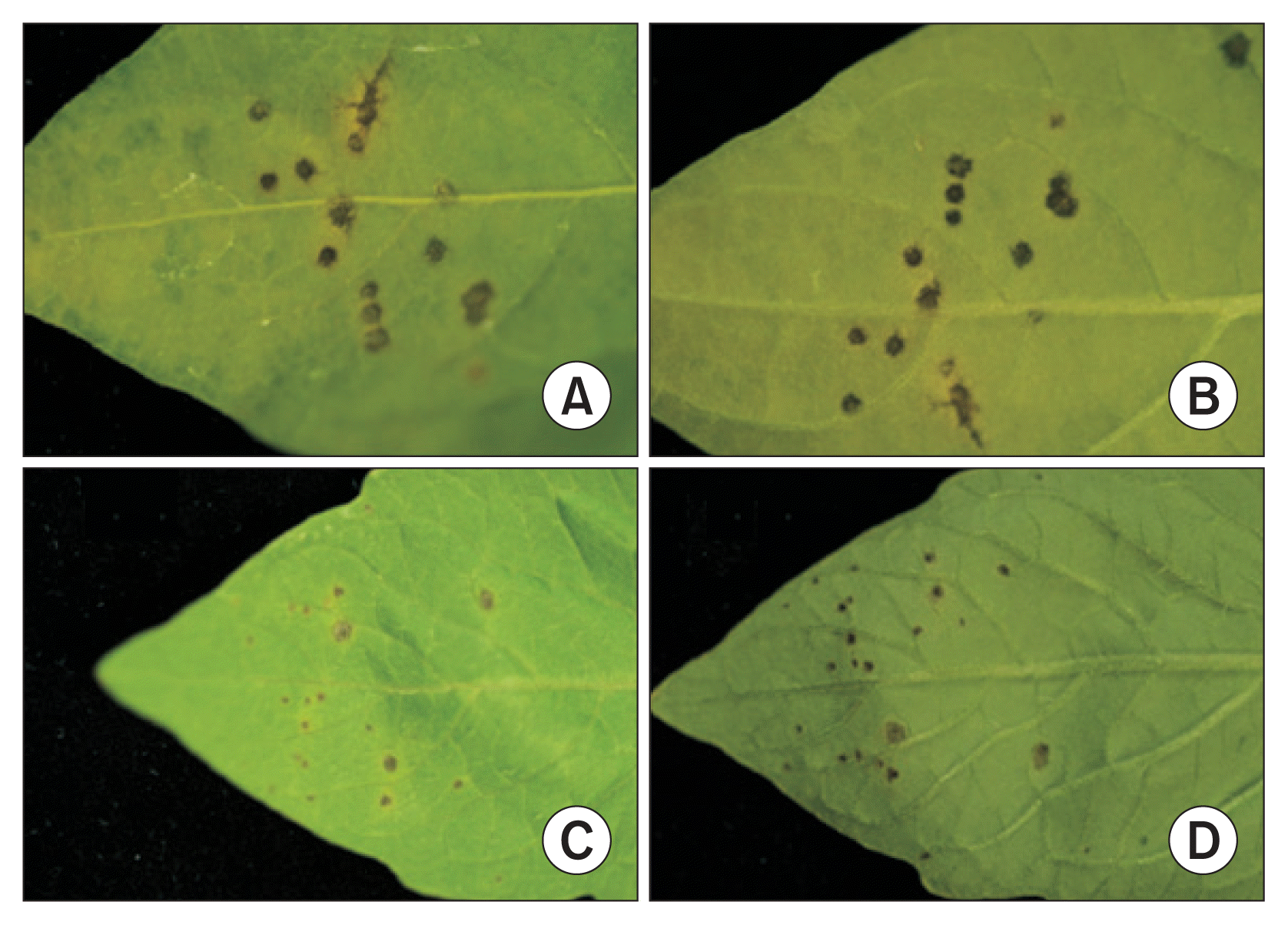
Bacterial spot symptoms on pepper and tomato leaves inoculated with CNUPBL 2039, a pathogenic isolate from bacterial spot lesion of pepper plant. Lesions on the upper epidermis of pepper leaf (A), lower epidermis of pepper leaf (B), upper epidermis of tomato leaf (C), and lower epidermis of tomato leaf (D).
Phenotypic characteristics
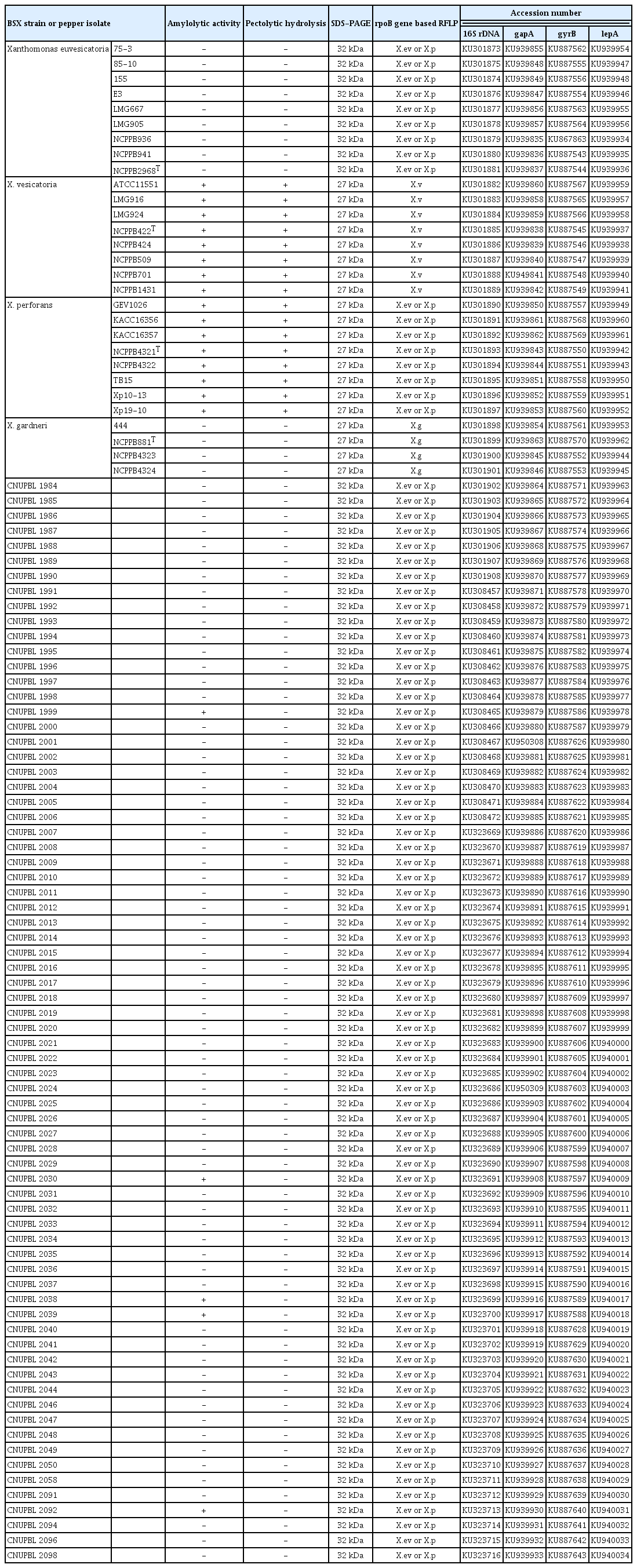
All of the X. vesicatoria and X. perforans reference strains were positive for amylolytic and pectolytic activities, whereas all of the X. euvesicatoria and X. gardneri reference strains were negative for both enzyme activities (Supplementary Fig. 1, Supplementary Fig. 2, Table 3). Five isolates (CNUPBL 1999, 2030, 2038, 2039, 2092) of the Korean pepper pathogens were positive for amylolytic activity and the rest were negative. All of the Korean pepper pathogens were negative for pectolytic activity (Table 3). As for the unique protein of BSX species, all of the X. euvesicatoria reference strains had a 32 kDa protein band and the other reference strains had a 27 kDa protein band (Supplementary Fig. 3, Table 3). All of the Korean pepper pathogens had a 32 kDa protein that is unique to X. euvesicatoria (Table 3).
Genotypic characteristics
In rpoB gene-based RFLP, all of the X. euvesicatoria and X. perforans reference strains had the same DNA band pattern with DNA bands of 339 bp, 154 bp, and 153 bp. X. vesicatoria and X. gardneri had DNA band patterns different from those of X. euvesicatoria and X. perforans, and also had different patterns from each other. X. vesicatoria had DNA bands of 216 bp, 123 bp, and 106 bp, and X. gardneri had DNA bands of 215 bp, 156 bp, 154 bp, and 123 bp (Fig. 2). All of the Korean pepper pathogens had the same DNA band pattern as that of X. euvesicatoria and X. perforans (Table 3).

Result of rpoB gene based restriction fragment length polymorphism. Lanes 1–3, Xanthomonas euvesicatoria 75-3, LMG667, LMG905; Lanes 4–6, X. vesicatoria LMG916, LMG924, ATCC11551; Lanes 7 and 8, X. perforans KACC16356, KACC16357; Lane 9, X. gardneri NCPPB881; Lane 10, negative control.
A phylogenetic tree of the 16S rDNA sequences showed that all of X. vesicatoria and X. gardneri reference strains were grouped into their own clade. All X. euvesicatoria and X. perforans reference strains, however, were grouped into a different clade. All of the Korean pepper pathogens were grouped together with the reference strains of X. euvesicatoria and X. perforans (Fig. 3). In a phylogenetic tree of the concatenated sequences of gapA, gyrB, and lepA, all of the reference strains of each species were grouped into the same clade with strains of the same species. All of the Korean pepper pathogens were grouped together with the reference strains of X. euvesicatoria (Fig. 4).

Phylogenetic tree of 16S rDNA sequences of bacterial spot-causing xanthomonads strains and Korean pepper isolates using MEGA 6.0 program, neighbor-joining tree, Kimura 2-parameter model, and 3,000 bootstrap samples.
Discussion
In this study, 72 pathogenic isolates were collected from bacterial spot lesions on pepper plants throughout Korea in order to reidentify the causative pathogen. The 3 phenotypic characteristics of the Korean pepper pathogens and the BSX reference strains were compared for correct identification. The 3 major characteristics were used to separate the 4 species of BSX referred to by Jones et al. (2004). All of the Korean pepper pathogens were negative for pectolytic activity, and all except 5 isolates were negative for amylolytic activity. These traits were identical to those of X. euvesicatoria and X. gardneri. The 5 amylolytic-positive isolates are not considered to be typical strains of X. euvesicatoria or X. perforans. Recently, Stoyanova et al. (2014) argued that some phenotype characteristics such as amylolytic activity and the utilization of cis-aconitic acid cannot be species-separating criteria of the BSX group. However, all of the Korean pepper pathogens have a 32 kDa protein that is unique to X. euvesicatoria. Thus, our results of the analysis of the 3 phenotypic characteristics suggest that all of the Korean pepper pathogens are X. euvesicatoria.
The result of rpoB based RFLP showed that all of the Korean pepper pathogens have DNA band patterns identical to those of X. euvesicatoria and X. perforans. A phylogenetic tree of the 16S rDNA sequences also showed that all of the Korean pepper pathogens were grouped together with X. euvesicatoria and X. perforans. These results suggest that rpoB based RFLP and 16S sequences are not enough to separate the 2 BSX species, X. euvesicatoria and X. perforans. These results also indicate that the 2 are very closely related to each other. MLSA generally gives more detailed genotypic information than does 16S rDNA sequencing. Several previous MLSA studies have also differentiated the 4 species of BSX (Almeida et al., 2010; Hamza et al., 2012; Kebede et al., 2014; Timilsnia et al., 2015). A phylogenetic tree of 3 housekeeping genes (gapA, gyrB, and lepA) showed that all of the Korean pepper isolates were grouped together into the same clade as that of the reference strains of X. euvesicatoria.
The phenotypic and genotypic characteristics of the Korean pepper pathogens suggest that all of those collected in this study are in fact X. euvesicatoria. Neither X. vesicatoria, which is considered the causative pathogen of pepper bacterial spot in the List of Plant Diseases in Korea (Yoo, 2009), nor X. perforans, which was recently reported as the causative pathogen of tomato bacterial spot, was isolated. It might be erroneous to designate X. vesicatoria as a causative pathogen of pepper bacterial spot in Korea since we could not find literature references for this. Although there is one study on the isolation of X. perforans from pepper plant in the United States, this species does not cause disease on pepper plant in Korea. Nevertheless, bacterial spot caused by X. perforans was reported on nursery-raised tomatoes in Korea, but not on field-grown tomatoes (Myung et al., 2009). X. axonopodis pv. vesicatoria is another species identified as a causative pathogen of pepper bacterial spot according to the List of Plant Diseases in Korea (Yoo, 2009). It was renamed as X. euvesicatoria following the reclassification of the 4 BSX species.
The results of the present study suggest that the bacterial spot of pepper plant in Korea is caused exclusively by X. euvesicatoria. Recently, Myung et al. (2015) reported that the latest strain of pepper bacterial spot disease in Korea is caused by X. euvesicatoria. Based on our study, the pepper bacterial spot reported as a new disease is in fact not new, but rather it is caused by the same pathogen whose scientific name was revised by Jones et al. (2004).
Acknowledgments
This study was supported by the Animal and Plant Quarantine Agency and by a research grant from Chungbuk National University in 2014. We thank Dr. Jeffery Jones for providing the reference strains of BSX.
Notes
Articles can be freely viewed online at www.ppjonline.org.