Characterization of Pseudomonas syringae pv. syringae, Causal Agent of Citrus Blast of Mandarin in Montenegro
Article information
Abstract
Citrus blast caused by bacterium Pseudomonas syringae is a very important disease of citrus occuring in many areas of the world, but with few data about genetic structure of the pathogen involved. Considering the above fact, this study reports genetic characterization of 43 P. syringae isolates obtained from plant tissue displaying citrus blast symptoms on mandarin (Citrus reticulata) in Montenegro, using multilocus sequence analysis of gyrB, rpoD, and gap1 gene sequences. Gene sequences from a collection of 54 reference pathotype strains of P. syringae from the Plant Associated and Environmental Microbes Database (PAMDB) was used to establish a genetic relationship with our isolates obtained from mandarin. Phylogenetic analyses of gyrB, rpoD, and gap1 gene sequences showed that P. syringae pv. syringae causes citrus blast in mandarin in Montenegro, and belongs to genomospecies 1. Genetic homogeneity of isolates suggested that the Montenegrian population might be clonal which indicates a possible common source of infection. These findings may assist in further epidemiological studies of this pathogen and for determining mandarin breeding strategies for P. syringae control.
Introduction
In Mediterranean countries, phytopathogenic bacteria do not generally cause serious damage in citrus groves, excluding the incidence of the blast and black pit, caused by Pseudomonas syringae pv. syringae van Hall 1902. This bacterium infects more than 180 different plant species (Bradbury, 1986), among them Citrus spp. where it causes bacterial blast of orange (Citrus sinensis) and mandarin (Citrus rediculate) (Smith and Fawcett, 1930; Whiteside et al., 1988) and black pit of orange fruits (Mirik et al., 2005; Whiteside et al., 1988). P. syringae pv. syringae causes damage especially under cool and humid conditions in spring when the development and spread of the bacterial blast happens quicker and more easily (Gorlenko, 1965), and when the shoots or fruits are damaged by wind, hail or thorns. Characteristic disease symptoms appearing as water-soaked lesions extended to the mid-vein and to the twigs surrounding the base of the petiole. Ultimately, the leaves dry and curl, while still firmly attached, and eventually fall without petioles. The necrotic areas on twigs further enlarge and the twigs are eventually killed within three to four weeks.
P. syringae pv. syringae is an organism which has been researched on non-woody hosts extensively compared to wood plant like citrus. Only a few studies have been done that take this topic with citrus into account (Mirik et al., 2005; Shigeta and Nakata, 1995; Smith and Fawcett, 1930; Thomidis et al., 2005; Timmer et al., 2000; Vanarelli et al., 2010). The lack of genetic data that considers this citrus pathogen is evident in literature. Scortichini et al. (2003) ascertained genetic variability of P. syringae pv. syringae strains isolated from apple, pear, apricot, Citrus spp. and cereals and determined that no clear relation was observed between the host plant and the bacterial genomic fingerprint (BOX-PCR). Despite the evidence of the occurrence of P. syringae pv. syringae in citrus growing areas in different parts of the world, the lack of molecular studies of pathogen variation is also a factor that limits the better understanding of the pathogen. In Montenegro, the symptoms of citrus blast have been observed for the first time in 1987, when severe outbreaks were found in many citrus nurseries, with disease incidence approaching 10% to 30% (Vučinić, 1987). In a survey for bacterial blast in Montenegro during the spring seasons of 2013 and 2014, the incidence of disease was at least 10%. Given the ubiquity of P. syringae pv. syringae, a better understanding of the epidemiology and pathogen population structure could be used in predicting disease and developing predictive models of outbreaks. For this reason, a collection of P. syringae pv. syringae isolates obtained from diseased Montenegrian mandarin (Citrus reticulata cv. Owari) were studied using syrB analysis which determines the toxin involved in virulence of P. syringae pv. syringae (Sorensen, 1998), partial 16S rDNA sequencing (Gironde and Manceau, 2012; Hilario et al., 2004) and multilocus sequence analysis (MLSA) using three different housekeeping genes (Bull et al., 2011). Furthermore, pathogenicity testing was performed in order to conclude Koch’s postulates. This paper also shows the general similarity and certain differences to other P. syringae reference strains obtained from different hosts deposited in the Plant Associated and Environmental Microbes Database (PAMDB) data base (www.pamdb.org) and to bring together certain information regarding the aformentioned.
Materials and Methods
Bacterial isolates
For the pathogen isolation, 20 samples of necrotic buds from diseased twigs were taken. Small pieces of tissue were taken from the margins of necrotic lesions and were first disinfected with 70% ethanol, then cut and crushed using a mortar and pestle in sterile phosphate-buffered saline (0.27% Na2HPO4, 0.04% NaH2PO4, 0.8% NaCl). After 20 min, the suspensions were plated on King’s medium B (King et al., 1954). After 2 days of incubation at 26°C, a total of 43 fluorescent colonies were selected and purified on Sucrose Nutrient Agar (SNA) (Difco nutrient agar, supplemented with 0.5% sucrose; Difco Laboratories, Detroit, MI, USA) and maintained at −80°C in Luria Bertani medium supplemented with 20% glycerol (Bertani, 1951). The strains CFBP 11 (P. syringae pv. syringae), CFBP 1582 (P. syringae pv. syringae), and CFBP 2119 (P. syringae pv. morsprunorum), were used as reference strains (Collection Francaise de Bacteries Phytopathogenes [CFBP], Angers, France).
Pathogenicity
Pathogenicity testing of the obtained isolates were checked by artificial inoculation of unripe green lemon (Citrus limon) fruits, bean pods (Phaseolus vulgaris) and leaves of mandarin and liliac (Syringa vulgaris) using the methods described by Lelliott and Stead (1987) and Klement (1990). Each experiment was repeated twice for all tested isolates. The bacterial cells were grown for 2 days on SNA at 25°C, and bacterial suspensions were prepared in sterile distilled water (SDW) and photometrically adjusted to the final concentration of 108 cfu/ml. Control fruits and leaves were also included in tests, but treated with reference strains and SDW as positive and negative controls, respectively.
The lemons were surface sterilized by being dipped into 2% NaClO solution for 2 min and then shortly rinsed with SDW. Four punctures were made on the fruits (1 mm wide, 3 mm deep), by using a sterile hypodermic needle and per 20 μl of the bacterial suspension (106 cfu/ml) was placed on each rupture. Bean pods were inoculated by injection of a bacterial suspension (107 cfu/ml) into young bean pods with a hypodermic syringe. Inoculated fruits and pods were kept in plastic bags and incubated at 25°C for 7 days under conditions of high humidity (70–80%). For each isolate, 12 sites involving three fruits/pods were inoculated. Pathogenic reactions were assessed seven days after inoculation (DAI).
Lilac (Syringa vulgaris cv. Sensation) and mandarin (cv. Owari) were ruptured with a sterile hypodermic needle by pricking the leaf midrib and into areas on the wing of the petiole of fully expanded leaves, and inoculated with a 20 μl drop of bacterial suspension (106 cfu/ml) deposited on a newly made rupture. Inoculated leaves and the petioles were placed in plastic bags and kept under room temperature conditions and high humidity (80%). For each tested isolate and controls, five different leaves and petioles were inoculated. Pathogenic reactions were assessed 14 DAI.
In addition, twigs of mandarin trees were longitudinally ruptured (1 cm) using a sterile scalpel, and per 100 μl of the bacterial suspension (106 cfu/ml) was placed onto the cuts. Twigs were kept in clear polyethylene bags for 24 h under room temperature conditions and high humidity (70%–80%). For each tested isolate and controls, three stems were inoculated. Symptoms were assessed 28 DAI.
Biochemical characteristics
Bacterial isolates and reference strains were tested for LOPAT (levan production, oxidase reaction, potato soft rot, arginine dihydrolase activity, and tobacco hypersensitivity) tests (Lelliott and Stead, 1987). Levan production was estimated by growing isolates on SNA medium. For oxidase reaction test, bacteria were grown on SNA medium for 24 h at 25°C until colonies were obtained. Colonies were picked with a loop and rubbed on filter-paper impregnated with 1% (w/v) aqueous tetramethyl-p-phenylene diamine dihydrochloride solution. Production of a purple colour within 10 s was recorded as positive, its development in 10 to 60 s as delayed positive, and the absence of coloration or its still later development as negative (Kovacs, 1956). Pectinase activity was tested by adding bacterial suspension (106 cfu/ml) onto the potato slices and incubated for 24 h at 25°C. Each isolate was tested for arginine dihydrolase production by the method of Thornley (Thornley, 1960), by growing in medium with L-arginine. They were then covered with a 5-mm layer of sterile liquid paraffin, tightly capped, and incubated at 25°C for 7 days. A positive test for the presence of arginine dihydrolase was indicated by a deep pink to red color, whereas a negative test was demonstrated by an absence of color change in the medium. Tobacco hypersensitivity, was tested on leaves of the tobacco. Inoculum were prepared in SDW as described for the pathogenicity test. Reaction to Gram stain, oxiditive/fermentative metabolism of glucose (O/F) and fluorescenent pigment production on the King’s medium B, were also tested (Lelliott and Stead, 1987; Schaad et al., 2001). In addition, GATT tests (Gelatine and Aesculine hydrolysis, formation of Tyrosinase activity and Tartrate metabolism) for the differentiation within the species P. syringae were carried out (Kaluzna et al., 2012). Gelatin liquefaction was done by growing bacteria in tubes containing 12% (w/v) gelatin, which were incubated up to 15 days at 25°C and then held at 4°C for 15 min before determining liquefaction. β-Glucosidase activity of tested isolates was estimated by growing isolates on 0.1% (w/v) aesculin agar medium and incubating four days at 25°C. Positive reaction is dark brown color of the medium after incubation. Tyrosinase activity of isolates were estimated on agar medium containing 0.1% (w/v) L-tyrosine by incubating 4 days at 25°C. A positive test is red dark brown color of the medium after incubation. Utilization of tartarate as a sole carbon source on Simmons basal medium (Holding and Collee, 1971) containing 0.2% (w/v) sodium tartarate. After 21 days of ingubation at 25°C if reaction is positive medium turn color from green to blue.
Genomic DNA extraction
Total genomic DNA was extracted using a modification of the procedure of Ausubel et al. (1992). All tested 43 Montenegrian mandarin isolates and P. syringae reference strains were grown on SNA solid medium for 48 h at 25°C. Bacterial cells were washed in 1.5 ml of SDW and centrifuged at 4,000g for 10 min at 4°C. The resulting pellet was washed twice in 500 μl 0.85% NaCl, recentrifuged and washed once in 0.1 M NaPO4 buffer, pH 6.8. Cells were treated with 10% sodium dodecyl sulfate (SDS) and with proteinase K at 37°C for 1 h. DNA was purified by adding 100 μl 5 M NaCl and 80 μl solution of 10% hexadecyltrimethyl ammonium bromide (CTAB) in 1 M NaCl at 65°C for 10 min extracted with an equal volume of chloroform and centrifuged for 10 min at 8,000g. The aqueous phase was transferred to a new tube and precipitated by adding 0.6 vol of isopropanol. The DNA was then washed with 70% ethanol and re-dissolved in 100 μl Tris-EDTA (TE; 10 mM Tris, 1 mM EDTA, pH 8.0), and quantified spectrophotometrically at 260 nm.
Detection of the syrB gene
In order to detect the possible presence of the syrB gene in all tested isolates, the primer pair B1 (5′-CTTTCCGTGGTCTTGATGAGG-3′) and B2 (5′-TCGATTTTGCCGTGATGAGTC-3′) was used. PCR amplification procedure was described by Sorensen (1998). For negative control, template was replaced with the same volume of SDW. Additionally, reference strains of P. syringae pv. syringae (CFBP 11 and CFBP 1582) and P. syringae pv. morsprunorum (CFBP 2119) were tested as templates for PCR amplification. The PCR was programmed for 35 cycles, denaturation at 94°C for 1.5 min, primer annealing at 60°C for 1.5 min, DNA extension for 3.0 min at 72°C and additional extension at 72°C for 10 min. PCR amplifications were performed in a Mastercycler personal model (Eppendorf, Hamburg, Germany).
Repetitive PCR genomic fingerprinting
An enterobacterial repetitive intergenic consensus (ERIC)-PCR with primer pair ERIC1R (5′-ATGTAAGCTCCTGGGGAT TCAC-3′) and ERIC2 (5′-AAGTA AGTGACTGGGGTGAGCG-3′) have been used for differentiation among the isolates (Lupski and Weinstock, 1992). ERIC-PCR conditions were as previously described by de Bruijn (1992). Amplification of PCR was performed with a Mastercycler personal model (Eppendorf) by using the following cycles: 1 initial cycle at 95°C for 7 min; 30 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 65°C for 8 min with a single final extension cycle at 65°C for 15 min and a final soak at 4°C. The PCR amplifications were performed in triplicate. Amplified PCR products were separated by gel electrophoresis on 1% agarose gels in 0.5× TAE buffer for 1 h at 5 V/cm, stained with 0.05 μl/ml ethidium bromide and visualized under UV illumination. Fingerprints generated from different strains were compared visually.
16S rDNA sequence analysis
Amplicons for the partial 16S rDNA sequences were generated using universal primers (5′-GAGAGTTTGATCCTGGCTCAG-3′) and P6 (5′-CTACGGCTACCTTGTTACGA-3′) (Grifoni et al., 1995) and reaction conditions described by Scortichini et al. (2005). PCR products were purified with the QIAquick PCR purification kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. The sequencing was performed by Macrogen Inc. (Seoul, Korea) and the sequences were deposited in the GenBank database. The MEGA6 software package (Tamura et al., 2013) was used to align sequences from forward and reverse strands and generate consensus sequences. A basic local alignment search tool BLAST (Altschul et al., 1997) was used to compare unidentified sequences with sequences in public databases. Neighbor joining (NJ) analyses were performed using 16S rDNA sequences which were assembled and edited using FINCHTV v.1.4.0 (http://www.geospiza.com). Multiple alignments and comparisons with reference strains were performed using CLUSTALW integrated into MEGA6 software (Tamura et al., 2013).
Multilocus sequence analysis
MLSA was performed in order to reveal the relationship between the P. syringae pv. syringae isolates obtained from mandarin and the P. syringae reference strains. Gene fragments of three housekeeping genes such as, gyrB, rpoD, and gap1 coding for DNA gyrase B, sigma factor 70 and glyceraldehyde-3-phosphate dehydrogenase, respectively, were amplified from genomic DNA and sequenced with primers described by Sarkar and Guttman (2004). PCR products were purified with the QIAquick PCR purification kit (Qiagen) following the manufacturer’s instructions. Sequencing was performed by Macrogen Inc. and the sequences were deposited in the GenBank database. Sequences were edited, aligned, and compared with other Pseudomonas sp. sequences as is described in the study given by Sarkar and Guttman (2004).
Data analysis
In order to estimate the evolutionary relation between P. syringae pv. syringae isolates obtained from mandarin and the pathotype reference strains obtained from PAMDB database, phylogenetic trees were built using NJ analyses. Gene sequences were assembled and edited using FINCHTV v.1.4.0 (http://www.geospiza.com). Multiple alignments and comparisons with reference strains for each of the genes were performed using CLUSTAL W integrated into MEGA6 software (Tamura et al., 2013). Pseudomonas fluorescens was used as out-group.
Minimum spanning tree (MST) was generated, where isolates are the nodes of a tree using SplitsTree software (Huson and Bryant, 2006). The method estimates a probability of ancestry for each individual isolate from each of the groups. The individual isolates are assigned to one cluster or jointly to two or more clusters if their genotypes indicate that they were admixed.
Results
Bacterial isolation and characterization
During the spring of 2013 and 2014, severe outbreaks of citrus blast were observed in mandarin (cv. Owari) in the regions of Bar and Ulcinj in Montenegro (Fig. 1). Isolations of bacteria from necrotic buds yielded almost pure cultures of whitish, translucent bacterial colonies that were fluorescent on King’s medium B after 48 h of incubation. A total of 43 separate bacterial colonies were selected and purified on SNA for further analysis (Table 1). On SNA, isolates formed large (2–3 mm in diameter), convex (levan positive), cream-whitish, mucoid colonies 2–3 DAI.

Symptoms of citrus blast disease caused by Pseudomonas syringae pv. syringae on mandarin natural infection.
P. syringae strains isolated from diseased mandarin tissues induced typical symptoms of P. syringae pv. syringae on an inoculated unripe lemon, appearing as deep, black necrotic pits. On dwarf bean pods, isolates produced water-soaked lesions with reddish margins 3 DAI. In syringe-inoculated lilac leaves, necrotic areas were seen 2–4 DAI. Inoculated petioles showed water-soaked or black areas, which expanded along the mid vein of the leaves, 7–10 DAI. Symptoms on mandarin leaves and petioles were observed 2 DAI in the form of a brown necrotic area at the point of inoculation. Lesions enlarged slightly on leaves until 10 DAI and brown-black necroses were recorded. On inoculated mandarin twigs there was typical and extensive tissue necrosis, while wilting and dieback were observed 4 weeks after inoculation. No symptoms were observed on SDW inoculated plants.
All isolates showed similar biochemical and physiological characteristics that were typical of P. syringae. They were Gram-negative, fluorescent on King’s medium B, showed strictly aerobic metabolism of glucose, produced levan, and none produced oxidase, arginine dehydrolase and pectinase. All bacterial isolates caused a hypersensitive reaction on the tobacco leaves. These reactions correspond to LOPAT Ia group which includes P. syringae pathovars.
All isolates were positive for gelatin and aesculin hydrolysis, but negative for tyrosinase activity and metabolism of tartrate, demonstrating typical characteristics for bacteria P. syringae pv. syringae.
Genetic characterization
PCR amplification with primers B1 and B2 amplified the specific 752 bp band for the syrB gene in all tested 43 isolates and two reference P. syringae pv. syringae strains, CFBP 11 and CFBP 1582. The strain of P. syringae pv. morsprunorum CFBP 2119 failed to show amplification and hence did not possess the syrB gene.
To determine genetic diversity of the 43 P. syringae pv. syringae isolates, ERIC-PCR genomic fingerprints were generated. DNA banding patterns were identical among tested isolates and P. syringae pv. syringae reference strains, and did not show any genetic diversity among them. Considering the fact that there was no difference among the isolates, a further genetic study was performed with a reduced number of isolates (12 in total) on the basis of the geographic origin of the isolates.
Molecular identification of isolates from mandarin was performed using 16SrDNA gene sequencing and MLSA. The sequence analysis of the conserved 16SrDNA gene deposited in NCBI GenBank database under accession numbers KP099969 to KP099980 (Table 2), confirmed that isolates originated from mandarin belong to P. syringae pv. syringae showing 100% homology with P. syringae pv. syringae strain ICMP 3023 from GenBank (accession number NR_117820).
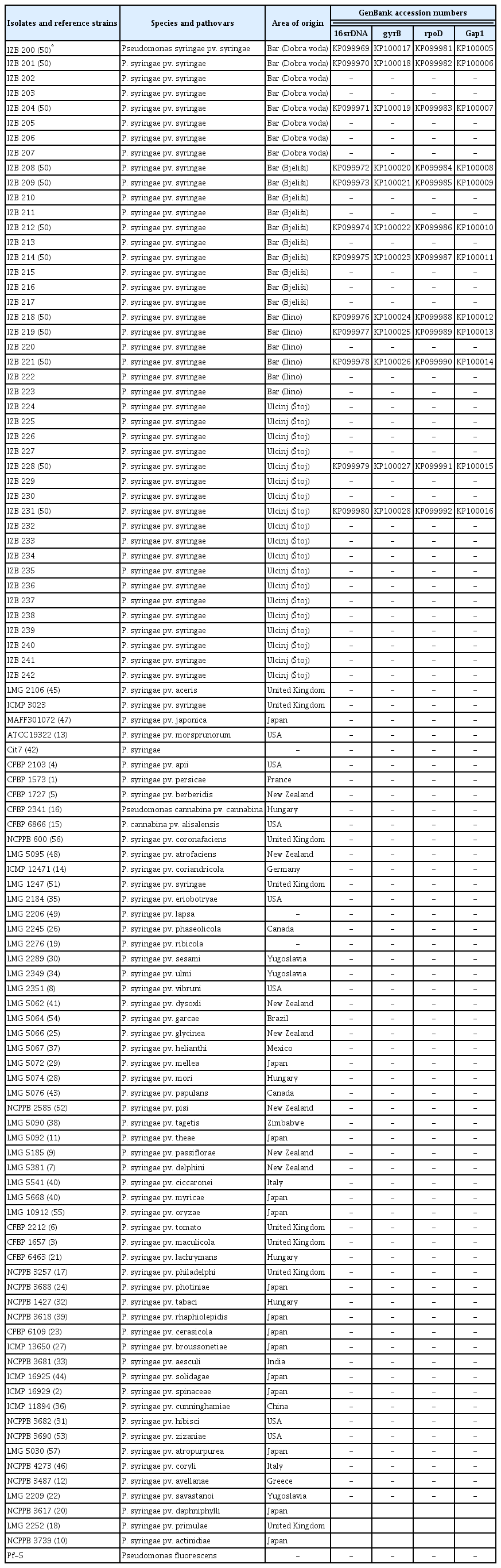
Bacterial isolates and strains used in this study and their geographic origin and GenBank accession numbers
The sequences generated with three housekeeping genes (gyrB, rpoD, and gap1), were deposited in the GenBank database under accession numbers KP099981 to KP100028 (Table 2). A NJ tree constructed for the concatenated data set showed that the mandarin isolates were grouped together with the following reference strains from the database; P. syringae pv. aceris LMG 2106, P. syringae pv. atrofaciens LMG 5095, P. syringae pv. coryli NCBI 4273, P. syringae pv. japonica MAFF 301072, P. syringae pv. lapsa LMG 2206, P. syringae pv. pisi NCPPB 2585, P. syringae pv. solidagae ICMP 16925, and P. syringae pv. syringae LMG 1247 (Fig. 2).
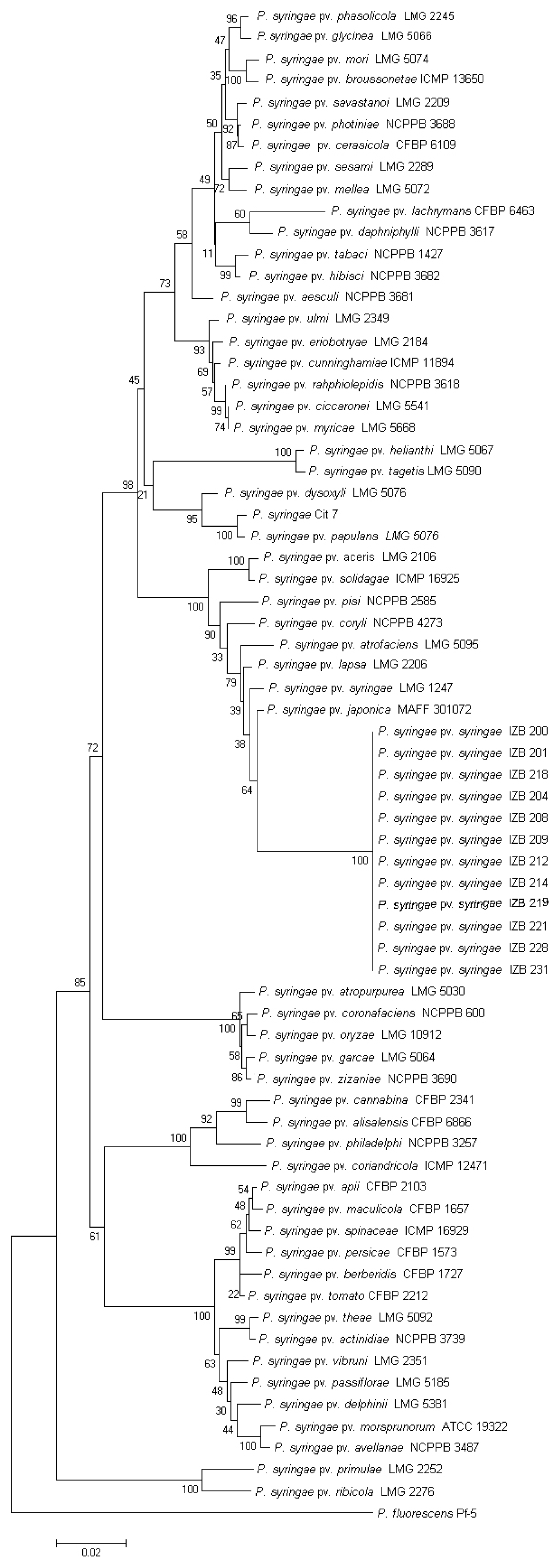
Neighbor joining phylogenetic tree of Pseudomonas syringae pv. syringae isolates from mandarin and reference strains derived from a concatenation of three housekeeping genes (gyrB, rpoD, and gap1). Bar–estimated nucleotide substitutions per site is 0.02.
An MST using the MLSA data using statistical parsimony with split network revealed no ambiguous connections between pathovars and confirmed clustering of testing isolates and reference strains associated within the same genomospecies 1 (Fig. 3). The first cluster (colored in black) included all of our 43 mandarin isolates and members of genomospecies 1. The second cluster included members of genomospecies 2 (colored in blue). The other six clusters present the strains which belong in genomospecies 3, 4 and 6–9 (represented in red, green, light blue, orange, white-green, and white-red, respectively).

Mini spanning tree splits genealogical networks for 12 Pseudomonas syringae pv. syringae isolates originating from mandarin colored in white-black (50) and representative pathotype P. syringae strains. Colored in black genomospecies 1, colored in blue genomospecies 2, colored in red genomospecies 3, colored in green genomospecies 4, colored in light blue to genomospecies 6, colored in orange genomospecies 7, colored in white-green genomospecies 8, colored in white-red genomospecies 9.
Discussion
The occurrence of citrus blast disease, caused by P. syringae pv. syringae in Montenegro was reported for the first time by Vučinić (1987), but a detailed analysis of the pathogen population structure was not performed until now. Based on the observations during epidemics on mandarin in Montenegro and on earlier findings in literature related to environmental influences of citrus blast disease, it is postulated that P. syringae pv. syringae probably easily colonizes buds when spring is cool and moist and enters through wounds when the tissue is previously damaged by frost (Cao et al., 1999; Crosse, 1966; Gross et al., 1983; Klement et al., 1984; Nejad et al., 2004; Sule and Seemuller, 1987).
The relationship of frost damage and P. syringae pv. syringae diseases of woody plants has been demonstrated previously in pear blossom blight (Panagopoulos and Crosse, 1964), leaf spot of sour cherry (Sule and Seemuller, 1987), and bacterial canker of peach (Cao et al., 1999), apricot (Klement et al., 1984), prune, almond (Cao et al., 1999), and willow (Nejad et al., 2004). It has been suggested that winter damage is a contributing factor to outbreaks on many host plants (Buttner and Amy, 1989; Cambours et al., 2005; Gross et al., 1988; Kennelly et al., 2007; Lindow et al., 1982; Sule and Seemuller, 1987). In cases where frost damage incites a greater degree of blast and black pit citrus diseases, cultural management strategies against frost damage may be a viable method of minimizing losses. If fruits and leaves were kept damp using sprinklers, it would allow thin layers of ice to form on plant surfaces rather than inside them where the tissue can get damaged. As ice crystals form, the latent heat of water is released and prevents frost damage (Cambours et al., 2005; Gross et al., 1988; Kennelly et al., 2007; Lindow et al., 1982).
In our study 43 isolates originated from diseased necrotic mandarin buds were identified and characterized using pathogenicity, biochemical tests and for the first time molecular analysis of pathogen. Based on obtained results, they could be assigned to the phytopathogenic bacterium P. syringae pv. syringae. This bacterium has been previously reported as the causal agent of citrus blast of mandarin in Italy, Japan, Iran and Turkey (De Cicco et al., 1978; Mirik et al., 2005; Salerno and Cutuli, 1985; Shams-Bakhsh and Rahimian, 1997).
Using biochemical and GATT differentiation tests our isolates could be discriminated as P. syringae pv. syringae, showing reactions typical for P. syringae pv. syringae as it is described by Lelliott and Stead (1987).
All isolates produced black pit lesions on lemons by needle pricks. The characteristics that positive lesions produced suggest that lemons might be readily inoculated with all strains of this bacteria obtained from any host (Bryan, 1928; Lelliott and Stead, 1987; Smith and Fawcett, 1930). Typical black pit lesions were also formed on lilac and mandarin leaves and leaf petioles and on mandarin twigs as is described by Bryan (1928).
P. syringae pv. syringae is a known bacterium which produces different phytotoxins depending on the host origin. Isolates from the stone fruits or peach produced syringomycin, isolates from citrus trees yielded syringotoxin while isolates from lilac blight produced syringostatin (Fukuchi et al., 1990; Gross and Cody, 1985; Isogai et al., 1989; Mitchell, 1981; Mo and Gross, 1991). Syringomycin was detected in our tested isolates originating from mandarin.
It should be noted that the lack of molecular data in literature sources concerning the genetic identity and structure of P. syringae pv. syringae populations is evident. The present study based on ERIC-PCR and 16SrDNA gene sequencing showed a genetically homogeneous population of P. syringae pv. syringae causing citrus blast disease in Montenegro. A phylogenetic tree obtained from MLSA, showed that our isolates belong to the previously described genomospecies 1 of P. syringae (Berge et al., 2014; Gardan et al., 1999; Sarkar and Guttman, 2004).
Results of MST showed that P. syringae pv. japonica is defined as a central node of the cluster and point of diversifiaction for other strains. P. syringae pv. syringae isolates from mandarin were closely related to other P. syringae pvs. such as atrofaciens, lapsa, syringae, japonica, and pisi. We consider that hybridization events could have occurred between them or their ancestors. MST shows, from the clustering of the P. syringae pv. syringae isolates originated from mandarin, that these isolates are genetically diversified from P. syringae pv. syringae pathotype strain LMG 1247. P. syringae pv. syringae isolates from mandarin represent a clearly clonal complex and further investigation is needed to understand the origin of this pathogen. In conclusion, our study revealed that P. syringae pv. syringae isolates originated from mandarin which caused citrus blast in Montenegro and that they belong to genomospecies 1. The homogeneity found in all used tests suggest that tested isolates may have originated from a single inoculum source such as planting material.
Further detailed biological studies, using more strains of P. syringae pv. syringae which cause citrus blast in other parts of the world should be performed to elucidate their origin and to further resolve their taxonomic status. This might be very important for efficient management of outbreaks and the prevention of further spreading of this pathogen.
Acknowledgments
This work was supported by the Ministry of Education and Science, Republic of Serbia (Grants No. TR31018) and the Ministry of Science, Montenegro (Grant “Invasive species”).
