Selection of Small Synthetic Antimicrobial Peptides Inhibiting Xanthomonas citri subsp. citri Causing Citrus Canker
Article information
Abstract
Citrus canker disease decreases the fruit quality and yield significantly, furthermore, emerging of streptomycin-resistant pathogens threatens the citrus industry seriously because of a lack of proper control agents. Small synthetic antimicrobial peptides (AMPs) could be a promising alternative. Fourteen hexapeptides were selected by using positional scanning of synthetic peptide combinatorial libraries. Each hexapeptide showed different antimicrobial spectrum against Bacillus, Pseudomonas, Xanthomonas, and Candida species. Intriguingly, BHC10 showed bactericidal activity exclusively on Xanthomonas citri subsp. citri (Xcc), while BHC7 was none-active exclusively against two Pseudomonas spp. at concentration of 100 μg/ml suggesting potential selectivity constrained in hexapeptide frame. Three hexapeptides, BHC02, 06 and 11, showed bactericidal activities against various Xcc strains at concentration of 10 μg/ml. When they were co-infiltrated with pathogens into citrus leaves the disease progress was suppressed significantly. Further study would be needed to confirm the actual disease control capacity of the selected hexapeptides.
Bacterial plant diseases are less prevalent than diseases caused by fungi and viruses. However, once bacterial infection occurs, the economic loss caused by bacterial diseases is often devastating (Vidaver, 2002). Only a few number of agricultural antibiotics for treatment of bacterial diseases of plants are available in the market such as streptomycin and oxytetracycline. The limitation of the number of antibiotics for treatment of bacterial disease makes farmers to use the same antibiotics for long time, repeatedly, and it will facilitate the development of antibiotics resistance. Once the antibiotics resistance is established in a population it becomes widespread rapidly. In spite of urgent demand, a new commercial agricultural antibiotics has not been developed for decades due to high cost of development, regulatory constraints, and environmental and human health concern. Effective alternatives to conventional agricultural antibiotics should be developed soon to fight against emerging antibiotics resistant pathogens. This study is designed to develop new antibacterial small synthetic antimicrobial peptides (ssAMPs) against citrus canker pathogen and evaluate the potential as an alternative of streptomycin to control wild type as well as antibiotics resistant citrus canker pathogen.
Citrus canker is caused by Xanthomonas citri subsp. citri (Xcc). When the pathogen infects citrus trees, erumpent pustules are formed in leaves, stems, and fruits, drastically reducing crop quality and quantity. Because of the rapid spread and high potential for Xcc to cause damage, the export of citrus fruit to foreign countries is strictly regulated by international quarantine programs. Citrus has been cultivated in Jeju Island for over 100 years, and copper and streptomycin have been intensively used to control the disease for decades with success; however, they are now facing a serious challenge because of development of resistance in Xcc populations, which is resulting in a reduction of disease control.
There have been many reports of the development of resistance to streptomycin in citrus canker pathogens (Behlau et al., 2012; Hyun et al., 2012). Recent field study revealed that 492 of 686 Xcc isolates (72%) were streptomycin-resistant (SR) strains in Jeju Island (Hyun et al., 2012). When citrus orchids were sprayed with streptomycin three times in a season, the number increased to 88.7%. In addition to the high percentage of streptomycin resistance, the stability of streptomycin resistance in the Xcc population can be another main concern because the persistence of resistance even after application of antibiotics was halted long time before (Loper et al., 1991; Moller et al., 1981). This persistence could be explained by a phenomenon known as compensatory mutation, in which the initial fitness cost of antibiotics resistance due to chromosomal mutation could be compensated as the bacterium evolves in the absence of selection where it undergoes mutations that compensate the fitness burden. The output of the secondary compensatory mutation is long term stability of SR strains in the absence of streptomycin (Levin et al., 2000).
The rapid emergence of antibiotic resistant populations strongly suggests that new control agents are needed. Antimicrobial peptides (AMPs) are promising alternatives to conventional antibiotics for controlling plant disease. AMPs can eliminate a broad spectrum of microorganisms, including viruses, bacteria and fungi (Montesinos and Bardají, 2008).
Innate AMPs are found in all types of living organisms; therefore, they are considered part of the fundamental host defense system (Hancock, 2001; Maróti et al., 2011). In contrast to conventional antibiotics, which often develop resistance, resistance to AMPs is rare, possibly because its primary target is the cell membrane (Hancock, 2001). Most known native AMPs are cationic and amphiphilic, and vary in size (12 to 50 amino acids) and structure (α- and β-secondary structure). In terms of amino acid composition, two groups of amino acids are relatively conserved. One group contains positively charged basic residues such as arginine (R), lysine (K), and/or histidine (H), which initiate interactions with negatively charged cell membranes, while the other group contains bulky hydrophobic residues such as tryptophan (W), proline (P), and/or phenylalanine (F), which are involved in cell membrane disruption (Liu et al., 2007).
Although native AMPs have good advantages, practical application has been delayed because of high minimum inhibitory concentrations (MICs), high cytotoxicity to animal cells, limited availability in nature to purify from as well as high cost to be synthesized (Gordon et al., 2005). Given the limits of currently available AMPs, ssAMPs (usually less than 10 amino acids) could be a good alternative since the cost of synthesis is much lower than that of long native AMPs. The sequences of ssAMPs can be selected by quick screening of synthetic oligo-peptide libraries (Hong et al., 1998) or through logical designing based on available data, while still maintaining most of the advantages of native AMPs. The designed ssAMPs have been studied intensively in pharmacological sectors. Synthesized short peptides with R and W repeats revealed that tetrapeptides or capped tripeptides were sufficient to kill bacteria, regardless of the amino acid order (Strøm et al., 2003). When the effects of chain length on the activities of AMPs with repeating (RW)n-NH2 (n = 1, 2, 3, 4, or 5) were analyzed, hexapeptides (RW)3-NH2 were the optimal chain length since longer chains had more antibacterial potential but stimulated increased hemolysis of red blood cells (Liu et al., 2007). Selecting new ssAMPs using positional scanning of a synthetic peptide combinatorial library (PS-SPCL) could provide a more diverse pool of potential AMPs than a simple logical design approach (Blondelle and Lohner, 2000; Choi and Moon, 2009).
In this study, we performed several PS-SPCLs against a wild type as well as three SR mutants of Xcc. The amino acid information for each position was first integrated into 14 complex hexamers (XC series) that were designed to allow more than one amino acid residue at a position, and then the final 14 individual hexapeptides (BHC series) were synthesized based on information obtained from the assay results of the XC series. The antimicrobial spectrum of the BHC series was addressed at 100 μg/ml with variety of microbes including a food pathogen, Bacillus cereus (Bc), two plant pathogenic pseudomonads, Pseudomonas syringe pv. actinidiae and P. syringe pv. tomato (Psa and Pst), and a candida strain, Candida glabrata (Can). Interesting selectivity was observed depending on the sequence of peptides. The antimicrobial activities of the BHC series on Xcc were tested with two wild types and three SR mutant strains. Three novel hexapeptides (BHC02, BHC06, and BHC11) were selected for further study. They showed antimicrobial activity against various Xcc strains at concentration of 10 μg/ml. When BHC06 or BHC11 was applied to citrus leaves, the disease progress was suppressed significantly.
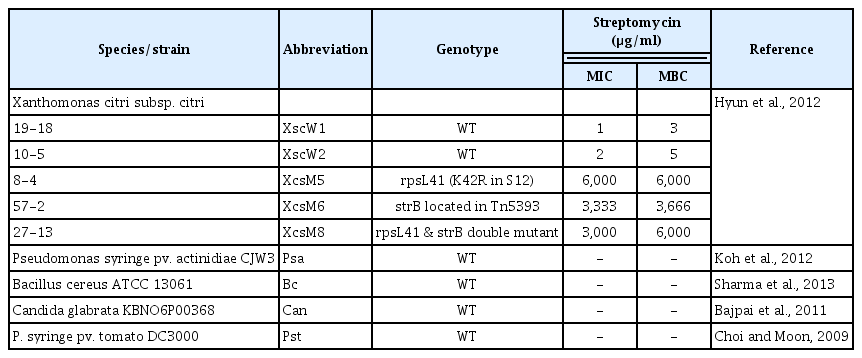
The strains used in this study are listed in Table 1 (Bajpai et al., 2011; Choi and Moon, 2009; Hyun et al., 2012; Koh et al., 2012; Sharma et al., 2013). The fungal strain, Can, was grown in potato dextrose medium (4 g potato starch and 20 g dextrose per liter of water) at 37°C, while all the wild type bacterial strains were grown in yeast nutrient (YN) medium (5 g yeast extract, 8 g nutrient broth/l) at 30°C. The MICs of wild type strains of Xcc for streptomycin were 1–2 μg/ml, while the MICs of SR strains were significantly higher (Table 1). The SR strains were grown in YN media amended with 100 μg/ml streptomycin. The three SR mutants have genetically different resistant mechanism for each other. A SR mutant, XccM5, contains a chromosomal point mutation at ribosomal protein S12. That blocks the initial interaction of streptomycin to its target site, ribosome, resulting in tolerance to streptomycin. Another SR mutant, XccM6, can inactivate streptomycin itself by phosphorylation enzyme encoded in an acquired gene (strA-strB) that originated from a transposable element, Tn5393. XccM8 is a double mutant that has the chromosomal mutation as well as transposon-based gene acquisition (Chiou and Jones, 1993; Hyun et al., 2012).
The PS-SPCL packages were purchased from the Peptide Library Support Facility at Pohang University of Science and Technology (Pohang, Korea). One set of the library consisted of 114 tubes (6 positions × 19 amino acids). Each tube contained a mixture composed of one fixed amino acid at a specific position and random amino acid residues at the remaining five positions. The random amino acid pool was composed of a mixture of 19 amino acids (excluding cysteine).
Positional scanning assays were performed as previously described (Choi and Moon, 2009). The wild type strain, XccW1, as well as three SR mutant strains were subjected to PS-SPCL assays. The relative growth inhibition rates (RGIR) were calculated using the following equation: [1 – (treated culture OD/culture control OD)] × 100. For the wild type strain, XccW1, the typical cationic amino acid residues, K and R, were dominant at position 1 and 2 with over 95% RGIR, indicating that positively charged cationic acids are required at the N-terminal region for its action. At position 3, Y and W residues were identified with relatively high RGIRs of 88% and 70%, respectively. Y is a polar hydrophobic amino acid not usually found in ssAMPs suggesting that it plays a unique role in antimicrobial activity against Xcc cells. At position 4, W, D, and I residues were identified with RGIR values ranging from 71% to 63%, while at position 5, D, I, and W residues were identified with RGIR values ranging from 60% to 52%. At position 6, I, W, Y, and F residues were identified with RGIR values of 73% to 56%. Over all a few dominant amino acids with high RGIR were identified in position 1–3, while several semi-dominant amino acid residues were identified in position 4–6. When the same PS-SPCL assays were performed with SR mutants, similar amino acid residues such as R, W and Y were identified with some variations. With the mutant XccM5, S residue was identified at position 3, while D residue was identified at position 2 and 5 with mutant XccM6. Amino acid residues, F and Y, were identified at position 3, 4 and 5 with XccM8.
The appearance of negatively charged amino acids D or E at position 3–5 was interesting, but unexpected considering the general mechanism of AMP in which cationic AMP usually interacts with negatively charged cell membranes. The appearance of D or E residues at position 3–5 suggests that Xcc either has unique membrane structure or composition that makes it partially positive and allows interactions with negatively charged amino acids, or the integrated structure of hexapeptide containing D or E residue forms a unique structure that does not hinder the electrostatic interactions with negatively charged cell membranes.
PS-SPCL identifies the effective amino acid residues at each position. However, it is not easy to identify effective hexapeptides at once from the peptide library that contains over 47,000,000 different hexapeptides. If three amino acid residues are identified at each position, the numbers of candidates to be synthesized are theoretically 36 peptides, which are out-numbered for practical synthesis. To narrow down the number for synthesis, the first round hexapeptide series (named XC) were synthesized where two or more amino acids were allowed at each position. The antimicrobial activities of the XC series were tested with two different concentrations (10 μg/ml and 100 μg/ml) against XccW1. Among 14 complex XC series, four hexapeptide complexes (XC01, XC02, XC 11, and XC12) showed strong activities at 100 μg/ml, with K, R and N residues dominant at position 1–2, and W, F, Y and R residues dominant at position 4–6 (data not shown). Based on this information, 14 new hexapeptides (BHC series) that allowed only one amino acid at a position were designed and synthesized. BHC hexapeptides with a purity of 90% were purchased from Peptron Co. (Daejeon, Korea) and dissolved in 50% dimethyl sulfoxide and stored at −20°C until use.
The antimicrobial activities of 14 BHC peptides as well as previously identified KCM21 (Choi and Moon, 2009) were addressed in YN agar plate with two different concentrations (10 μg/ml and 100 μg/ml). Most of 14 BHC peptides except BHC13 and BHC14 (85.7%) were bactericidal against XccW1 at the concentration of 100 μg/ml (Fig. 1). BHC14 had a negatively charged E residue at position 3. Even though E residue was identified as a potential antimicrobial residue during PS-SPCL, its location and nearby amino acid context appeared to be critical for its activity. BHC13 contained almost identical sequences with BHC9 but the K residue of BHC9 at position 1 was replaced with similar cationic residue N. The simple amino acid change knocked out the activity completely at 100 μg/ml. Considering that most of the 14 BHC series peptides share similar residues such as K, R, W and Y, these results emphasized that certain sequences or context is critical for their activity. Among the 12 BHC peptides that were active at 100 μg/ml, only three hexapeptides, BHC02, BHC06 and BHC11, were able to maintain the bactericidal activity at 10 μg/ml.

Sensitivity of Xanthomonas citri subsp. citri wild type 1 (XscW1) against 14 selected hexapeptides and previously identified KCM21 at the concentrations of 10 μg/ml and 100 μg/ml.
The antimicrobial spectrum of each hexapeptide was monitored by checking the colony formation in solid media at the concentration of 100 μg/ml (Fig. 2). Fresh grown overnight culture cells were adjusted to 5 × 105 cfu/ml with 2× YN broth, and mixed with the same volume of 200 μg/ml peptide solution in a 96 well microtiter plate (total 150 μg/ml) and incubated for 28–36 h with gentle shaking (80 rpm). The culture of each well was replicated to agar plates by using a 48 well Replica Plater (Sigma-Aldrich, St. Louis, MO, USA) and incubated for 24–36 h depending on strains. For Can, the same procedures were applied; however, the initial concentration was adjusted to 1 × 105 cfu/ml. Arbitrary units were applied to score the antimicrobial spectrum. The scale 3, 2, 1 and 0 were inverse-related to the number of grown colony, respectively (scale 3 = no growth of any colony, scale 2 = growth of one colony, scale 1 = growth of two colonies, scale 0 = growth of all three colonies). If there was a colony with intermediate growth, 0.5 was subtracted from the total score. All experiments were conducted independently at least two times with three replicates.
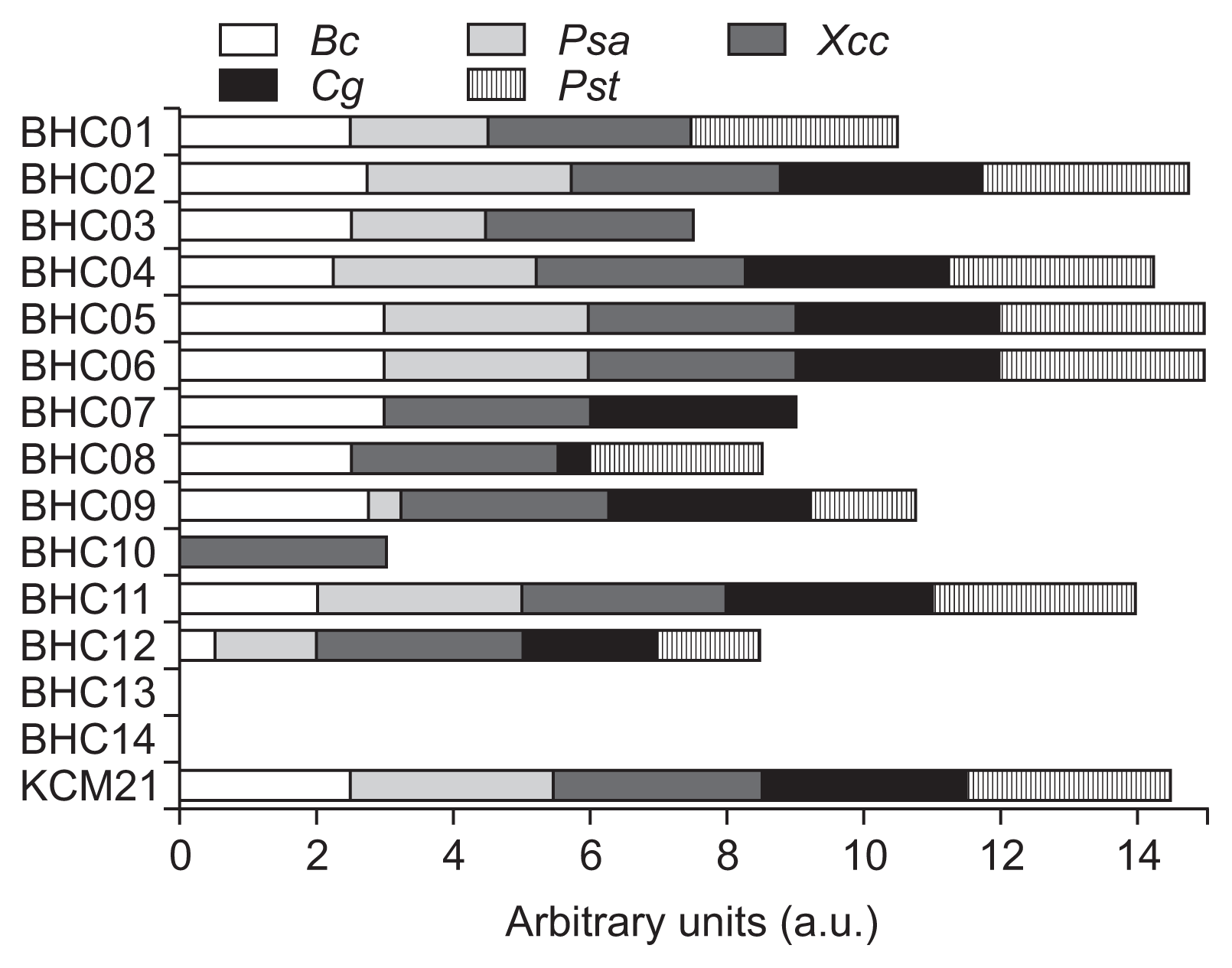
Antimicrobial spectrum of each hexamer against 5 different microorganisms including Xanthomonas citri subsp. citri wild type 1 (Xcc), Bacillus cereus (Bc), Pseudomonas syringe pv. actinidiae and P. syringe pv. tomato DC3000 (Psa and Pst), and Candida glabrata (Cg). Each strain was grown in the liquid culture amended with each hexapeptides (100 μg/ml) for 24 h in 96 well plate and transferred to solid agar plate by using 48 pin replicator. The arbitrary unit represents average none-grown colonies for each strain in the agar plate. If no colonies were grown within 3 replications, it was counted as 3. If there was a colony with intermediate growth 0.5 was subtracted.
As shown in Fig. 2, most of the selected hexapeptides had antimicrobial spectrum against variety of microorganisms. BHC13 and BHC14 showed no antimicrobial activities against the five-tested microorganisms. In contrast, BHC02, BHC04, BHC05, BHC06 and KCM21 had a wide range of antimicrobial activity with over 14 combined arbitrary units (a.u.). The BHC11 showed strong bactericidal activity against XccW1 at 10 μg/ml; however, its antimicrobial spectrum score was below 14 a.u. since Gram-positive strains Bc was not fully sensitive to it at 100 μg/ml. The BHC10 showed the most interesting characteristics in terms of species selectivity since it inhibited the growth of XccW1 completely at 100 μg/ml but it cannot inhibit the growth of any other testing organisms. Even though hexamer BHC10 showed strong selectivity against XccW1 at 100 μg/ml, the antibacterial activity was not retained at low concentration (10 μg/ml). It is not clear why BHC10 was so specific to XccW1 at 100 μg/ml; however, it is noteworthy that BHC10 was the only one that contained anionic residue D at position 4. Another interesting selectivity was observed with BHC07, which was active against Xcc, Bc, and Can, but not against the two tested pseudomonads, Psa and Pst. It is not clear if BHC7 may have inverse-selectivity against other pseudomonads. Further study will be needed to understand the antimicrobial selectivity as well as activities in different hexapeptide context.
The MICs of three selected hexapeptides as well as KCM21 were tested against two wild type and three SR mutant strains of Xcc (Fig. 3). The MIC was measured using the modified microtiter broth dilution assay (Choi et al., 2014; Zhang et al., 2005). The MIC was taken as the lowest peptide concentration at which more than 90% of culture growth was inhibited after 24 to 36 h of incubation. The growth was measured by the optical density (OD) at 600 nm using a microplate reader (Infinite 200 Pro; Tecan, Männedorf, Switzerland). All four hexapeptides showed highly activity against the five different strains of Xcc, even though the antimicrobial activities were different for each other against each strain of Xcc. When the MICs against various Xcc strains were averaged, BHC11 showed the lowest MIC of 9.4 μg/ml (Fig. 3). All hexapeptides except BHC02 had lower MIC values against single SR mutants (e.g., XccM5 and XccM6) than wild type or double mutants (XccM8).

Minimum inhibitory concentrations (MICs) of four-selected hexapeptides against two wild (XccW1 and W2) and three mutant (XccM5, M6, and M7) strains of Xanthomonas citri subsp. citri. Each strain showed different sensitivity to four different hexapeptides. Data are expressed as the mean ± standard deviation.
The disease progresses of wild type strain XccW2 as well as SR strain XccM8 were monitored in fully expanded leaves of 5-year-old citrus plants (On-ju Milgam; Citrus unshiu var. sathuma mandarin) in the glass house at Citrus Research Station, Jeju island following the procedures previously described (Ahmad et al., 2014; Amaral et al., 2010; Li and Wang, 2011). About 0.1 ml of bacterial suspension adjusted to 1 × 108 cfu/ml (ca. OD 0.55 at 600 nm) was infiltrated into the abaxial side of leaf by using a needless syringe. Sterile tap water was used as negative control. The disease suppression by selected hexapeptides was tested by co-infiltration of mixture of 2× concentration of hexapeptide with bacterial suspension. The symptom development was monitored for 17 days after inoculation by using a disease index scaled 0–5 (Fig. 4). Final hexapeptide concentrations of 50 μg/ml and 100 μg/ml were used to see dose-dependent effects. Streptomycin was used alone as a positive control and also used together with hexapeptides to see synergic effects.
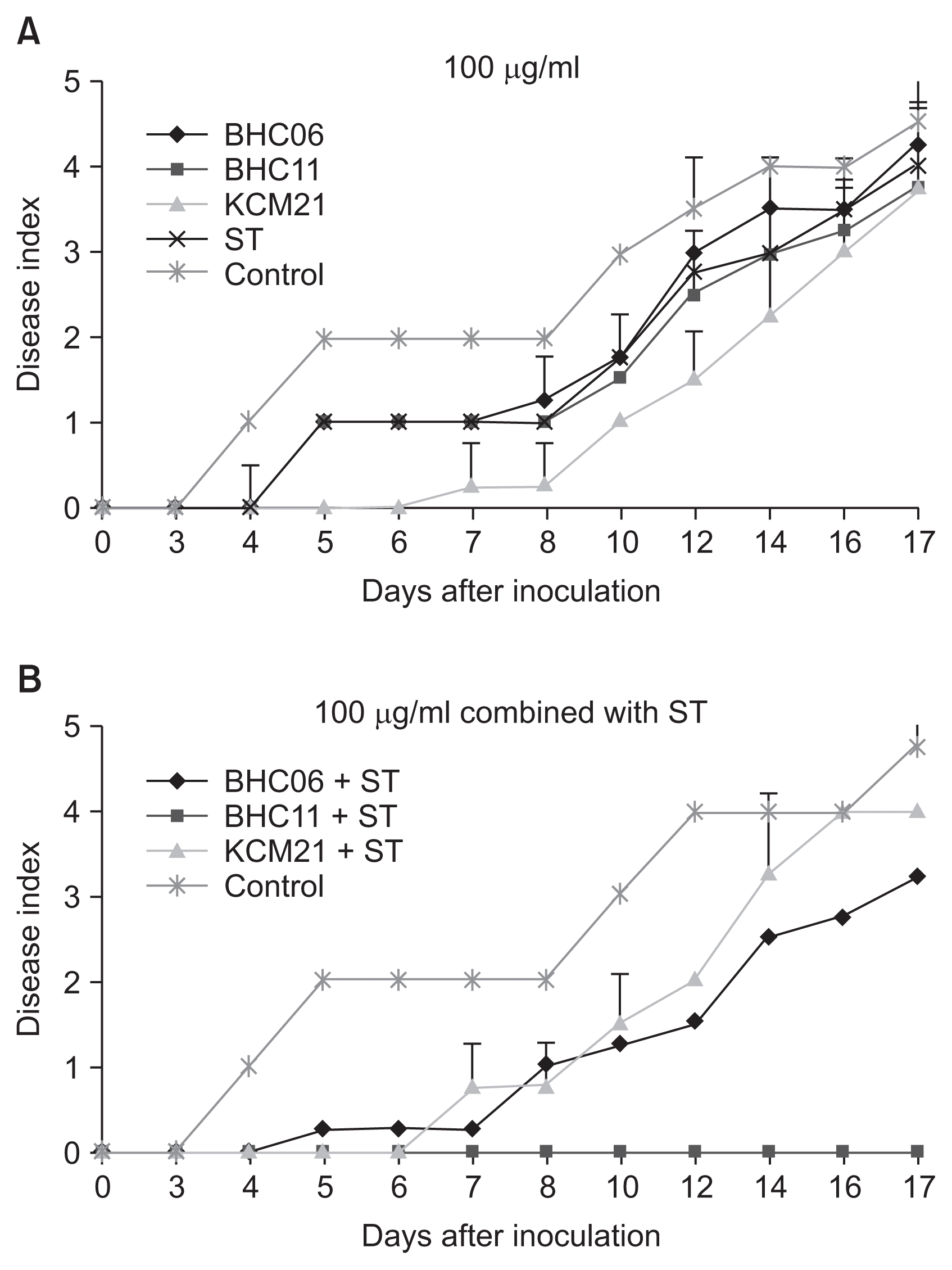
The suppression of citrus canker disease caused by wild type strain XccW1 upon treatment of hexapeptides and streptomycin (ST). Effects of three hexapeptides and ST on the disease progress were evaluated for 17 days by using a disease index: 0 = no symptom, 1 = first symptom, 2 = yellowish halo around the pustule, 3 = prominent pustule, 4 = reddish or brownish pustule appearance of the necrotic tissue, 5 = leaf distortion or leaf drop. The hexapeptides were treated individually (A) or simultaneously with ST (B).
The streptomycin treatment (50 μg/ml) did not stop the symptom development completely but it slowed down the first pustule formation about 1 day (data not shown). Considering that streptomycin has been used for decade successfully to control the wild type Xcc strains the delayed symptom development could be a good indicator of disease control in this experimental setting. For wild type Xcc strain the symptom development was delayed more when the streptomycin concentration was increased from 50 to 100 μg/ml. It took 10 days to form the first yellowish halo around the pustule (index 2) when 100 μg/ml of streptomycin was treated (Fig. 4). The hexapeptide BHC06 and BHC11 took 10 days for the halo formation and it took 14 days for KCM21 with 100 μg/ml treatment. The symptom development was suppressed up to 17 days after inoculation either by streptomycin or the selected hexapeptides. The selected hexapeptides were also co-treated with streptomycin to see if they showed better disease suppression effects. BHC06 and BHC11 showed higher control efficiency with combined treatment with streptomycin. It was noteworthy that no symptom was observed at all up to 17 days by combined treatment of BHC11 with streptomycin suggesting their potential synergic interactions. In contrast, the hexapeptide KCM21 showed stronger disease control effect when it was treated alone than it was co-treated with streptomycin suggesting antagonistic or at least no synergistic interaction of KCM21 with streptomycin.
Streptomycin did not suppress the disease progress at concentration of 50 μg/ml or 100 μg/ml against SR mutant XccM8 as expected. All three hexapeptides showed strong disease suppression effects when they were treated individually at the concentration of 100 μg/ml and the disease progress was suppressed up to 17 days after inoculation against SR mutant Xccm8 (Fig. 5). The pustule symptoms were not formed for 8 days after inoculation with KCM21 and 10 days with BHC11. When streptomycin was co-treated, the suppression efficiency was not changed with BHC11 or slightly increased with BHC06 against XccM8. No synergistic interaction of KCM21 with streptomycin was also observed against XccM8.

The suppression of citrus canker disease caused by streptomycin-resistant (SR) strain XccM8 upon treatment of hexapeptides and streptomycin (ST). Effects of three hexapeptides and ST on the disease progress were evaluated for 17 days by using a disease index: 0 = no symptom, 1 = first symptom, 2 = yellowish halo around the pustule, 3 = prominent pustule, 4 = reddish or brownish pustule appearance of the necrotic tissue, 5 = leaf distortion or leaf drop. The hexapeptides were treated individually (A) or simultaneously with ST (B).
Previously selected KCM21 and the selected three hexapeptides showed the antibacterial activity against all the tested citrus canker pathogens regardless of their genetic background. Considering that each strain had slightly different sensitivity to the hexapeptides, combined treatment with different hexapeptides could enhance the control efficiency by compensating for the weak activity of certain hexapeptides against a certain strain, even though each hexapeptide showed sufficient potential to control all of the Xcc strains under the concentration of 25 μg/ml.
Interestingly, selectivity depending on the hexapeptide sequences was observed suggesting that the selectivity could be constrained in hexapeptide context. During the antimicrobial spectrum assay with 5 different microorganisms, hexapeptide specific for Xanthomonas and reverse selective hexapeptide against Pseudomonas were identified. Hexapeptide BHC10 was active only against Xcc, while BHC07 was active against three different species, but not against two pseudomonads at 100 μg/ml. These data suggest that genus-selective or species-selective AMP could be selected in short peptide context. Considering the application of AMP in agricultural ecosystems, it would be desirable if they have a certain degree of selectivity against target pathogens.
The selected hexapeptides effectively suppressed the citrus canker disease in citrus leaves when they are co-infiltrated with the wild type Xcc. The control effect of BHC11 was drastically increased with co-treatment with streptomycin. The SR mutant XccM8 caused citrus canker as effectively as wild type but cannot be controlled by streptomycin. The selected hexapeptides suppressed the disease progress effectively caused by the SR mutant. Somehow the selected hexapeptides seemed to control the mutant strains more effectively than streptomycin did for the wild type strain. Co-treatment of streptomycin enhanced the activity of BHC06 and BHC11 but not with KCM21 suggesting sequence-dependent synergic interactions with streptomycin. Further study would be need to understand the potential synergic interactions of selected hexapeptides with streptomycin or by themselves. Overall, the selected hexapeptides showed good potential to suppress the disease development in this experimental setting. However it should be mentioned that the infiltration inoculation is an artificial methods and the hexapeptides would have direct effect to pathogens when they were co-treated. Ultimately field test would be needed to confirm the disease control effects of hexapeptides in plant.
Recently, outbreaks of two serious plant bacterial diseases were reported in Korea; apple fire blight caused by Erwinia amylovora and kiwi canker caused by P. syringe pv. actinidiae, which is also considered as worldwide pandemic. Emerging of antibiotics resistant strains for these disease was well documented worldwide (Stockwell and Duffy, 2012). In Korea there is no official reports about emerging of SR strains for fire blight pathogens and only one report for kiwi canker pathogens (Han et al., 2004). However, emerging of the antibiotics resistant strains in Korea could be matter of time either by introducing from foreign or by autonomous establishment of antibiotics resistance through continuous use of the same antibiotics. Once the antibiotics resistant strains are introduced in field, control of them would be very difficult due to lack of proper control agents. There is great demand for alternatives to conventional agricultural antibiotics to control bacterial disease.
In this study we showed that selection of ssAMPs through PS-SPCL can provide us a rich pool of novel antimicrobial peptides with semi target selectivity. And the selected hexapeptides showed good potential to suppress the disease development in citrus leaves when they were co-infiltrated with SR strains. However, further studies will be needed to develop alternative agricultural antibiotics based on hexapeptides such as testing the cytotoxicity, the environmental effects on non-pathogenic microbes and the residual periods of hexapeptides in addition to confirm the disease control efficiency in field condition.
Acknowledgments
This work was carried out with the support of the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ010934), Rural Development Administration, Republic of Korea.