MoJMJ1, Encoding a Histone Demethylase Containing JmjC Domain, Is Required for Pathogenic Development of the Rice Blast Fungus, Magnaporthe oryzae
Article information
Abstract
Histone methylation plays important roles in regulating chromatin dynamics and transcription in eukaryotes. Implication of histone modifications in fungal pathogenesis is, however, beginning to emerge. Here, we report identification and functional analysis of a putative JmjC-domain-containing histone demethylase in Magnaporthe oryzae. Through bioinformatics analysis, we identified seven genes, which encode putative histone demethylases containing JmjC domain. Deletion of one gene, MoJMJ1, belonging to JARID group, resulted in defects in vegetative growth, asexual reproduction, appressorium formation as well as invasive growth in the fungus. Western blot analysis showed that global H3K4me3 level increased in the deletion mutant, compared to wild-type strain, indicating histone demethylase activity of MoJMJ1. Introduction of MoJMJ1 gene into ΔMojmj1 restored defects in pre-penetration developments including appressorium formation, indicating the importance of histone demethylation through MoJMJ1 during infection-specific morphogenesis. However, defects in penetration and invasive growth were not complemented. We discuss such incomplete complementation in detail here. Our work on MoJMJ1 provides insights into H3K4me3-mediated regulation of infection-specific development in the plant pathogenic fungus.
Introduction
In eukaryotes, development and organogenesis are often associated with epigenetic changes. Such epigenetic changes include DNA methylation, histone modifications, and nucleosome remodeling. Among them, the most diverse and complex are histone modifications, which are mediated by histone-modifying enzymes that covalently add acetyl-or methyl groups to lysine residues within histone proteins (Jeon et al., 2014; Kouzarides, 2007). It is known that the combination of histone modifications is indicative of the transcriptional status of the underlying genes (Li et al., 2007). Therefore, histone-modifying enzymes play important roles in transcriptional regulation by controlling tissue and/or stage-specific gene expression during normal development and organogenesis. Furthermore, the histone marks can serve as recognition and binding sites of other proteins responsible for various biological processes, such as DNA replication and repair (Kouzarides, 2007).
Specific histone lysine methylation/demethylation, catalyzed by histone methyltransferases (HMTs) and histone demethylases (HDMs), respectively, are commonly associated with either the active or the repressed chromatin state. Histone H3K4, K36, and K79 methylation are generally considered to be a mark for active transcription, while H3K9, K27, and H4K20 methylation acts as transcriptional repressive marks. Unlike histone acetylation considered as being reversible process regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), it was previously thought that histone methylation is an irreversible modification that can only be removed through dilution during DNA replication or histone eviction (Kooistra and Helin, 2012). Shi et al. (2004) were first to identify lysine-specific demethylase 1 (LSD1) enzyme that can demethylate mono/dimethyl H3K4 via flavin adenine dinucleotude (FAD)-dependent amine-oxidation. Later, JmjC domain-containing histone demethylase 1 (JHDM1) was shown to specifically demethylate H3K36, thereby corroborating a previous hypothesis that JmjC domain-containing protein could act as histone lysine demethylase via an oxidative mechanism involving Fe(II) and α-ketoglutarate as cofactors (Tsukada et al., 2006). Eventually, numerous studies had demonstrated the involvement of JmjC domain-containing demethylases in development, diseases, and organogenesis in human and plants. Examples include human JMJD3, which is associated with lung and liver carcinomas, and several haematological malignancies (Agger et al., 2009; Kooistra and Helin, 2012). Similarly, REF6 is related to early flowering in Arabidopsis and JMJ706 is required for proper development and organogenesis in rice (Lu et al., 2011; Sun and Zhou, 2008).
JmjC demethylases are generally classified into seven groups according to their protein sequence homology and presence of additional domains. Among them, JARID1 contains JmjN, BRIGHT/ARID (AT-rich interactive domain, one of the defining features of JARID group), C5HC2-zinc-finger and PHD domains, and specifically demethylases H3K4 (Klose et al., 2006). In mammalian, four JARID1 subgroups, JARID1A/RBP2, JARID1B/PLU-1, JARID1C/SMCX and JARID1/SMCY, are present and associated with several cancer types and neurological disorders (Kooistra and Helin, 2012). In Drosophila, it was shown that little imaginal discs (LID), the sole JARID group protein, may be an important component of development and viability as a histone lysine demethylase (Eissenberg et al., 2007). In budding and fission yeast, Jhd2 (Yjr119c) and Jmj2 (Spac1002.05c) were identified as HDMs belonging to JARID group. All of the previously studied enzymes in JARID group have been shown that they target the me3- and me2-modification states of H3K4 (Eissenberg et al., 2007; Huarte et al., 2007; Kooistra and Helin, 2012; Seward et al., 2007).
The filamentous fungus Magnaporthe oryzae, is a destructive fungal pathogen that causes blast disease in rice (Dean et al., 2012). Rice is an economically important crop that supports approximately one-half of the world’s population (Khush, 2005). Due to the socioeconomic impact of rice blast disease, M. oryzae has been extensively studied as a model system (Talbot, 1995; Wilson and Talbot, 2009). Infection of host plants usually starts with dissemination of asexual spores, called conidia onto leaf surface. In the presence of water, conidia germinates and develop a specialized infection structure called appressoria at the tip of germ tube upon recognition of environmental cues such as surface hydrophobicity (Howard and Valent, 1996). Using turgor pressure generated inside appressorium, the fungus penetrates into plant cells and colonizes tissues leading to the development of disease lesions. Several studies have used M. oryzae as a model system to understand genetic regulation of plant-microbe interactions, thereby yielding considerable amount of information on genetic components involved in cAMP, Ca2+, and MAP kinase signaling pathways (Lee and Dean, 1993; Lee and Lee, 1998; Xu and Hamer, 1996).
Our understanding of epigenetic phenomena has been facilitated by identification and characterization of histone modifying enzymes such as HATs, HDAC, HMTs, and HDMs. In Neurospora crassa and Aspergillus nidulans, histone acetylation is an important step for transcriptional activation of the genes associated with light response signaling and production of secondary metabolites (SM) (Grimaldi et al., 2006; Shwab et al., 2007; Strauss and Reyes-Dominguez, 2011). Analysis of histone methylation in N. crassa had demonstrated that H3K9me3 and DNA methylation are required for heterochromatin formation (Smith et al., 2011; Tamaru and Selker, 2001). In addition, it was shown that methylation of H3K36, regulated by SET-2 methyltransferase has important role in fungal growth, conidiation and female sterility (Adhvaryu et al., 2005). In Aspergillus, histone methylations are related to SM production. For instance, CclA, a member of COMPASS complex involved in H3K4 methylation, and LaeA, affecting methylation level at H3K9, regulate SM gene clusters (Bok et al., 2009; Strauss and Reyes-Dominguez, 2011). Recently, in M. oryzae, it was shown that MoSET1, catalyzing dimethyl H3K4, is associated with substrate-induced transcriptional activation of a cellulose gene (Vu et al., 2013). Although role of histone modification in various biological processes of filamentous fungus is well studied, the functional aspects of fungal HDMs are yet to be explored. In this study, we set out to investigate roles of a JmjC histone lysine demethylases for fungal development and pathogenicity via genetic analysis combining gene knockout and transcriptome profiling.
Materials and Methods
Fungal strains and culture condition
M. oryzae wild-type strain KJ201 used in this study was obtained from the Center for Fungal Generic Resources (CFGR; hrtp://genebank.snu.ac.kr). All strains including wild-type strain and mutants constructed in this study were incubated on V8 juice agar medium (8% V8 juice [w/v], 1.5% agar powder [w/v], 10 N NaOH) or oatmeal agar medium (OMA, 5% oat meal [w/v], 2.5% agar powder [w/v]) at 25°C under constant fluorescent light. Mycelia used for RNA, DNA, and protein extraction were incubated in liquid complete media (LCM, 0.6% yeast extract [w/v], 0.6% casamino acid [w/v] and 1% sucrose [w/v]) at 25°C with 150 rpm shaking. Selection of hygromycin-resistant transformants was carried out using TB3 agar plates (0.3% yeast extract [w/v], 0.3% casamino acid [w/v], 1% glucose [w/v], 20% sucrose [w/v] and 0.8% agar powder [w/v]) supplemented with 200 ppm hygromycin B.
Sequence and phylogenetic analysis
Nucleotide and protein sequences were procured and analyzed at Comparative Fungal Genetic Platform (CFGP; http://cfgp.snu.ac.kr) and National Center for Biotechnology Information (NCBI). Sequences of JmjC domain-containing protein were obtained from ChromDB database (www.chromdb.org). Analyses using Hidden Markov Model (HMM) profiling (http://hmmer.janelia.org) were performed to search for additional JmjC proteins in M. oryzae and to compare the protein sequence homology in other organisms. Protein sequences were aligned using ClustalW and T-coffee algorithm. Phylogenetic tree was constructed with MEGA 5.2 using neighbor-joining method with a Poisson correction model and a bootstrap of 1,000 replicates. Information about domain architectures was obtained from InterProScan.
Nucleic acid isolation and manipulation
Fungal genomic DNA was extracted by two different methods according to the purpose. For large-scale screening of transformants, fungal DNA was extracted from mycelia on TB3 agar plates using quick extraction protocol (Chi et al., 2009). For Southern blot analysis genomic DNA was extracted using phenol-chloroform method. Total RNA was extracted with the Easy-Spin total RNA extraction kit (iNtRON Biotechnology, Seoul, Korea) according to the manufacturer’s instruction. cDNA for expression analysis by quantitative real-time PCR (qRT-PCR) was synthesized by using ImProm-II Reverse Transcription System kit (Promega, Madison, WI, USA) following the manufacturer’s instruction.
Targeted deletion of MoJMJ1 and construction of complementation strains
To construct gene deletion mutant, homologous recombination strategy was used. The original sequence of MGG_04878.7 was replaced with a knock-out construct containing 1.4 kb of hygromycin B phosphotransferase (HPH) cassette fused with ~1.2 kb 5′- and 3′-flanking regions by double joint PCR. Protoplast isolation and fungal transformation were carried out using PEG mediated transformation. Transformants resistant to hygromycin B within TB3 media were selected. Disruption of genes was verified by PCR using primers for screening (Supplementary Table 1). Southern blot analysis using genomic DNA was performed to confirm correct integration of HPH cassette. Genomic DNA fragments, digested with restriction enzyme, were separated by agarose gel electrophoresis and transferred to nitrocellulose membrane, Hybond-N+ (GE Healthcare, Piscataway, NJ, USA). The blot was hybridized with probe labeled with P32 by using Rediprime II Random Prime Labeling System kit (Amersham Pharmacia Biotech, Piscataway, NJ, USA) and the hybridized membrane blot was exposed to imaging plate, BAS-2040 (Fuji Photo Film, Tokyo, Japan) and read by phosphorimage analyzer, BAS-1000 (Fuji Photo Film).
Complementation strain for MoJMJ1 was generated by the following procedures. DNA fragment containing ORF and upstream sequences were amplified from KJ201 genomic DNA. The resulting PCR product and pII99 plasmid containing geneticin-resistant gene as a selection marker were co-transformed into protoplast of deletion mutant ΔMojmj1. Complementation strains were selected on geneticin-supplemented media and verified by PCR using primer pairs ORF_F_screening and ORF_ R_screening (Supplementary Table 1) for existence of the whole MoJMJ1 ORF, which was absent in the deletion mutant.
Developmental phenotypic assays—mycelial growth, conidiation, conidial germination, and appressorium formation
Mycelial growth was measured as colony diameter of 9-day-old culture on modified complete agar media (1% glucose [w/v], 0.2% peptone [w/v], 0.1% yeast extract [w/v], 0.1% casamino acid [w/v], 0.1% trace element [v/v], 0.6% sodium nitrate [w/v], 0.05% potassium chloride [w/v], 0.05% magnesium sulfate [w/v], 0.15% potassium dihydrogen phosphate [w/v], 1.5% agar powder [w/v]) and 4 mm diameter inoculums were taken from 5-day-old minimal agar media in marginal region.
Conidiation was measured by counting the number of asexual spores in 15 μl of conidia suspension on a haemacytometer. For conidiation assay, conidia were harvested from 7-day-old V8 juice agar media using 5 ml of sterilized distilled water. Conidiophore development was monitored by observing agar block (~1 × 1 cm) from 9-day-old culture following scrape-off.
For measuring the rates of conidial germination and appressorium formation, conidia were harvested from 7-day-old V8 juice agar media with sterilized distilled water and filtered with two-layers of miracloth (Calbiochem, San Diego, CA, USA). Thirty microliters of conidial suspensions (3 × 104 conidia/ml) was dropped on hydrophobic coverslips (W. Knittel Glass, Braunschweig, Germany) with three repeats. The droplets were incubated in moist-lasted airtight containers at room temperature. Samples were incubated for 12 and 8 h for measuring conidial germination and appressorium formation/development rates respectively. For nikkomycin Z sensitivity test, conidia from wild-type, deletion mutant, and complementation strain were incubated with or without 100 μM of nikkomycin Z, and examined after 2.5 h.
Pathogenicity test, wound inoculation, and sheath assay
For pathogenicity tests and wound inoculation, conidia formed on 7-day-old V8 juice agar media were rubbed using sterilized cotton swabs dipped in distilled water containing Tween 20 (250 ppm) and filtered with two-layers of autoclaved miracloth. For sheath assay, the abovementioned collection procedure was followed, except that Tween 20 was not used. Final concentration of spore suspensions was maintained at 5 × 104 conidia/ml for pathogenicity test and wound inoculation, and 2 × 104 conidia/ml for sheath assay. Three- or four-week-old rice seedlings of Oryza sativa cv. Nakdongbyeo were used for above tests.
For pathogenicity test, spore suspension (10 ml of total volume) was sprayed onto 3-week-old rice seedlings which was then placed in a dark, humid dew chamber for 18 h at 25°C. Next, the seedlings were transferred to a growth chamber maintained at 80% humidity and 25°C. Leaves of the rice seedling were collected on day 7 after the inoculation and scanned.
Leaves of Nakdongbyeo were used for wound inoculation. Conidial suspensions (5 × 104 conidia/ml) were dropped at wounds, which were picked about 10 times by tip of a sterilized pin. Inoculated leaves were incubated in a humid container at 25°C. Lesions were observed and scanned at 6 days post inoculation (dpi).
Sheathes of Nakdongbyeo in 5–6 leaf stage were used for rice sheath assay. Rice sheathes were inoculated with conidial suspension (3 × 104 conidia/ml) and incubated in a moist container at 25°C. The sheath samples were sliced by razor blade and observed with light microscope.
Western blotting analysis
Frozen fungal mycelia were homogenized by homogenizer and added to PRO-PREP protein extraction solution (iNtRON Biotechnology). The homogenates were centrifuged (13,000 rpm, 10 min, 4°C) and the supernatant was collected. Equal amounts of homogenized protein (15 μg) were applied to 12% SDS-PAGE gel (Bio-Rad, Hercules, CA, USA), and electro-blotted onto an Immunobilon-P PVDF membrane (Bio-Rad). For the gel running and transfer procedures, ReliaBLOT® running buffer (20×) and transfer buffer (10×) (Bethyl, Montgomery, TX, USA) were used, respectively. The blots were probed with polyclonal histone H3 and H3K4me3 antibody (Active Motif, Carlsbad, CA, USA), using Pierce® Fast Western Blot Kit, ECL Substrate (Bio-Rad).
Results
MoJMJ1 encodes JmjC domain-containing histone demethylase in JARID group
As a first step toward elucidating importance of histone demethylation in M. oryzae, we searched the proteome for putative HDM in M. oryzae using hidden Markov model built on known HDM sequences, and found that there are a total of seven genes encoding putative HDM among eleven proteins containing JmjC-domain. The following phylogenetic analysis showed where the seven genes could be placed in reference to known JmjC protein sequences of human, rice, Arabidopsis and yeast. Among the predicted seven genes in M. oryzae, MGG_04878 was predicted to belong to JARID group; MGG_09186 belonged to JMJD2; MGG_01543 and MGG_04401 to JmjC domain only groups, and MGG_09841, MGG_02045 and MGG_01068 appeared to be fungal specific through phylogenetic analysis (Supplementary Fig. 1). Among these genes encoding putative JmjC domain-containing proteins, we focused on a gene encoding JARID group histone lysine demethylase (MGG_04878) for further analysis, since we were able to obtain deletion mutants for the gene, and designated it as MoJMJ1 (M. oryzae jumonji 1).
Several orthologs MoJMJ1 have been characterized in species as diverse as human, Drosophila melanogaster, Caenorhabditis elegans to Saccharomyces cerevisiae and Schizosaccharomyces pombe. From these studies, it was shown that majority of the JARID1 group proteins share the same target histone residues (di- and tri-methyl H3K4) and contain highly conserved cofactor-binding sites required for histone lysine demethylase activity (Agger et al., 2008; Huarte et al., 2007; Klose et al., 2006; Secombe et al., 2007). When we compared domain architecture, we found that MoJMJ1 protein was predicted to have JmjN (IPR003349), JmjC (IPR003347), BRIGHT/ARID (IPR001606), C5HC2-zf (IPR004198), and PHD domain, all of which are typical domains of human JARID1 orthologs (Fig. 1A). Multiple sequence alignment shows that all the key residues for cofactor binding is conserved in MoJMJ1 (Fig. 1B).
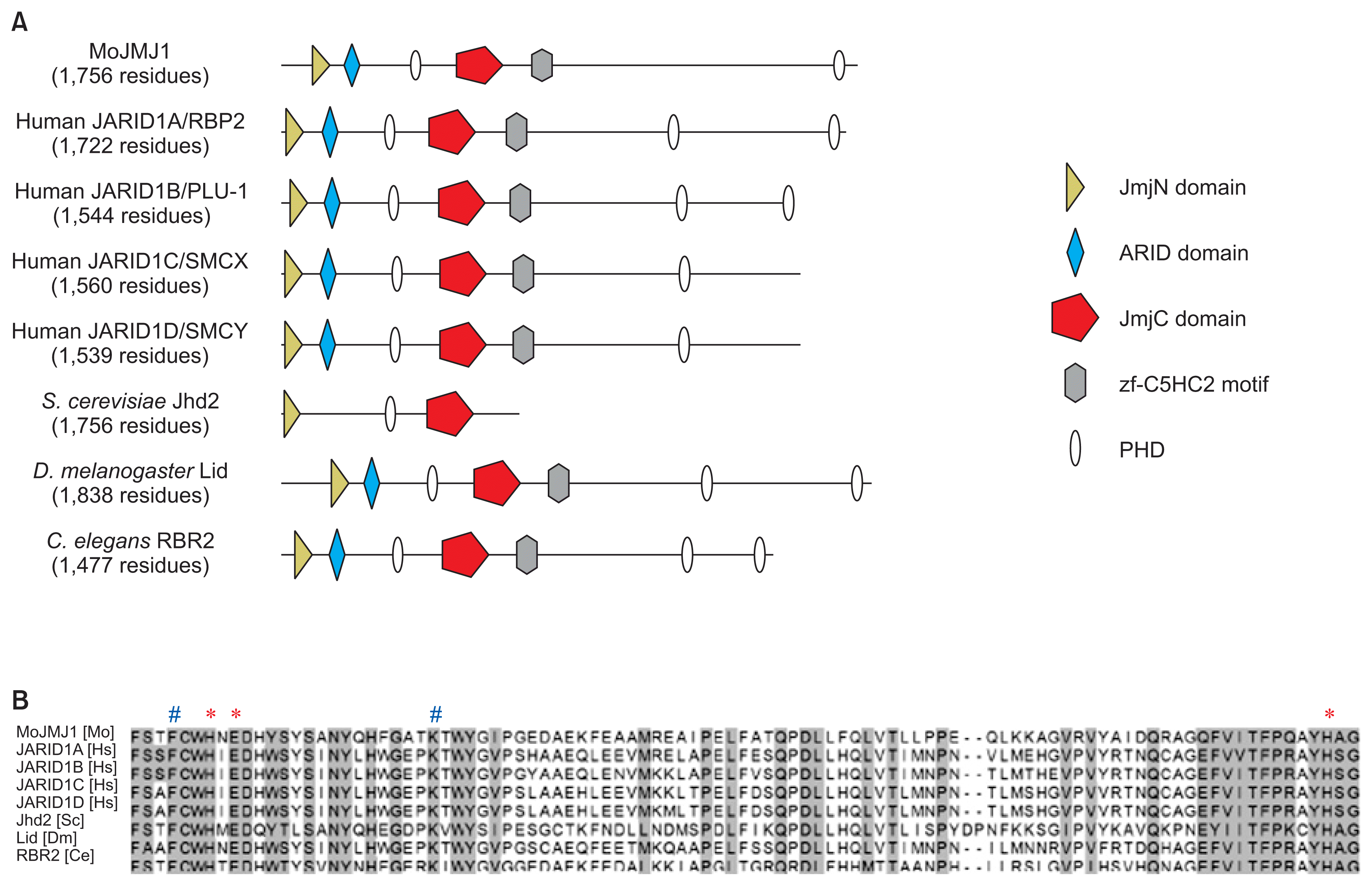
Domain architectures and conserved key amino acids for demethylase activity in JmjC domain. (A) Schematic representation of domains in MoJMJ1 and JARID group proteins in other organisms. Domain architecture was obtained from InterProScan. (B) Protein sequence alignment of JARID group histone demethylases including MoJMJ1. Proteins of other organisms were previously shown to have histone lysine demethylase activity. Asterisks (*) and pound signs (#) indicate Fe(II) and α-ketoglutarate binding site, respectively. Mo, Magnaporthe oryzae; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegan.
Deletion of MoJMJ1 and immunoblot assay
To investigate the role(s) of a JARID group demethylase in fungal development and pathogenicity, we generated deletion mutants in which the native MoJMJ1 sequence in wild-type strain, KJ201, was replaced by HPH cassette (Fig. 2A). Correct gene replacement event was verified through PCR screening and following Southern blot analysis in the mutants (Fig. 2B). Absence of transcript from Mo-JMJ1 locus in ΔMojmj1 mutant strain was confirmed by RT-PCR (Fig. 2C).

Targeted gene replacement of MoJMJ1 in Magnaporthe oryzae. (A) Schematic representation for targeted gene deletion of MoJMJ1. Knockout construct was made by fusion PCR. MoJMJ1 was replaced with HPH cassette by homologous recombination. (B) Southern blot analysis performed to confirm insertion of single copy DNA construct. Genomic DNA was digested with SalI and probed with 5′ flanking fragment of MoJMJ1. (C) Reverse transcriptase PCR for checking expression of MoJMJ1. RNA was extracted from mycelia of wild-type, ΔMojmj1 mutants. (D) Western blot analysis. Proteins were extracted from mycelia of wild-type and ΔMojmj1 mutants. Blot was probed with polyclonal anti-H3 antibody and polyclonal anti-trimethyl H3K4 antibody. Relative H3K4me3 level was quantified using ImageJ software.
Based on similarity at primary amino acid sequences, we predicted that MoJMJ1 is a histone lysine demethylase targeting H3K4me3, which serves as a mark for active transcription (Kooistra and Helin, 2012; Lee et al., 2007b). To test if deletion of MoJMJ1 affects global H3K4me3 level, we performed western blot analysis using antibody raised against H3K4me3. The western blot analysis showed that global H3K4me3 level increased by at least 70% in the mutant compared to the wild-type strain, suggesting that MoJMJ1 possesses HDM activity for H3K4me3 (Fig. 2D, Supplementary Fig. 2).
Vegetative growth, asexual reproduction, and appressorium formation of ΔMojmj1 mutants
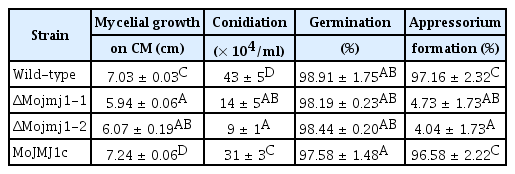
Phenotypes of ΔMojmj1 were examined to investigate the roles of Mo-JMJ1 in vegetative and developmental stages. First, effects of MoJMJ1 deletion on fungal vegetative stage were monitored. The diameter of ΔMojmj1 mutants showed reduction of 20%, compared to wild-type at 9 dpi on complete agar media (Table 1). In addition, autolysis was observed in both deletion mutants (Fig. 3A). These observations indicate that MoJMJ1 is important for normal vegetative growth in M. oryzae.
Four phenotypes of wild-type, ΔMojmj1 mutants, and complementation strain MoJMJ1c in Magnaporthe oryzae

Vegetative growth, conidiation, and appressorium formation in ΔMojmj1. (A) Colony morphology of wild-type, ΔMojmj1 mutants, and MoJMJ1c on complete agar media. Photographs were taken at 9 day post inoculation. Arrows indicate regions in which autolysis is observed. (B) Light microscopic images of conidia and conidiophores development on oat-meal agar block were captured after 24 h incubation. Scale bars = 100 μm. (C) Appressorium formation in ΔMojmj1. Appressorium formation from germinating conidia was observed at 8 h after drop and incubation on hydrophobic coverslips (left panels). Conidia were harvested from 7-day-old V8 juice agar media. Hyphal-driven appressorium development was monitored by placing and incubating mycelia blocks on hydrophobic surface for 24 h (right panels). Scale bars = 20 μm.
When we compared asexual reproduction between the wild-type and mutant by measuring the ability to produce conidia on axenic culture, we found that concentrations of conidial suspension obtained from the colony of ΔMojmj1 mutants were 25–40% of those of wild-type, indicating that MoJMJ1 is involved in asexual reproduction as well (Table 1). To further investigate the reason why asexual reproduction of the mutant was reduced in the mutant, we monitored conidiophore development on agar blocks. Although it appeared that the mutant was able to form at least three conidia in a sympodial pattern at a conidiophore, overall number of conidiophores decreased in ΔMojmj1 mutants (Fig. 3B). This suggests that MoJMJ1 may be associated with efficient development of conidiophore from hyphal tissue.
Normal development of appressoria is a pivotal step for host infection in M. oryzae. To investigate whether MoJMJ1 affects conidial germination and appressorium formation, a drop of conidial suspension was incubated on an artificial hydrophobic surface (coverslip). Most of conidia germinated in both the wild-type and deletion mutants (Table 1). Unlike the wild-type conidia developing a single appressorium at a germ tube tip, conidia of ΔMojmj1 showed bipolar germ-tubes that started to branch at 8 hours post incubation (Fig. 3C, left panels), and 95% of the conidia in ΔMojmj1 failed to develop appressoria, suggesting requirement of MoJMJ1 in appressorium formation (Table 1). Interestingly, abnormal appressorium started to develop at the tip of excessively elongated germ-tubes of ΔMojmj1 at around 20 hours post inoculation (hpi) and proportion of the mutant conidia that developed abnormal appressoria increased up to 30–40% at 36 hpi on coverslips (Supplementary Fig. 3). Such delay in appressorium formation prompted us to investigate the implication of MoJMJ1 in regulating signaling pathways involved in appressorium formation. To this end, we employed chemical complementation using cAMP, CaCl2, and 1,16-hexadecanediol. However, adding those chemicals separately or in combination into conidial suspension of ΔMojmj1 on hydrophobic surface was not able to complement the defects in appressorium formation of the mutants. It was previously shown that M. oryzae could develop appressoria not only at the germ tube but also at hyphal tips (Kim et al., 2009). When we tested the effect of MoJMJ1 deletion on the formation of hyphal-driven appressoria, we found that the mutant lost the ability to develop appressoria at the hyphal tip as well (Fig. 3C, right panels). These results suggest that MoJMJ1 is essential for normal development of appressorium from two different types of cells, germ-tubes and hyphae. When we ectopically introduced MoJMJ1 gene plus 1.2 kb putative promoter sequences back to the deletion mutant, all the defects in vegetative growth, conidiation, and appressorium formation were restored to the wild-type level (Fig. 3, Table 1).
Pathogenicity of ΔMojmj1 mutants and a complementation strain
Next, we tested whether MoJMJ1 is indispensable for fungal pathogenesis, given the inability of the mutant to develop appressoria on artificial hydrophobic surface. To evaluate pathogenicity of the mutant, we spray-inoculated the rice plants with conidial suspensions of wild-type and ΔMojmj1 strain, and compared the symptom development at 5 dpi. While the leaves challenged with wild-type strain showed typical spindle-shaped lesions, leaves inoculated with the deletion mutants remained asymptomatic (Fig. 4A). To our surprise, the complementation strain MoJMJ1c, however, was not able to cause disease at all on the rice plants (Fig. 4A). Such inability of complementation strains to cause disease was consistently observed in other complementation strains that were generated by a number of independent experiments. Upon failure of complementation for pathogenicity defect, we examined the mutant and complementation strain in detail to elucidate the nature of complementation failure by monitoring infection processes at the single cell level using sheath assay method. Unlike wild-type strain penetrating host cells via appressorium at 24 hpi and colonizing neighboring cells at 48 hpi, most of the conidia from deletion mutant was not able to form appressoria, which is consistent with our observation for in vitro appressorium formation of the mutant. Mishap appressoria, which developed at the tip of abnormally elongated germ tube of the mutant, appeared to have lost the ability to penetrate underlying host cells. Similarly, MoJMJ1c appressoria did not show any sign of host cell penetration even at 48 hpi, although the complementation strain developed appressoria that are indistinguishable from those of wild-type (Fig. 4B). One possibility of such penetration defect is disruption of cell wall integrity. We tested this possibility using Nikkomycin Z, an inhibitor of cell wall biosynthesis. While wild-type conidia were able to develop appressoria when treated with 10 ng/ml of Nikkomycin Z, both ΔMojmj1 and MoJMJ1c conidia exhibited ballooning of the germ tube tip and were not able to develop appressoria at the same concentration of Nikkomycin Z, suggesting defective cell wall in the mutant and complementation strains (Supplementary Fig. 4A). We then went further to test effect of such defect in cell wall on generation of turgor pressure within appressorium using cytorrhysis assay. The assay result showed that appressoria of both the mutant and complementation strains tend to collapse in less frequency at 3 M glycerol than the wild-type, indicating problem(s) in cell wall integrity of appressoria, which is important for appressorium functionality (Supplementary Fig. 4B).

Pathogenicity of ΔMojmj1 and MoJMJ1c. (A) Pathogenicity test of ΔMojmj1, MoJMJ1c mutants was performed using spray inoculation of spore suspensions. Conidial suspensions (5 × 104 conidia/ml) were sprayed onto 3-week rice seedlings, and leaves of the rice seedling were collected at 7 days post inoculation. (B) Rice sheath was inoculated with conidial suspension (2 × 104 conidia/ml). hpi, hours post inoculation. Arrowheads indicate appressorium. Scale bars = 30 μm. (C) Wound inoculation was observed at 5 days after drop of conidial suspensions (5 × 104 conidia/ml) on wounded spots of rice leaves.
We then ask if the mutant and complementation strain retain the ability to grow inside plant tissues by allowing direct entry of the fungus into host tissues through a wound site without appressoirum-mediated penetration process. To test this, the conidial suspension of ΔMojmj1 or MoJMJ1c was injected into plant tissues using a 22-guage syringe. In our assay, the wild-type strain developed large disease lesions but MoJMJ1c as well as ΔMojmj1 strains were unable to develop disease lesions. These data indicate that introduction of MoJMJ1 gene plus 1.2 kb upstream sequences into ΔMojmj1 strain can complement pre-penetration stage developments, but not penetration and post-penetration development.
In-depth analysis of complementation defects
For partial complementation of phenotypes, we first considered the possibility that unsolicited mutations occur during preparation and/or transformation of protoplast. To check this, we evaluated pathogenicity of two independent ectopic mutant strains (MoJMJ1e_52 and MoJMJ1e_185) that are obtained from the same batch of transformants as the deletion mutant strain. The ectopic mutants harbor knockout construct with intact MoJMJ1 gene. Therefore, if there is additional mutation(s) during protoplast preparation or transformation of protoplasts with knockout construct, they will share the mutation(s) and will be non-pathogenic as well. Spray-inoculation with these strains clearly showed that ectopic mutants are fully pathogenic on the rice plants, allowing us to rule out the possibility of presence of unsolicited mutation(s).
The second possibility that we considered is the length of 5′ upstream region required for complete complementation. To explore this, we extended 5′ upstream region of the gene included in a new complementation construct up to 2.1 kb. When we introduced the new complementation construct into ΔMojmj1 and screened for geneticin-resistance, we found that at least 80% of the resulting transformants were able to form appressoria on plastic coverslip (Fig. 5A). Among them, we randomly chose three transformants (MoJMJ1c_2-1, MoJMJ1c_2-2, and MoJMJ1c_2-3) and carried out spray-inoculation on rice plants using spores from the three strains as inoculum. Our data showed that none of them are pathogenic, suggesting that failure to complement pathogencity defect of the deletion mutant is unlikely to be associated with promoter length (Fig. 5B).
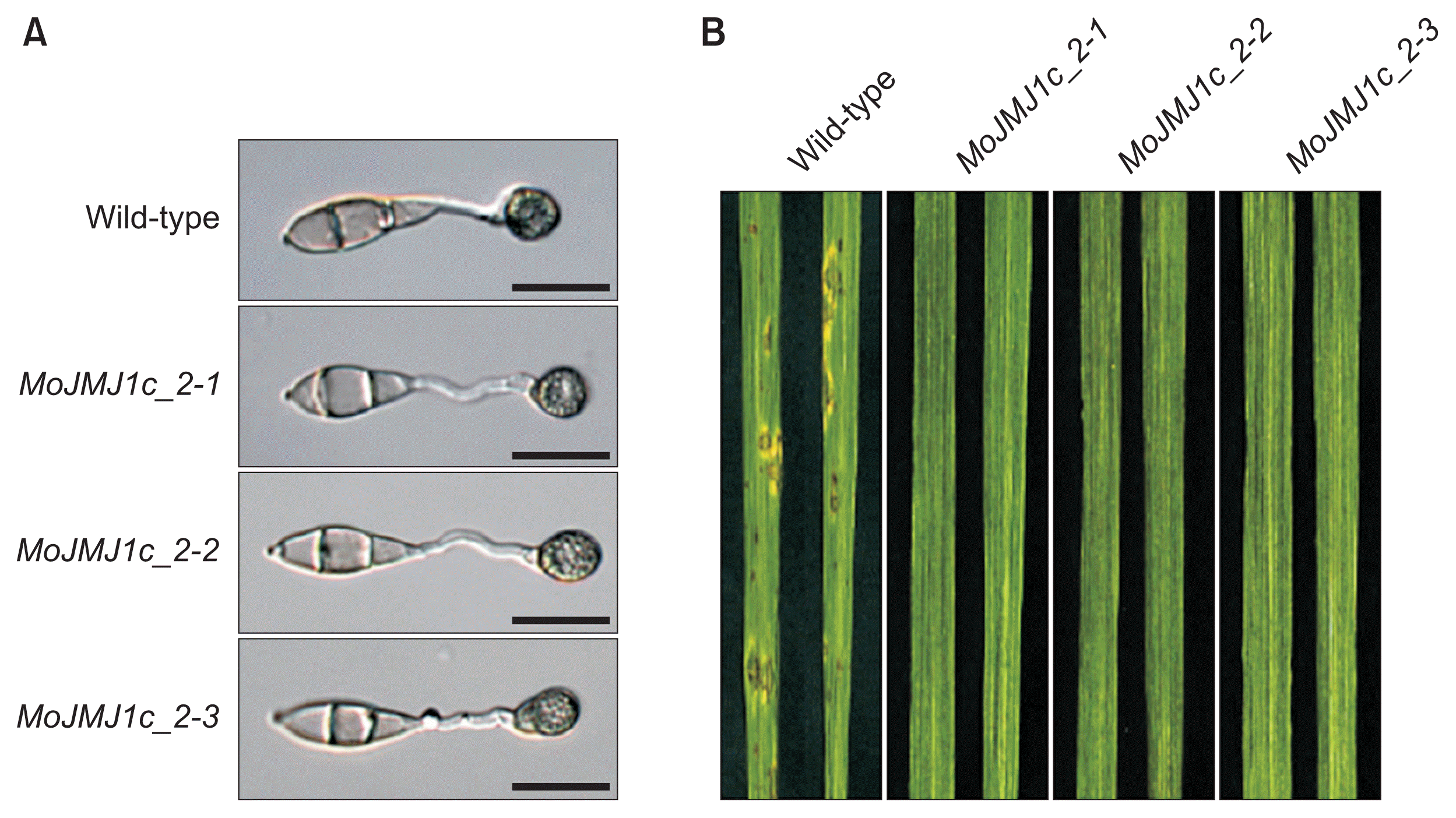
Appressorium development and pathogenicity test of regenerated complementation strains. (A) Appressorium formation of wild-type and regenerated complementation strains (MoJMJ1c_2-1, 2-2, and 2-3) transformed with construct of 2.1 kb 5′ flanking region. Scale bars = 20 μm. (B) Pathogenicity of wild-type and regenerated complementation strains, MoJMJ1c_2-1, 2-2, and 2-3. Pathogenicity test was performed using spray inoculation of spore suspensions. Conidial suspensions (5 × 104 conidia/ml) were sprayed onto 3-week rice seedlings and leaves of the rice seedling were collected at 7 days post inoculation.
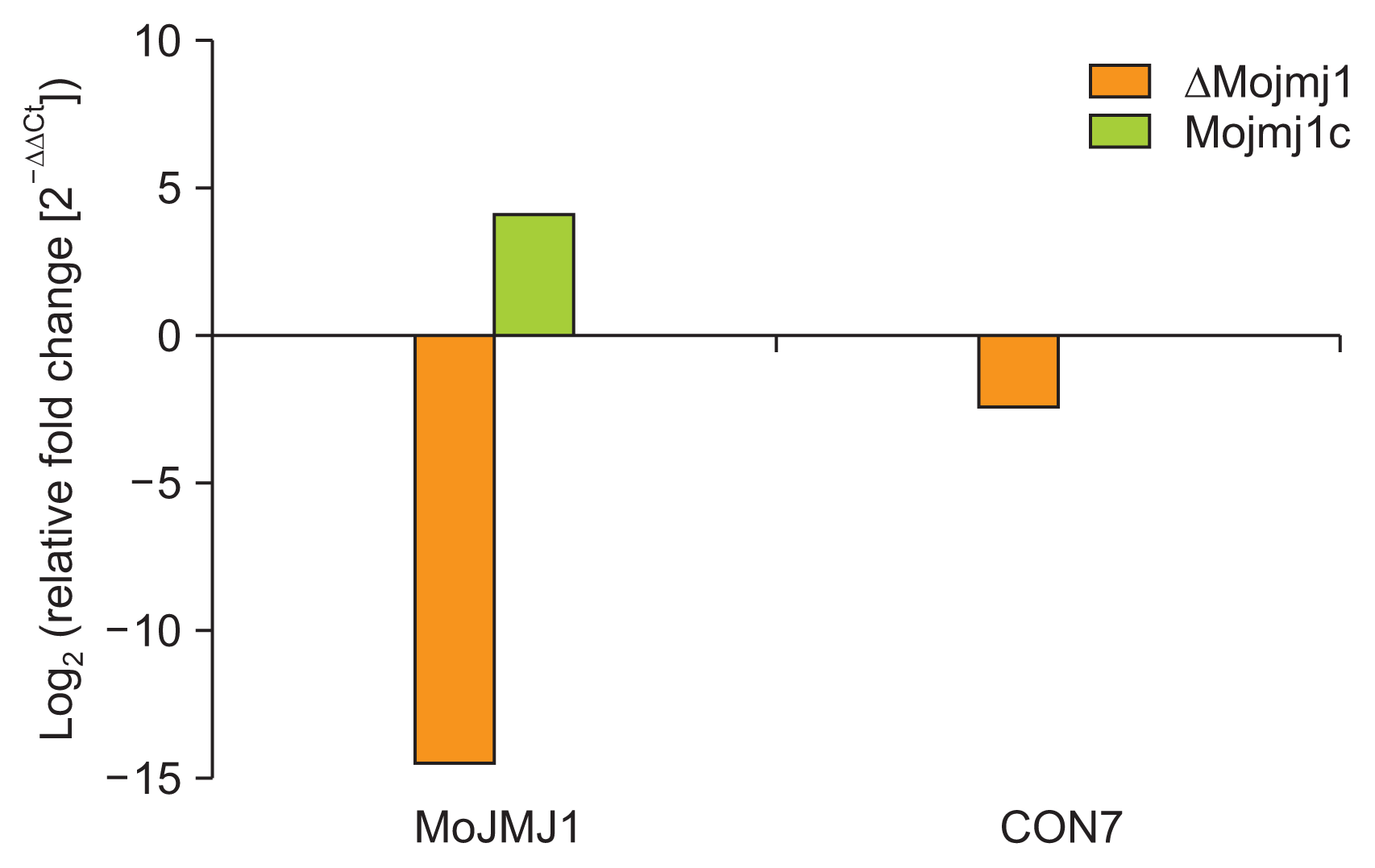
Lastly, we checked out expressions of two genes using qRT-PCR: (1) MoJMJ1 gene that is ectopically introduced into the complementation strain (construct having 1.2 kb promoter sequences) and (2) CON7 gene, which is known to be related to Nikkomycin Z sensitivity (Odenbach et al., 2007). Interestingly, MoJMJ1 gene was shown to be up-regulated in the complementation strain, while expression of CON7 was down-regulated in the mutant and restored to the wild-type level in the complementation strain (Fig. 6).
Discussion
Conidiation and appressorium development are essential steps for many fungal pathogens to infect host plant. A considerable number of studies have contributed towards establishing the knowledge of genetic components implicated in these processes, but the aspect of epigenetic regulation, especially that of histone demethylation still remains largely unexplored. It is well known that histone methylation/demethylation is involved in cell development and various other biological processes of plants and mammals. Recently, MoSET1, which is a histone methyltransferase that targets H3K4, was shown to be important for regulation of genes during infection-related morphogenesis of M. oryzae (Pham et al., 2015). In this study, we carried out genetic analysis for MGG_04878, a gene encoding putative HDM, using gene knockout in order to ascertain the roles of JmjC HDM in development and pathogenicity of a model plant pathogenic fungus, M. oryzae.
Our analysis of sequence homology and phylogeny indicated that MGG_04878 is a JARID group HDM. Conservation in domain architecture and sequences suggested that MoJMJ1 is likely to be a bona-fide HDM. Like other known H3K4 demethylases, MoJMJ1 possessed cofactor binding sites in JmjC domain and additional domains such as JmjN, BRIGHT/ARID, and C5HC2-zf domains that are shown to be essential for enzymatic activity (Iwase et al., 2007; Lee et al., 2007a, 2007b; Yamane et al., 2007). Overall, these results suggested that target substrates of MoJMJ1 are dimethyl/trimethyl H3K4. Our western blot analysis examining global H3K4me3 level for the wild-type and mutant lacking MoJMJ1 showed that H3K4me3 increased in the mutant by at least 70%, compared to the wild-type, corroborating that MoJMJ1 functions as a HDM targeting H3K4. It should be noted that knocking out JARID1B/PLU-1 in mammals did not increase global H3K4 methylation levels. Rather it resulted in increased trimethyl H3K4 levels at the region near transcription start sites of specific target genes (Yamane et al., 2007). Therefore, increase in global H3K4me3 level observed in ΔMojmj1 may suggest that MoJMJ1 targets higher proportion of genes in the genome than its mammalian ortholog does, and that disruption of the gene would result in pleiotropic phenotypes.
In accordance with this, we found that deletion mutants of MoJMJ1 are defective in all the phenotypes we examined except germination: vegetative growth, asexual reproduction, appressorium development and pathogenicity. Most notably, the mutant conidia were not able to develop appressoria within 8 h. Instead, they abnormally elongated germ tubes that at some point started to branch as vegetative hyphae do. At around 20 h, appressoria-like structures developed from some of the conidia, and proportion of conidia that developed such structures reached up to 30–40% of all the conidia at around 40 h. Furthermore, the mutant has lost the ability to differentiate hyphal-driven appressoria, which is the reminiscent of what was observed in ΔMohox7 (Kim et al., 2009). This suggests that defect in appressorium formation from both germ tube and hyphae might be attributed to faulty regulation of this important transcription factor. However, it should be noted that regulation and/or recruitment of transcription factors such as MoHOX7 could be influenced by appressorium stage-specific histone modifications, since our speculation is based only on the data from mycelia tissue. In addition, this defect in appressorium formation was not restored by addition of CaCl2 and/or cAMP, indicating that MoJMJ1 functions at the levels downstream of cAMP and calcium signaling pathways. Growth retardation, reduction in asexual spores, and appressorium formation defect were all complemented by introduction of MoJMJ1 gene into ΔMojmj1, suggesting that MoJMJ1-mediated histone demethylation is required for pre-penetration development.
In eukaryotes, H3K4 methylation is known as an epigenetic mark for gene activation (Kooistra and Helin, 2012). Recently, it was shown that deletion of MoSET1 (Pham et al., 2015), a histone methyltransferase targeting H3K4, resulted in 4,077 genes, which include some of the genes involved in cAMP signaling. In this work, severe reduction in appressorium formation of the deletion mutant was restored by addition of exogenous cAMP. This may suggest that MoJMJ1 regulates down-stream processes of MoSET1-dependent genes and/or plays important backup for MoSET1 for fine-tuning the expression levels of target genes.
To our surprise, complementation construct containing MoJMJ1 gene plus 1.2 kb upstream sequences was not able to complement pathogenicity defect of the deletion mutant in our work. In an effort to resolve the problem, we extended 5′ upstream flanking region from 1.2 to 2.1 kb. Despite longer upstream sequences, the regenerated complementation strains were also unable to restore the defects in penetration and invasive growth, indicating that promoter length may not be the reason for the partial complementation. Examination of ectopic transformants showed that ectopic mutants retained the ability to cause disease symptoms on rice plants, allowing us to rule out the possibility that there are unsolicited mutations that can result in penetration and pathogenicity defect. We reasoned that such failure for complete complementation should be associated with defect in cell wall, since both the deletion mutant and complementation strains showed hypersensitivity to Nikkamycin Z. However, expression level of CON7, which encodes a transcription factor involved in regulating cell wall integrity, was restored in the complementation strain. These observations suggest two things: (1) Molecular mechanisms underlying defects in cell wall might be different between ΔMojmj1 and Mo-JMJ1c and (2) Defects in cell wall could be one of main reasons why appressoria of complementation strain cannot penetrate and infect host plants (Supplementary Fig. 4).
It should be also noted that, in the complementation strain, MoJMJ1 was up-regulated (Fig. 6). It is not clear why ectopic insertion of MoJMJ1 gene and its upstream sequences can cause over-expression of the gene. One explanation would be chromosomal location of and epigenetic environment around MoJMJ1 gene might be important for proper regulation of the gene. It is also possible that deletion of MoJMJ1 and subsequent genome-wide perturbation of histone modifications may have long-lasting effects on the following generations of fungal cells. Over-expression of HDM was shown to cause developmental defects (Nottke et al., 2009). Similarly, overexpression of MoJMJ1 in the complementation strain could lead to mis-regulation of many genes. To explore this possibility, we attempted to test if we can recapitulate the defect in appressorium function by over-expressing MoJMJ1 in wild-type strain using strong constitutive promoter. Unfortunately, we repeatedly failed to produce an over-expression construct of MoJMJ1. However, it is still tempting to speculate that expression level of MoJMJ1 may have more profound impact on genes involved in appressorium-mediated penetration and subsequent host infection than genes implicated in pre-penetration development.
In this study, we demonstrated that MoJMJ1 encoding JmjC HDM is required for fungal development, especially pre-penetration phase development. It thus appears that reversible lysine methylation of histones is important for regulation of genes involved in infection-specific morphogenesis, and that those genes are likely to be functioning downstream of major signaling pathways. Last but not least, in-depth analysis of complementation strain provided insights into complexity and subtlety of phenotypes and transcriptional program that can be entailed in the genetic study of a histone modifying enzyme.
Supplementary Information
Acknowledgments
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2012R1A6A3A04038022). This work was also supported by the National Research Foundation of Korea grant funded by the Ministry of Science, ICT & Future Planning (NRF-2014R1A2A1A10051434), the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ011154), Rural Development Administration, Republic of Korea.