Enterobacter cloacae, an Emerging Plant-Pathogenic Bacterium Affecting Chili Pepper Seedlings
Article information
Abstract
A previously unreported bacterial disease on chili pepper (Capsicum annuum L.) seedlings affecting as many as 4% of seedlings was observed in greenhouses in Chihuahua, Mexico (Delicias and Meoqui counties). Initial lesions appeared as irregular small spots on leaves and brown necrosis at margins tips were observed. Later, the spots became necrotic with a chlorotic halo. Advanced disease was associated with defoliation. A Gram negative, rod-shaped bacterium was isolated from diseased chili pepper seedlings. Three inoculation methods revealed that isolated strains produce foliage symptoms, similar to those observed in naturally infected seedlings. Pathogenic strains that caused symptoms in inoculated seedlings were re-isolated and identified to fulfill koch’s postulate. Polyphasic approaches for identification including biochemical assays (API 20E and 50CH), carbon source utilization profiling (Biolog) and 16S rDNA, hsp60 and rpoB sequence analysis were done. Enterobacter cloacae was identified as the causal agent of this outbreak on chili pepper seedlings.
Introduction
Enterobacter cloacae is a ubiquitous Gram-negative, facultative anaerobic, rod-shaped bacterium belonging to the Enterobacteriaceae family. E. cloacae is found as a commensal in the gut flora of many humans and animals (Fanaro et al., 2003). Under certain conditions, E. cloacae can lead to infections of the urinary or respiratory tract or of the bloodstream (Mezzatesta et al., 2012). As an occasional contaminant of medical devices and solutions, E. cloacae has been linked to nosocomial outbreaks, which are of particular concern in neonatal units (Dalben et al., 2008). Moreover, frequent isolation of multidrug-resistant strains is going to confront antibiotic treatment of patients and silver sterilization of wounds and medical devices (Davin-Regli and Pagès, 2015).
More recently, E. cloacae has increasing importance as a plant pathogen. For instance, E. cloacae has been associated with internal decay of onion (Schroeder et al., 2009; Bishop and Davis, 1990) and internal yellowing disease of papaya (Nishijima et al., 1987). Moreover, E. cloacae and other species of this complex are reported as pathogenic in mulberry in China (Wang et al., 2010), dragon fruit in Malaysia (Masyahit et al., 2009), macadamia in Hawaii (Nishijima et al., 2007), lucerne seeds in China (Zhang and Nan, 2013), odontioda orchids in Japan (Takahashi et al., 1997) and cassava in Venezuela (Santana et al., 2012). Among others, the main symptoms observed in these plants are wilted leaves that turned dark brown, and defoliated plants at late stage of the infection.
Precise methods for identification and taxonomic classification are necessary, since its first description as E. cloacae underwent several taxonomic changes. The taxon E. cloacae was introduced in 1960 (Hormaeche and Edwards, 1960). Biochemical and molecular studies revealed a high degree of genomic heterogeneity among the species of Enterobacter. As a consequence, whole-genome DNA-DNA hybridizations and phenotypic characteristics led to the formulation of the taxon “E. cloacae complex” (also called “E. cloacae nomenspecies”), which comprises six species of Enterobacter (Mezzatesta et al., 2012). Phenotypic identification of species within the E.cloacae complex is difficult and not always reliable. Unambiguous identification of Enterobacter species can be achieved by DNA sequencing of the 16s rRNA gene, combined with genotyping based on gyrB, hsp60 and rpoB genes (Mezzatesta et al., 2012). Sequence information from hsp60 and/or rpoB genes allows to assign isolates to one of the 12 genetic clusters, nine of which correspond to species, or to a 13th less defined “sequence crowd” (Hoffmann and Roggenkamp, 2003). Higher resolution is possible by including more housekeeping genes in a multilocus sequence typing scheme (Paauw et al., 2008) or using phylogenomics (Liu et al., 2013).
The aim of this work was to characterize and identify the causative agents responsible for a new disease on chili pepper (Capsicum annuum) that is affecting as many as 4% of seedlings grown in commercial greenhouses in northern, Mexico.
Materials and Methods
Isolation of plant-pathogenic bacteria from greenhouse-grown pepper seedlings
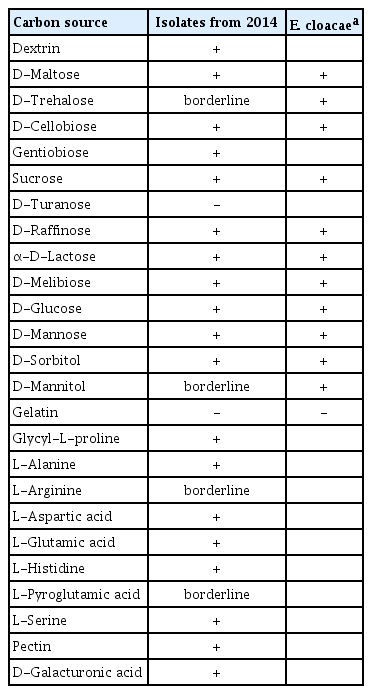
In three consecutive years, 2013 to 2015, diseased seedlings of pepper were observed in two commercial greenhouses located on Delicias (28°11′ 36″ N, 105°28′16″W, 1171 masl) and Meoqui (28°16′ 04″N, 105°33′ 37″W, 1100 masl) countries of Chihuahua State in Mexico. The concerned greenhouses displayed relatively warm (36–38°C) and humid (80–90% RH) conditions, while seedlings remained healthy in other greenhouses with lower RH. Symptoms were small necrotic lesions on leaves and brown necrosis at margins tips (Fig. 1). Assuming that the disease could be caused by bacteria, they were isolated from leaves of diseased seedlings of Jalapeño chili pepper cultivars (cvs.) Imperial, Perfecto, Tula and Maraja; Anaheim chili pepper cvs. Maverick and Cayenne; and a commercial hybrid (Table 1).

Symptoms observed in chili (Capsicum annuum L.) pepper during seedlings production in commercial greenhouses in Mexico. (A) Initial symptoms in jalapeño pepper seedlings showing water-soaked in the middle of the leaves. (B) Chili pepper leaves with irregular brown spots and slightly yellowing. (C) Yellowing on the leaves with darkening in the affected area, apical necrosis is observed in some leaves. (D) Eventually, seedlings got completely defoliated after 60 days.

Bacterial strains isolated of Jalapeño, Anaheim and hybrid pepper (Capsicum annuum L.) seedling leaves from 35 days-old (10–18 cm high) planted in greenhouses during 2013–2015 in Chihuahua, México
Leaves of symptomatic seedlings were collected in the greenhouse, then they were rinsed with sterile distilled water (SDW), disinfested with 0.3% sodium hypochlorite for 30 s and rinsed with SDW three times. Then leaves were drained and dried before aseptically sectioning. Small (3 × 3 mm) leaf sections were then excised from the edge of the diseased tissue and macerated in 3 ml of SDW for 90 s in a sterile mortar. The suspensions were streaked onto nutrient agar (NA) plates and incubated at 30°C for 24–48 h. Single colonies were isolated and re-streaked at least twice to purify the bacterial strains. Isolated bacteria were identified using API 20E strips and were stored in 15% glycerol at −80°C for long-term conservation.
Pathogenicity tests and re-isolation of bacteria upon seedlings inoculation
In 2015, different isolates were activated on Nutrient Agar (NA) medium and keep at 30°C for 48 h. Plate-grown bacteria were resuspended in 30 ml of SDW and adjusted to an optical density (OD) of 0.2 at A600, corresponding to approximately 107 colony-forming units (CFU) per milliliter. As control, SDW was used.
Pathogenicity tests were performed on 50 day-old healthy seedlings, with four to six leaves, approximately 15 cm tall at the time of inoculation. Fourteen isolates were inoculated into Jalapeño pepper seedlings of the varieties Autlan and Rio grande, and Anaheim pepper seedlings of the variety Maverick.
Three different inoculation methods were evaluated. In the first one, three leaves of a seedling were puncture-wounded with a hypodermic needle, followed by pipette-inoculation of 4 μl of the bacterial suspension onto the wound. In the second one, the bacterial suspension was sprayed on the leaves until they were uniformly wet. In the third one, the bacterial suspension was injected with a needle-less 1-ml syringe, pressing the syringe on the lower side of three leaves per seedling. Four seedlings were inoculated per bacterial strain and SDW treatment controls, and each experiment was conducted two times.
Inoculated seedlings were held in polyethylene moist chambers within the greenhouse at 37°C and 80 to 90% relative humidity (RH) and seedlings were watered daily. Following inoculation, seedlings were monitored to observe symptom development, such as dark spots with brown necrosis at the margins and tips of the leaves. Seedlings were maintained in the moist chambers until they were completely defoliated, as it had occurred in natural conditions. For re-isolation of the strains, symptomatic leaf tissue of inoculated seedlings was treated as described above. Re-isolated bacteria were identified by morphological characteristics and by using API 20E strips (Table 2). This identification allowed to know the re-isolated bacteria are identical to the original, thus fulfilling Koch’s postulates.
Physiological and biochemical characterization
Re-isolated bacteria from 2013, 2014 and 2015 were grown on NA plates at 30°C for 24 to 48 h. Selected bacterial cultures were used for the biochemical tests. Gram staining was performed and cell morphology was investigated by microscopic examination. Biochemical identification was conducted using API 20E and API 50CH strips (BioMerieux Inc., Durham, NC, USA), according to the manufacturer’s instructions.
Carbon substrate utilization was assessed using the Biolog® system (Biolog Inc., Hayward CA, USA). Bacterial suspensions were inoculated into the GEN III Microplate® (Biolog Inc.) and incubated according to the manufacturer’s instructions. The color reaction indicating utilization of each carbon substrate was determined visually and results were analyzed using the Biolog® database (http://www.biolog.com). A taxon was identified when the similarity index (SIM) was > 0.5.
Molecular biological techniques
Genomic DNA was extracted by a modified hexadecyl trimethyl ammonium bromide (CTAB) method (Wang et al., 2010). One loop of 48 h-old bacterial culture grown on NA plates were lysed. The lysis solution consisting of 600 μl of CTAB/NaCl (10% CTAB [Sigma-Aldrich, MO, USA] in 0.7 M NaCl) at 65°C for 40 min. Proteins were removed by extraction using chloroform/isoamyl alcohol (24:1) and phenol/chloroform/isoamyl alcohol (25:24:1). DNA was recovered by isopropanol precipitation and washed with 70% ethanol. The pellet was resuspended in 50 μl of ultrapure water (Invitrogen, Carlsbad, CA, USA) and stored at 4°C. DNA concentration and quality was measured using a spectro-photometer NanoDrop 2000C (Thermo Fisher Scientific Inc., Waltham, MA, USA).
For molecular diagnosis, an internal portion of the 16S rRNA gene was amplified by polymerase chain reaction (PCR) with 8F/1492R and sequenced using U514F/800R primers (Li et al., 2010). In addition, two housekeeping genes, hsp60 and rpoB, were analyzed by PCR and DNA sequencing, they showed to be polymorphic among Enterobacter isolates (Hoffmann and Roggenkamp, 2003; Miyoshi-Akiyama et al., 2013) (Table 4). Amplification reactions were carried out in a volume of 25 μl, started after a preheating time of 4 min at 95°C. Samples for 16S rRNA gene were subjected to 35 cycles of 2 min denaturation at 95°C, 1 min annealing at 56°C, and 1.3 min of extension at 72°C. Cycling was completed by a final elongation step of 5 min at 72°C. Amplification conditions for hsp60 and rpoB genes were 30 cycles of 30 s denaturation at 95°C, 30 s annealing at 57.5°C, and 3 min of extension at 72°C. Final elongation was at 72°C for 15 min.
DNA amplicons was checked by gel electrophoresis at 100 V for 40 min on a 1.5% agarose gel. PCR products remained were sequenced using a commercial service (Beckman Coulter Genomics, Roissy, France). For species identification, DNA sequences were compared with the non-redundant (nr) and whole-genome shotgun contigs (wgs) databases at the National Center for Biotechnology Information (NCBI) using the BLASTN option with default parameters (http://blast.ncbi.nlm.nih.gov) (Altschul et al., 1990). The strain GAQ130 have been deposited at GenBank under accession numbers KX822725 (16S rRNA), KX822726 (hsp60), and KX822727 (rpoB) as representative sequences.
Bioinformatic analyses
Sequence comparisons were performed on the websites of the European Bioinformatics Institute (http://www.ebi.ac.uk/services). DNA sequences were aligned using the Needleman-Wunsch algorithm (Needleman and Wunsch, 1970). For multiple sequence alignments, the MUSCLE algorithm was used (Edgar, 2004).
The platform Phylogeny.fr was selected to perform the phylogenetic analyses (Dereeper et al., 2008). MUSCLE (v3.7; Mill Valley, CA, USA) configured for highest accuracy (MUSCLE with default settings) was used to align the sequences. Regions containing gaps or poorly aligned were deleted with Gblocks (v0.91b; Castresana Lab, Barcelona, Spain). The phylogenetic tree was rebuilt on the PhyML program (v3.0; Oregon University, USA) with maximum likelihood, assuming the number of invariant sites and 4 gamma-distributed rate categories, to account for rate heterogeneity across sites, the HKY85 substitution model was used. The gamma shape parameter was calculated from the data. Internal branch reliability was estimated by the aLRTtest (SH-Like). The phylogenetic tree graphic and edition was performed with TreeDyn (v198.3; Montpellier, France).
Results
Plant pathogenic bacteria
Symptoms similar to those that had initially been observed in greenhouses appeared within four to 23 days after inoculation. No symptoms were observed on control plants that had been inoculated with SDW.
All 14 tested isolates were pathogenic to chili pepper seedlings. Inoculated seedlings exhibited irregular small spots on leaves. Later, these spots became necrotic with a chlorotic halo. Brown necrosis at margins tips was also observed. Finally, the seedlings were defoliated (Fig. 2). Control seedlings remained healthy. Bacterial strains were re-isolated from symptomatic inoculated seedlings. Our experiments represent a completion of the steps of Koch’s postulates, demonstrating that E. cloacae is the causal agent of the new disease on chili pepper.
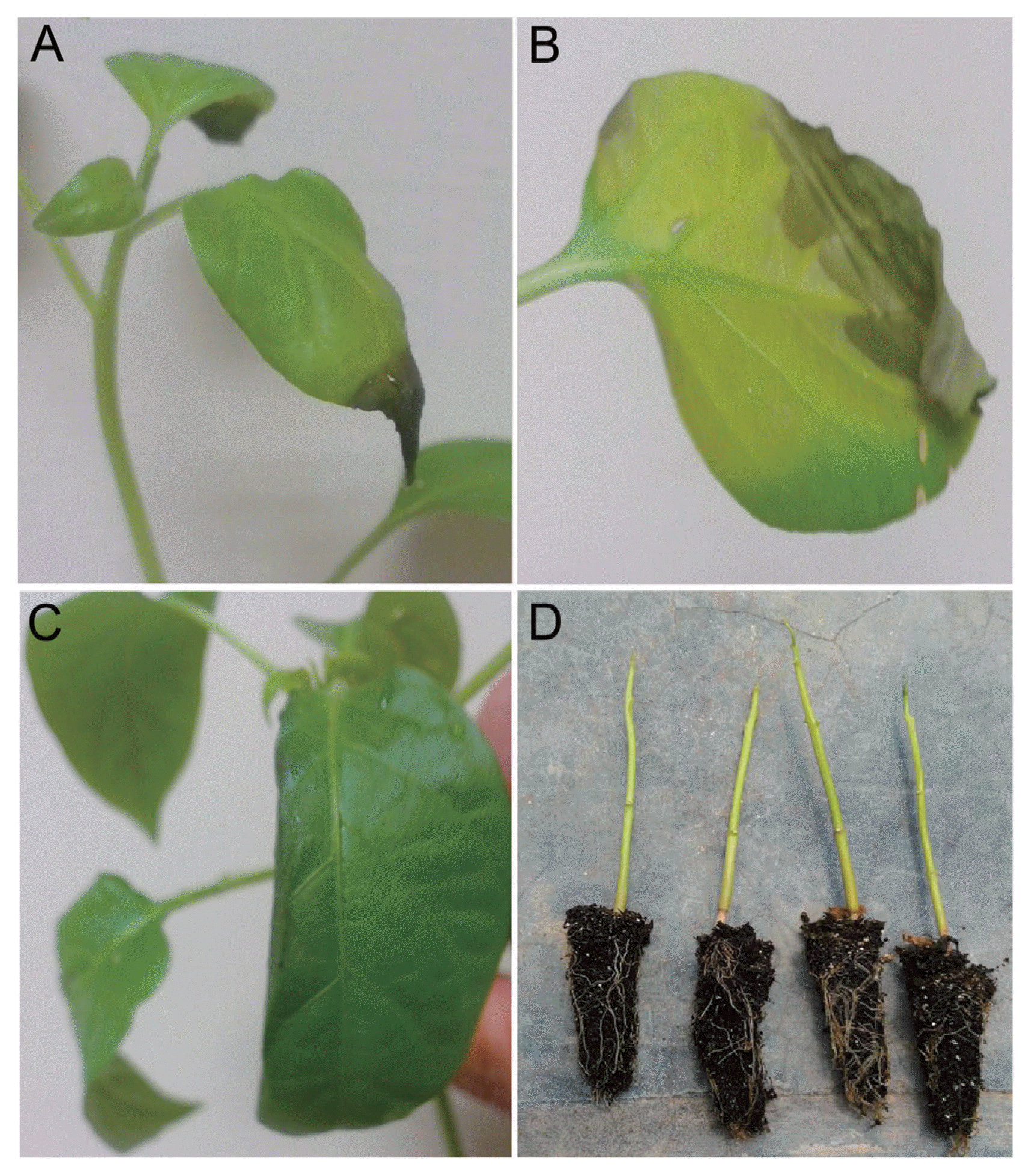
Pathogenicity test in healthy jalapeño pepper seedlings from 50 days-old. (A) Apical necrosis observed in jalapeño pepper, 23 days after inoculation by injection method. (B) Foliar water-soaked necrosis with chlorotic halo. (C) Reduction on the foliar lamina. (D) Defoliated seedlings from 90 days-old.
Physiological and biochemical characterization of bacteria isolated from pepper seedlings
The isolates formed creamy colonies on NA plates. Bacterial cells were rod-shaped. Biochemical characteristics of three selected strains from 2013, four strains from 2014 and four strains from 2015 were evaluated using the API 20E and API 50CH systems and are summarized in Table 2. Comparison with published profiles and the E. cloacae ATCC 13047 reference strain suggested that the pepper-pathogenic bacteria belong to the E. cloacae. Identical taxonomic assignments were obtained with API 20E and API 50CH systems. The same results were observed with the inoculated and re-isolated strains, suggesting that the strains were identical.
Carbon substrate utilization profiles were obtained using the Biolog system® (Biolog Inc.). Bacterial isolates from 2014 were tested and shown to be able to utilize 69 carbon sources (Table 3). A comparison with data from Bergey’s manual (Bergey and Holt, 1994) and MacFaddin (2003) confirmed that the strains in study belonging to the E. cloacae. Based on the calculated similarity index (0.624) our isolates were identified as E. cloacae subsp. dissolvens.
Molecular identification of bacteria isolated from pepper seedlings
To further support the assignment of the pepper-pathogenic bacteria to the E. cloacae, portions of the 16S rRNA genes were amplified by PCR and sequenced. The obtained 16S rRNA gene fragments were identical to portions of the 16S rRNA genes of several E. cloacae strains (e.g., GenBank accession numbers KT328458, KR061390, KF516245, CP006580).
To further refine the assignment of the pepper-pathogenic bacteria to distinct clades within the E. cloacae complex, portions of two housekeeping genes, hsp60 and rpoB, were PCR amplified and sequenced. A phylogenetic tree based on 264-bp fragments of the hsp60 gene showed that most isolates cluster with strains from the E. cloacae Cluster V (Hoffmann and Roggenkamp, 2003) (Fig. 3). Sequences of our isolates had 99% identity (1 mismatch) to sequences from E. cloacae strains P101 and EcWSU1 (accession numbers CP006580 and CP002886). A comparison with the hsp60 fragments used by others revealed that our sequences were 100% identical to E. cloacae strain EN_303 (accession number AJ543775; Fig. 3). The phylogenetic tree based on a 545-bp portion of the rpoB gene was concordant with the hsp60-derived tree, assigning the strains to the E. cloacae (Fig. 4). All strains with at least 99% identity to the rpoB gene belonged to clade 2 according to Miyoshi-Akiyama et al. (2013).
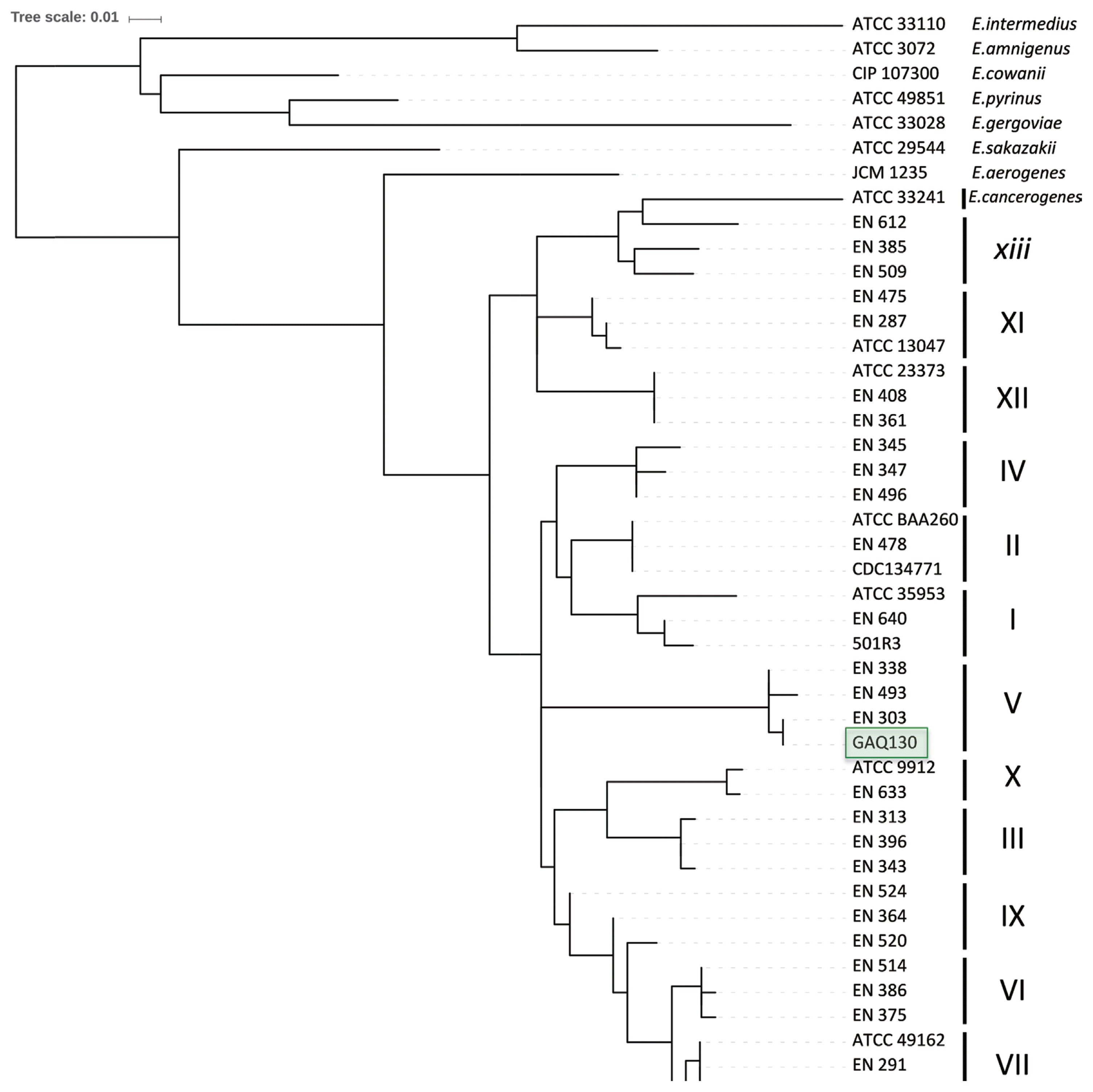
Neighbor-joining tree after analyzing 264 nucleotides of the hsp60 gene 429 of the 14 isolates and reference strains of the genus E. cloacae from Hoffmann and Roggenkamp (2003).
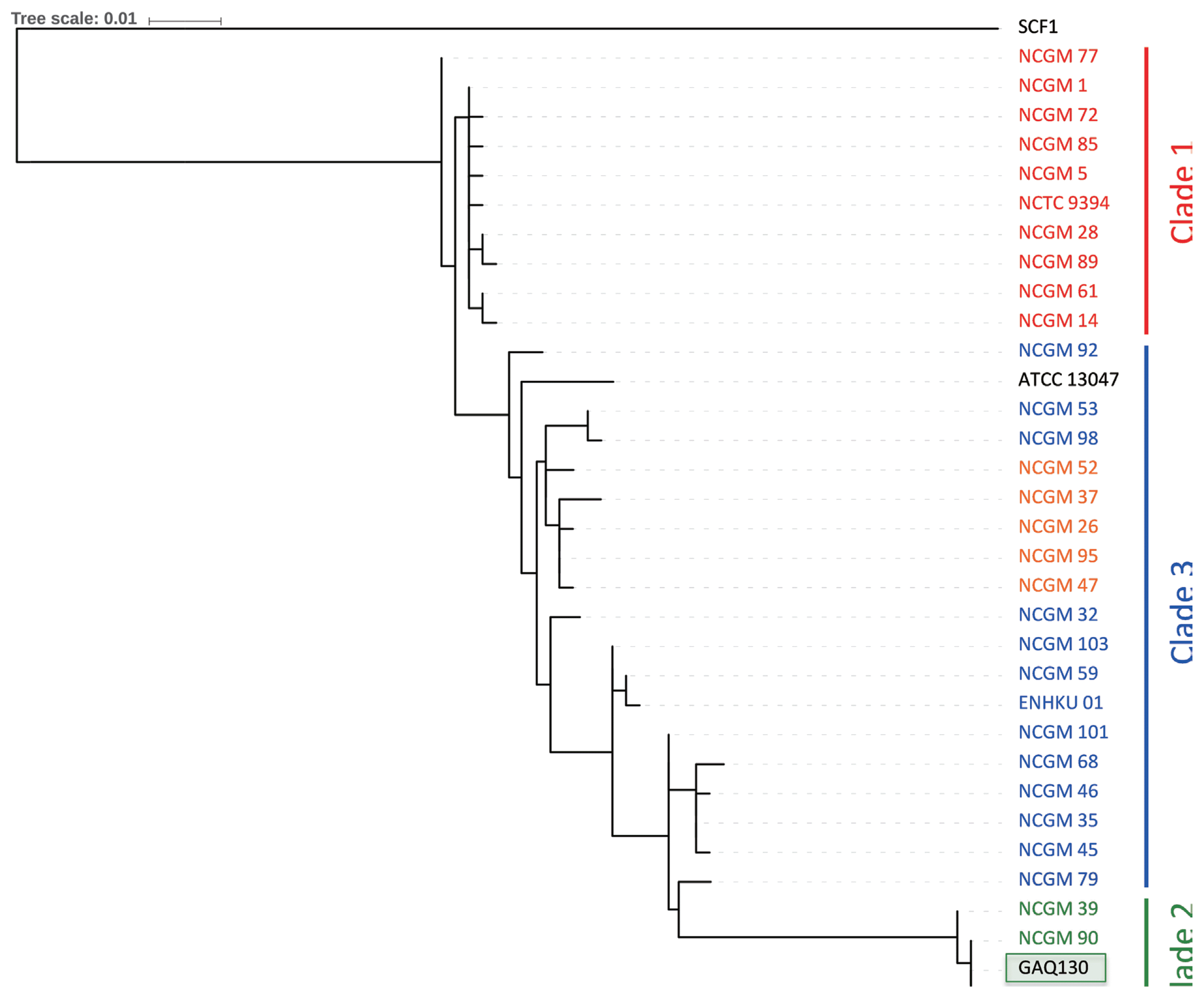
Neighbor-joining tree after analyzing 545 nucleotides of the rpoB gene and strains of the genus E. cloacae from Miyoshi-Akiyama et al (2013).
Discussion
In this study, we describe a new disease on chili pepper seedlings caused by bacteria of the E. cloacae. Biochemical, physiological and molecular analyses were performed for 14 bacterial isolates and re-isolated bacteria upon artificial infection, thus fulfilling Koch’s postulates. Compared to the Biolog system® (Biolog Inc.) with DNA-based methods, the latter showed to be more robust, reliable and resolutive for determination of the taxonomic position of the isolates in study. Yet, sequence information from the 16S rRNA gene alone was not conclusive. It did not allow discrimination among some of the Enterobacter species. However, the inclusion of sequence information from the hsp60 gene resulted in an accurate identification of the E. cloacae isolates, as reported by Hoffmann and Roggenkamp (2003) and Paauw et al (2008). Additionally, a partial rpoB sequence revealed genetic differences between the different isolates and allowed them to be compared with a large set of isolates, in agreement with a previous report that this gene provides good phylogenetic resolution (Case et al., 2007). Thus, sequences from rpoB and hsp60 allow to accurately assign the isolates as E. cloacae subsp. dissolvens.
Bacteria of the genus Enterobacter, including members of the E. cloacae complex are adapted to survive and proliferate in diverse environments (Sanders and Sanders, 1997). Even if the E. cloacae is recognized mainly as causing harmful diseases affecting humans (Mezzatesta et al., 2012), it has also an important role as a causal agent of several plant diseases. Indeed, since 1922, E. cloacae has been repeatedly reported to cause diseases on a surprisingly wide variety of plants, such diverse as maize (Rosen, 1922), elm tree (Carter, 1945; Murdoch and Campana, 1983), coconut (George et al., 1976), onion (Cother and Dowling, 1986; Bishop and Davis, 1990; Schroeder et al., 2010), papaya (Nishijima et al., 1987), orchid (Takahashi et al., 1997), macadamia (Nishijima et al., 2007), dragon fruit (Masyahit et al., 2009), and mulberry (Wang et al., 2010).
Bacteria of the genera Enterobacter may become an opportunistic pathogen when changes in host physiology occurs related with high temperature (Cother and Dowling, 1986; Sanders and Sanders, 1997). Some reports mention that E. cloacae can be present in symptomless kernels (Nishijima et al., 2007), thus the emergence of an infection can be latent until environmental conditions are favorable for the onset of the disease. For example, a previous study reported an association of diseased onion plants in the field with a period of extreme heat (40–45°C) (Bishop and Davis, 1990). This association has also been reported for mung bean sprouts (Wick et al., 1987) and for internal yellowing in papayas (Nishijima et al., 1987). In this study, we similarly observed that certain abiotic conditions provoked the production of symptoms. The disease caused by E. cloacae was severe at 38°C and 80% RH, whereas no symptoms were observed at 25°C and 30% of RH. In a preliminary field survey conducted in Mexican chili pepper fields to identify the E. cloacae symptoms on plants, no symptoms were observed on plants perhaps due to the relatively low humidity in this area of approximately 20 to 30%. It is still unknown how the chili pepper seedlings became infected with E. cloacae. More research is needed to understand the epidemiology of this new disease and to develop management strategies to reduce its expansion.
Acknowledgments
Research was supported by the Secretaría de Educación Pública (SEP) Mexico under the PRODEP grant UACH PTC 305. Work in RK’s laboratory was supported by IRD.
Notes
Conflict of Interest
The authors declare no conflict of interest.