Exploring the Potentiality of Novel Rhizospheric Bacterial Strains against the Rice Blast Fungus Magnaporthe oryzae
Article information
Abstract
Rice blast caused by Magnaporthe oryzae is a major disease. In the present study, we aimed to identify and evaluate the novel bacterial isolates from rice rhizosphere for biocontrol of M. oryzae pathogen. Sixty bacterial strains from the rice plant’s rhizosphere were tested for their biocontrol activity against M. oryzae under in vitro and in vivo. Among them, B. amyloliquefaciens had significant high activity against the pathogen. The least disease severity and highest germination were recorded in seeds treated with B. amyloliquefaciens UASBR9 (0.96 and 98.00%) compared to untreated control (3.43 and 95.00%, respectively) under in vivo condition. These isolates had high activity of enzymes in relation to growth promoting activity upon challenge inoculation of the pathogen. The potential strains were identified based on 16S rRNA gene sequencing and dominance of these particular genes were associated in Bacillus strains. These strains were also confirmed for the presence of antimicrobial peptide biosynthetic genes viz., srfAA (surfactin), fenD (fengycin), spaS (subtilin), and ituC (iturin) related to secondary metabolite production (e.g., AMPs). Overall, the results suggested that application of potential bacterial strains like B. amyloliquefaciens UASBR9 not only helps in control of the biological suppression of one of the most devastating rice pathogens, M. grisea but also increases plant growth along with a reduction in application of toxic chemical pesticides.
Rice (Oryza sativa) as a cereal grain, is the most widely consumed staple food for more than 50% of the world population, especially in Asia. In India, rice is grown over an area of 43.97 million hectares with an annual production of 104.32 million tonnes and productivity of 2177 kg/h (Datanet India, 2016). In rice, 30% of yields are lost due to blast disease caused by fungal pathogen Magnaporthe oryzae is the most significant disease by affecting rice cultivation and crop loss worldwide. Capitalizing on the abilities of naturally occurring rice rhizospheric bacteria to reduce M. oryzae infections could provide a sustainable solution to reduce the amount of crops lost (Prasanna Kumar et al., 2017; Wang et al., 2016). Cultivation of resistant cultivars and using fungicides are the main methods to control the disease (Ghazanfar et al., 2009; IRRI, 2010; Miah et al., 2017). However, research in past decades had establish another method for the disease management in exploiting the biocontrol agent against the diseases. Use of biocontrol organism is an eco-friendly and cost-effective strategy, could be supplemented with chemical fungicide to achieve a greater level of protection, sustain yields and value of the crop. Various group of bacteria are known to exist in the rhizospheric region of rice are useful biocontrol candidate due to their endospore formation, tolerant to heat and desiccation which are good characteristics necessary for long-term storage and field level application. Bacillus spp. including B. cereus, B. amyloliquefaciens, B. pumilus, B. subtilis, and B. sphaericus have been reported to significantly reduce the incidence of disease on different hosts. Antimicrobial peptides (AMPs) are interesting compounds produced by bacterial strains which are involved in the biocontrol of several plant pathogens causing seed and aerial-borne diseases and also helps in the promotion of plant growth (Kim et al., 2010; Souza et al., 2015). AMP biosynthetic genes include surfactin, iturin, fengycin and bacillomycin compounds are characterized by its wide antimicrobial and relevance surfactant activities. Specifically, the production of a combination of AMP genes by Bacillus amyloliquefaciens has been related to control M. oryzae in rice (Prasanna Kumar et al., 2017). The production of bacilysin, and iturin in B. subtilis ME488 to the suppression of Fusarium wilt of cucumber (Romero et al., 2007) and Phytophthora blight of pepper (Chung et al., 2008). The dominance of AMP genes helps in the management of diseases in natural environments (Breen et al., 2015; Mora et al., 2011). Rhizobacteria are directly involved in improving plant growth, synthesis of phytohormones, solubilization of minerals such as phosphorus, phosphate, and production of siderophores and iron chelates. Since biological control is a key component of integrated disease management, it is important to explore the novel bacterial strains to control rice blast and also to evaluate these antagonists for their application in field conditions (Amruta et al., 2016; Compant et al., 2010; Pathma et al., 2011).
Recent research is focused on the isolation of potential bacterial strains from the rice rhizospheres and then they could be applied further as inoculants (Babu, 2011; Raaijmakers and Mazzola, 2012). This strategy may help to adapt non-native bacterial species in a new environment (Cordero et al., 2012; Fischer et al., 2010; Mavrodi et al., 1998; Naureen et al., 2009). It is useful screening tools based on their in vitro and in vivo methods that allow selecting potential isolates. In this study, we aimed to isolate and characterize the potential bacterial strains from rice ecological niche for both bioantagonism and plant growth promotion activity in managing the rice blast disease caused by Magnaporthe oryzae.
Materials and Methods
Collection of rhizosphere soil samples and isolation
About 110 bacterial samples of the rhizosphere soil and roots were collected from two different commercial rice fields growing cultivars Jaya, BR-2566 and KRH-4 (1) ZARS, Mandya (latitude 12.5200° N and longitude 76.9000° E) and (2) GKVK, Karnataka (latitude 12.9667° N and latitude 77.5667° E), India during the year 2014, Kharif season. The plant roots with surrounding soil were shaken gently to separate soil and the remaining about 1cm thick soils tightly attached to the root system of plants was considered rhizosphere-enriched soil was collected from 40 days old rice plants showing five fully opened leaves and tillering under submerged conditions. The samples were brought to the laboratory, dried and further used for isolation. Ten grams of the rhizosphere soil was added to 90 ml of sterile saline (0.85%) and was diluted upto 10−7 following serial dilution method (Bharathi et al., 2004; Liu et al., 2008). Samples were kept at 4°C until further processing. About 0.1 ml of each dilution was spread on Luria Bertani (LB) agar and were incubated at 28 ± 2°C until colony development.
Purification and maintenance of bacterial culture
The suspected bacterial colonies were picked up with the help of sterilized inoculation loop and streaked onto the surface of nutrient agar by quadrant streak plate method to get well separate colonies. The isolated bacteria were was stored in the refrigerator at 4ºC on NA slants for routine use. The cultures were also stored as glycerol stocks stored in stocks (−80°C).
From 110 isolates, the most dominant bacterial colonies of sixty different strains were selected and each strain was designated as UASBR1, UASBR2, UASBR3 up to UASBR60 which indicates “University of Agricultural Sciences Rhizosphere”. The strains were initially studied for the colony and cell morphology using light microscopy and Gram staining (Vincent, 1970).
Anti-pathogenic activity against rice blast fungus, Magnaporthe oryzae
Identified and proven pathogen rice blast fungus (MG01) was obtained from Prof. Malali Gowda, cultures deposited at C-CAMP, NGG, NCBS, Bengaluru, Karnataka, India The obtained culture, were maintained on Potato Dextrose Agar (PDA) medium. The bacterial isolate was inoculated at the periphery and M. oryzae (5 mm disc) at the center of Petri plates containing LB (Luria Bertani) agar media and incubated for 7 days at 28ºC. A control plate of Magnaporthe oryzae was maintained throughout the study as a reference.
In vivo assay of bacterial strains against the rice blast fungus (M. oryzae)
Twenty-eight effective bacterial strains had high inhibitory effect under in vitro were selected to evaluate growth promotion and blast disease suppression under controlled greenhouse conditions. An experiment was conducted for mass screening of isolates with cultivar HR-12 (The seeds of the rice cv. HR-12, susceptible to blast obtained from ZARS, Mandya, Karnataka state, India was used throughout the study) were sown in pots (5 seeds/pot) containing sterilized soil (5 kg/pot) placed in the controlled greenhouse at a temperature of 28 ± 2ºC during the day and 25 ± 2ºC at night with relative humidity above 90%. The bacterial isolates grown in nutrient broth for 48 h at 28 ± 2ºC was centrifuged (6000 × g for 5 min), washed and resuspended in sterile water (suspension of 109 log10 cfu/ml) were individually treated with seeds before sowing. Each treatment had four replicates. Untreated plants were considered as control. The emerged seedlings were transferred to controlled greenhouse a day before the inoculation for acclimatization. The rice blast fungus M. oryzae spores were harvested in 5 ml sterile water containing 0.5% gelatin, filtered through 0.2 μm nylon mesh and immediately transferred to an Ice-cold thermostat container to prevent germination. The spore suspension (1 × 105 spores/ml) was sprayed on 20 days old (20DAS) rice plants (100 ml/plant). Immediately after inoculation, the plants were covered with a black polythene hood for 24 h to stimulate infection. Disease assessment was done after six days of inoculation and each plant was assessed for infection (Prasanna Kumar et al., 2017).
Leaf blast
Visual scoring was recorded on a percentage of Disease Severity by considering the three replications per each treatment, on the 14th and 21st days after sowing. Leaf blast scoring was followed as per Standard Evaluation System (SES) for Rice, IRRI, Manila, Philippines.
Where:
(n) = Number of plants in each category
(v) = Numerical values of symptoms category
(N) = Total number of plants
(9) = Maximum numerical value of symptom category
Growth promotion and enzymatic characterization of bacterial strains
The seed germination test was conducted in the laboratory environment using blotter paper method as per ISTA (2010), Zürichstrasse, Switzerland. One hundred treated seeds of four replications were placed on moist germination paper; the rolled towels were incubated in a germination chamber maintained at 25 ± 10ºC and 90% RH. The germinated seedlings were evaluated on the 14th day and the germination percent was expressed based on normal seedlings.
Mean seedling length (cm) and seedling dry weight (mg)
Ten normal seedlings were selected from each treatment. The seedling length was measured from point of attachment of seed to the growing meristematic tip and expressed in cm.
Ten seedlings from each treatment and replication were used for measuring the seedling dry weight and were kept in the hot air oven at 85 ± 1ºC for 24 h. The dry weight (mg) was measured and expressed as dry weight of 10 seedling (mg).
Enzymatic characterization of isolates
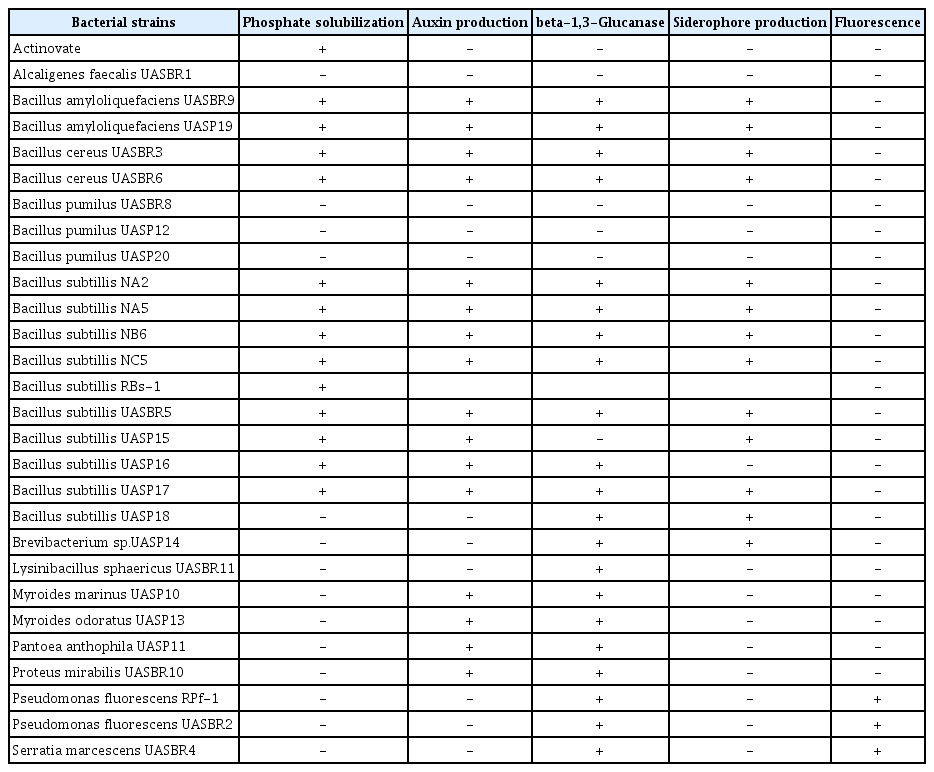
The 28 most promising isolates of rhizobacteria identified in previous bioassays (Section 2.4) were selected for the following assays using a completely randomized design with four replications. Phosphate solubilization (PS). Bacterial isolates were grown in Petri plates containing GY medium (glucose-yeast extract) and phosphorus was supplement to the medium according to Sylvester-Bradley et al (1982). The plates were incubated at 28ºC for 3 days and evaluated by identifying a translucent milky white growth around the bacterial colony and production of Indoleacetic Acid (IAA). The plates were evaluated by identifying a red halo formed on the GY medium (IAA positive). For ferric siderophore production (Sid). The isolates of rhizobacteria were evaluated by its ability to produce siderophore and to convert ferric ions (Fe III) to soluble forms (Fe II) under chelation. The bacterial isolates were transferred to Petri plates containing agar and chrome azurol S and were incubated at 28ºC for 48 h according to Schwyn and Neilands (1987). Colonies exhibiting a pink halo after incubation period were considered as positive. In vitro production of beta-1,3 glucanase by the rhizospheric bacteria was also evaluated. The glucanase activity was identified by adding beta-1,3-glucan in a semi-solid nitrogen-free medium according to Renwick et al (1991). The plates were incubated for 3 days at 28ºC and identifying as positive for the formation of an orange halo.
DNA isolation and PCR amplification
The bacteria were grown in Luria Bertani (LB) broth for 24 h and centrifuged to obtain a pellet. The pellet was mixed with 750 μl of suspension buffer (Amnion Biotech Pvt. Ltd., Bengaluru, India), and the addition of 5 μl of RNaseA (mg/ml) (Invitrogen) and incubated at 65°C for 15 min. After incubation, 1 ml of lysis buffer was added to the same tube and incubated at 65°C for 15 min. The tubes were centrifuged at 13000 RPM for 2 min to pellet the debris. The supernatant was transferred to fresh tubes and added 900 μl of isopropanol each tube for precipitating DNA. The DNA pellet was centrifugation at 13000 rpm for 15 min. The supernatant was discarded and the pellet was washed twice with 70% ethanol. The DNA was diluted with TE buffer. The PCR master mix was prepared using the reaction mixture consisting of (Thermo Scientific, MA, USA), 1× dNTPs (0.2 mM), primers (2 ng), template DNA (10 ng) and 1.2U of Taq polymerase (Fermentas). The final volume was made up to 50 μl with autoclaved water. The bacterial ITS 16-S rRNA was amplified using forward 16SBACF (5′ GAGTTTGATCCTGGCTCAG 3′) and 16SBACR (5′ CGGGTCCATCTGTAAGTGGT 3′) primers.
Sequencing and data analysis
Nucleotide sequencing of the PCR fragments was performed (Amnion Biotech Pvt. Ltd., Bengaluru, India) to sequences corresponding to 16s rRNA gene of the strains were reverse complemented using software Bio edit and aligned using the clustlX software. The identification was based on the best BLASTN match.
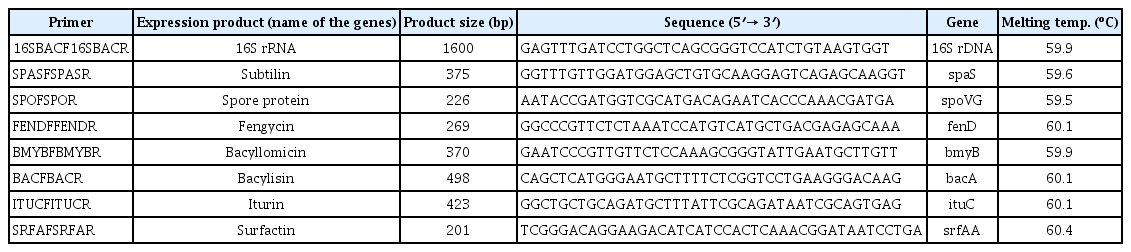
Screening of bacterial strains by PCR amplification and Antimicrobial Peptide (AMP) genes
Primers reported by Mora et al. (2011) were used to detect the genes bmyB, ituC, srfAA bacA, and spaS. The PCR amplification was carried out for all the AMPs gene with respective bacteria. The presence of AMP biosynthetic genes in Bacillus and non-bacillus isolates were determined (Table 1).
Statistical analysis
The results were represented as the mean ± SD. Statistical significance was analyzed with one-way analysis of variance followed by a Tukey’s HSD-post hoc test. Differences with a P value less than 0.05 were considered statistically significant. Duncan’s multiple range tests were also applied to separate the means.
Results
In vitro dual culture assay
Results revealed that the all 60 rhizospheric bacterial strains significantly inhibited the mycelial growth of M. oryzae (Fig. 1, 2). Out of sixty bacterial strains screened eleven were found effective against the M. oryzae. Among these bacterial strains B. amyloliquefaciens UASBR9 and B. cereus UASBR3 were highly inhibitory (84.14 and 76.83%) followed by B. pumilus UASBR8 (74.99%), B. cereus UASBR6 (74.44%), Proteus mirabilis UASBR10 (63.37%), Alcaligenes faecalis UASBR7 (62.73%), Alcaligenes spp. UASBR1 (51.18%). These bacterial strains were superior compared to reference positive controls RBs-1 (72.00%) and RPf-1 (55.24%).
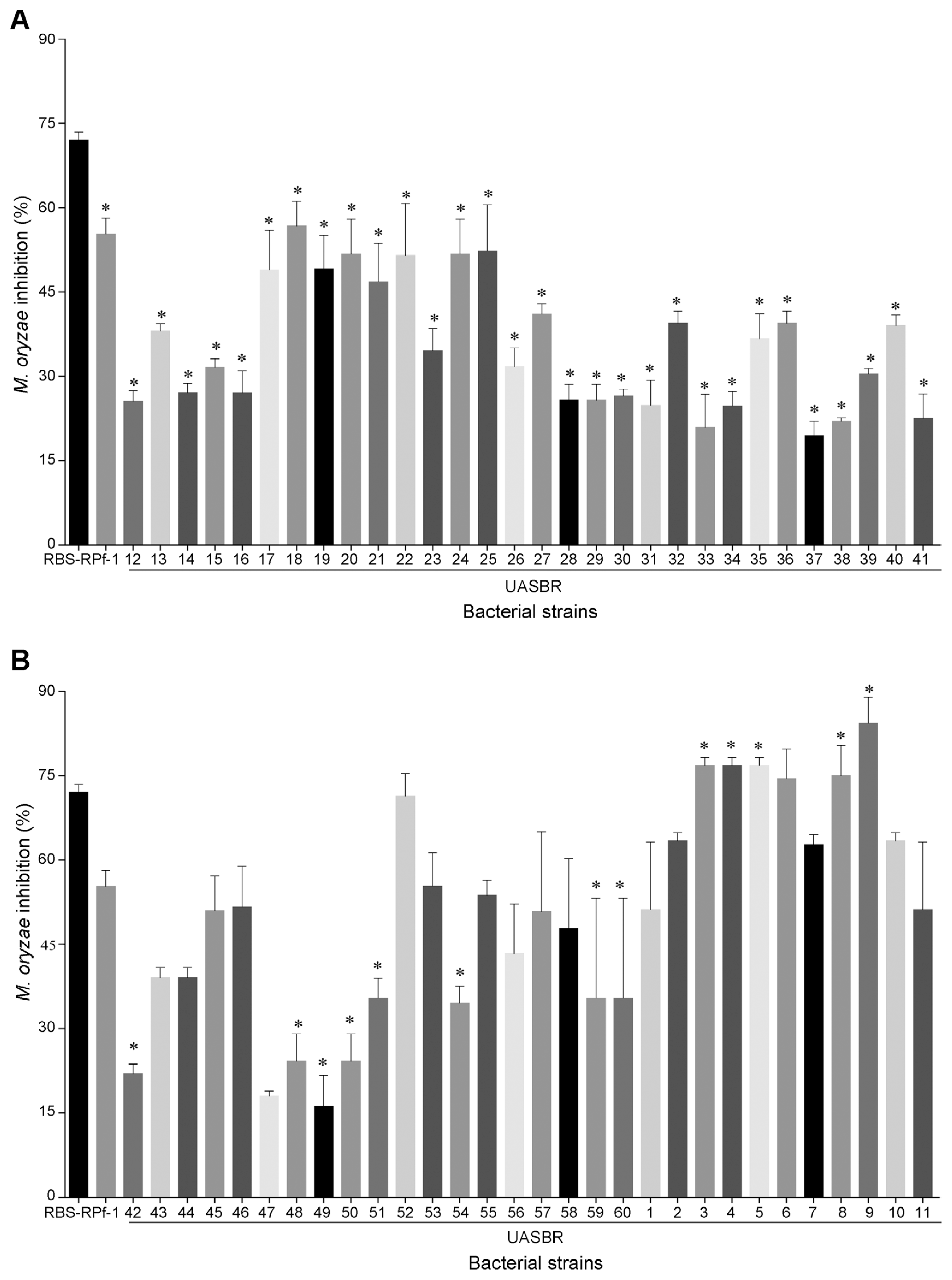
Effect of bacterial strains against Magnaporthe oryzae under in vitro evaluation. asterisk indicates a statistically significant difference of 95% (P < 0.05) when comparing with the positive control (Rpf-1). Bars indicate standard errors.

In vitro evaluation of rice rhizospheric bacterial isolates against M. oryzae. Note: UASBR1: Alcaligenes sp.; UASBR2: Pseudomonas fluorescens; UASBR3: Bacillus cereus; UASBR4: Serratia marcescens; UASBR5: Bacillus subtillis; UASBR6: Bacillus cereus; UASBR7: Alcaligenes faecalis; UASBR8: Bacillus pumilus; UASBR9: Bacillus amyloliquefaciens; UASBR10: Proteus mirabilis; UASBR11: Lysinibacillus sphaericus; RPf1: Bacillus subtilis; RBs1: Pseudomonas fluorescens and pathogen alone indicates control against M. oryzae, where pathogen culture in the centre of the plate and bioagent around the plate (perimeter).
Effect of bacterial isolates against rice blast severity under in vivo
In seed treated with all isolates, the disease severity was significantly reduced compared by the control (P < 0.05). Significant differences were observed in rice blast severity in case of seeds treated with selected bacterial strains (Table 2). Least disease severity was recorded in seed treated with B. amyloliquefaciens UASBR9 (0.96%) followed by NA5 (0.97%), while untreated control showed highest disease severity of 3.43%.
Plant growth promotion activity as influenced by seed treated with bacterial strains
Seed vigour enhancement treatments affected significantly (P < 0.05) the germination vigour of rice. Significant differences were observed on germination percent by seed treated with selected bacterial strains. The highest germination was observed in B. cereus UASBR6 (98%) which is on par with B. subtillis UASBR5, and Serratia marcescens UASBR4. However, delayed and scattered germination was observed in control (96.00%) (Table 2). Mean seedling length differed significantly due to seed treated with selected bacterial strains, the highest was observed in P. fluorescens UASBR2 (33.15 cm), followed by NA2 (17.64 cm) and compared to untreated control (28.05 cm) (Table 2).
Statistically highest seedling dry weight was observed in B. amyloliquefaciens UASBR9 (485 cm) followed by B. subtillis UASP15 (315.6 cm), however, comparatively lower seedling dry weight was observed in control (237 cm) (Table 2).
Enzymatic characterization of rhizobacterial isolates
Out of 28 isolates, 14 were positive for PS, 16 for auxin production and three for fluorescence, 20 for beta-glucanase and 13 for Sid (hydroxyquinoline and CAS) (Table 3). No single isolate was positive for all of the metabolites assessed.
Identification of bacterial isolates by partial 16S rRNA sequencing
Molecular characterization results revealed that amplification and sequencing of 16s rRNA (1300bp PCR product). All 11 bacterial strains and two reference strains (positive controls) were amplified and product of the expected 1300 bp was obtained (Fig. 3).
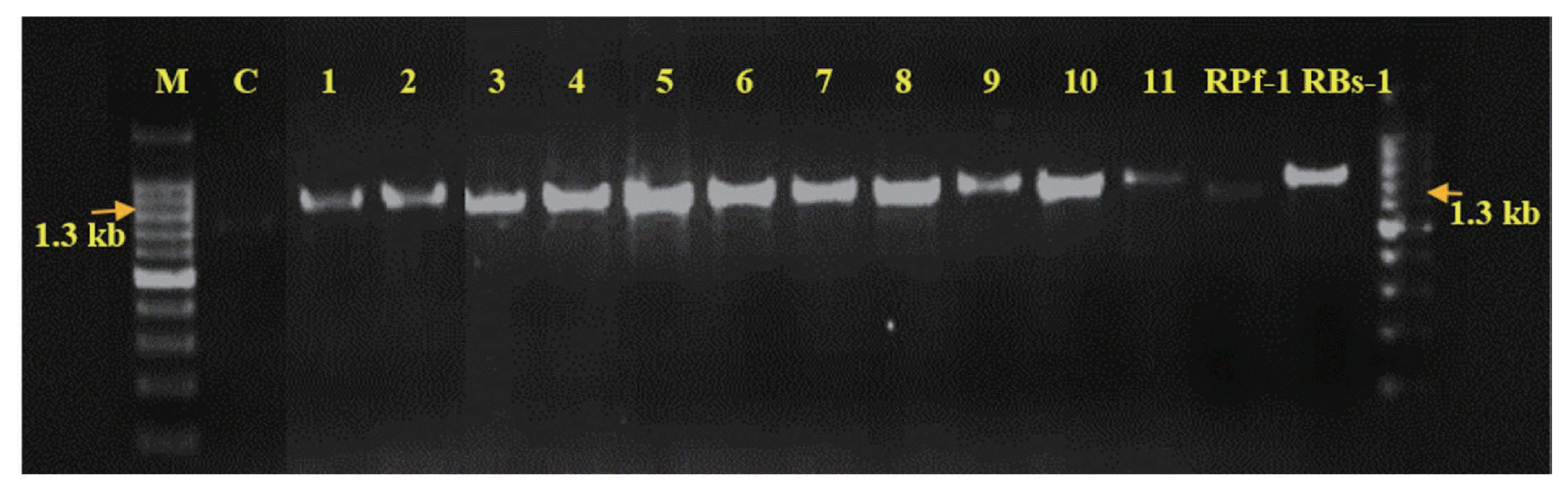
PCR Amplification of 16SrRNA gene of 13 bacterial isolates Note: M: Marker-1300 bp, 1: Alcaligenes sp.; 2: Pseudomonas fluorescens; 3: Bacillus cereus; 4: Serratia marcescens; 5: Bacillus subtillis; 6: Bacillus cereus; 7: Alcaligenes faecalis; 8: Bacillus pumilus; 9: Bacillus amyloliquefaciens; 10: Proteus mirabilis; 11: Lysinibacillus sphaericus; 12: Bacillus subtilis; 13: Pseudomonas fluorescens.
Sequence analysis
The sequences obtained from Sanger sequencing of 16S rRNA region identified the bacteria by comparing with those in public databanks on the basis of similarities with existing sequences, we were able to identify most strains to the species level, as shown in Table 4. The sequences were submitted to NCBI data Gen Bank for the accession numbers. The phylogenetic tree was constructed for bacterial strains to know the homology-based on results obtained from NCBI database along with the accession numbers (Fig. 4).

List of bacteria isolated from rhizosphere rice and identified by based on PCR amplification and 16S rRNA gene sequencing. Species or taxonomic group from the National Center for Biotechnology Information GenBank database showing a high degree of sequence similarity with the strains isolated in this study. UASBR indicates that these bacterial strains were isolated from the rhizosphere rice

Rooted neighbor-joining tree constructed by using partial 16S rRNA gene sequencing attributed to species of the (A) Bacillus group and (B) other than Bacillus group Phylogenetic analyses were conducted in MEGA6 (Tamura et al., 2013). The evolutionary history was inferred using the Neighbor-Joining method. Bootstrap values (expressed as percentages of 500 replications) of > 50% are shown at branch points. Accession numbers are given in parentheses. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. The scale bar indicates 0.5% nucleotide substitutions.
Screening of bacterial strains for antimicrobial peptides genes AMPs
AMPs gene markers results revealed that amplifications of the AMP genes (bmyB, ituC, srfAA, bacA, and spaS) in all five Bacillus strains whereas, genes like bmyB and bacA was present in two non-Bacillus strains (P. fluorescens UASBR2 and Serratia marcescens UASBR4). Nonspecific amplifications of the AMPs genes were not observed in the strains of the other Bacillus species. Specific amplification of AMP genes was observed in all bacterial species but not in P. fluorescens (UASBR2) which could amplify only spoVG, bmyB and bacA.
The strains B. cereus UASBR3, B. subtillis UASBR5, B. cereus UASBR6, B. pumilus UASBR8, and B. amyloliquefaciens UASBR9 were known to possess all the six genes representing the antagonist characteristics of the bioagent screened (Fig. 5). Other than Bacillus spp. biosynthetic genes bmyB was noticed in strains P. fluorescens UASBR2, Serratia marcescens UASBR4, Alcaligenes faecalis UASBR7, and Proteus mirabilis UASBR10. Gene ituC was found in strains UASBR2 and UASBR11. srfAA was reported in all strains except strain Lysinibacillus sphaericus UASBR11, whereas, bacA and spaS were noticed in all the strains except Alcaligenes faecalis UASBR1, Alcaligenes faecalis UASBR7, and Serratia marcescens UASBR4.

Representative electrophorograms of antimicrobial peptide (AMP) biosynthetic genes amplification products in different bacterial isolates Note: M: Marker-, 1: Alcaligenes sp.; 2: Pseudomonas fluorescens; 3: Bacillus cereus; 4: Serratia marcescens; 5: Bacillus subtillis; 6: Bacillus cereus; 7: Alcaligenes faecalis; 8: Bacillus pumilus; 9: Bacillus amyloliquefaciens; 10: Proteus mirabilis; 11: Lysinibacillus sphaericus; 12: Bacillus subtilis; 13: Pseudomonas fluorescens.
Discussion
A novel bacteria, including members of the genera Bacillus, Pseudomonas, and Enterobacter etc. are known to colonize the rhizosphere of most of the cereal crops, including rice, maize and wheat further these could be used as both plant growth promoters and biocontrol agents (Naureen et al., 2009). The scope of this investigation was to isolate, characterize and select potential bacterial strains from the rice rhizosphere beneficial in terms of biological suppression of one of the most devasting phytopathogenic fungi, M. grisea (Betelho et al., 1998; Naureen et al., 2005; Suprapta, 2012). The most active and dominant sixty strains were screened under in vitro condition against rice blast fungus M. oryzae from which the best inhibitory strains were selected for further characterization. Rhizosphere-associated bacteria could be a source of antagonistic bacteria (Cazorla et al., 2007; Ghazanfar et al., 2009; Miah et al., 2017). An attempt have been made to use the bacterial antagonists in the management of rice blast (Prasanna Kumar et al., 2017; Vidhyasekaran et al., 2001). Several Bacillus spp. including B. amyloliquefaciens, B. pumilus, B. subtilis, B. cereus, B. mycoides and B. sphaericus have been reported to significantly reduce the incidence of disease on a different host plants. The mechanism of inhibition of bacterial strains against M. oryzae under in vitro condition significantly inhibited mycelial growth over a inhibition period of 28 days. There were physical contact observed between any of the bacterial strains tested against M. oryzae; moreover, halo zone of inhibition observed in the case of few bacterial strains, suggesting the presence of fungistatic metabolites secreted by the rhizospheric bacteria (Fig. 2). The B. amyloliquefaciens UASBR9 strain, produced spores around the fungus causing complete fungal lysis and these appeared to be a strong association between siderophore productions. Which could be attributed due to the production of a variety of antibiotics like kanamycin and lipopeptides such as iturin, surfactin and fengycin. In the previous study Joshi and Gardener (2006) have reported that the antibiotic bacillycin and surfactin can inhibit the growth of pathogenic fungi whereas, in our study fengycin produced by B. amyloliquefaciens had inhibitory activity against fungus M. oryzae. Pathogenic fungi do not have the ability to produce the siderophore as beneficial bacteria. due to lack of iron element, the mycelial growth of pathogenic fungi was reduced. Significant differences were observed on rice blast severity in seed treated with selected bacterial strains under in vivo condition. The antagonistic effect of these Bacillus active strains against M. oryzae were more potential when compared to the untreated control. This could be due to the effect of Bacillus strains on plant pathogens by specifically targeting sugar linkages and release of extracellular enzymes such as cell wall degrading enzymes, proteases, lipases and siderophore production. Based on this mechanism, the active bacterial strains were selected to assess its performance for the management of rice disease both under laboratory and glasshouse conditions. Similarly, a large number of rhizospheric bacteria are known possess antagonistic activities which has a novel mechanism of blast reduction, including particular Bacillus strain, which catabolizes collagen and gelatin. Some of these strains were also able to reduce blast symptoms by disrupting the adhesion for the spore tip mucilage and extracellular matrix from the leaf surface, thus preventing the proper attachment by M. oryzae they were seed treated. The results obtained in the study are in concordance with the findings of Wang et al. (2009), Carla Spence et al. (2014), Shimoi et al. (2010) where they found that the Pseudomonas chlororaphis strain EA105, demonstrated the ability to resist the growth of M. oryzae, and therefore, characterize has a potential biocontrol agent against rice blast disease.
Seed treatment with bacterial strains improved the seed germination, seedling vigour, emergence and seedling stand when compared to the control, as bacterial strains triggered the production of phytohormones like auxins, gibberellins, and cytokinins. The plant growth promoting activities of B. amyloliquefaciens UASBR8 and P. fluorescens UASBR2 reported that the bacteria’s are free-living plant growth promoting. Similar improvement of seed vigour parameters has also been reported in cucumber by B. amyloliquefaciens IN937a and B. subtilis GBO3 which ultimately resulted in a significant plant growth promotion and reductions in disease severity (Raupach and Kloepper, 2000; Raju et al., 1999 in as sorghum; Niranjan Raj et al., 2004 in pearl millet). These results are in compliance with findings of Wu et al. (2005) in maize, reported minimum portion (2–5%) of rhizobacteria occupied by Plant Growth Promoting Rhizobacteria (PGPR), where they actively colonize around plant roots and thus increase plant growth and yield. The studies conducted on bacterial strains under in vitro condition demonstrated that only 22% of the tested isolates were positive for IAA production and 44% for Thirupal Reddy et al. (2010) and Amruta et al (2016). The phytostimulation can be related to the production of IAA, gibberellic acid compounds, and alterations in the signaling pathway.
Molecular characterization results revealed the identification of bacteria by amplification and sequencing of 16s rRNA (1300 bp PCR product) that the studied identified bacteria has been reported as the most dominant and active bacteria belonging to the genera Bacillus, Pseudomonas, Serratia, Alcaligenes and Proteus spp. respectively (Amruta et al., 2016; Prasanna Kumar et al., 2017). The Antimicrobial compounds produced were potential candidates for the biocontrol agents and these were dependent on the specific genes. The specificity of the genes were assessed by PCR amplification with specific primers (Mora et al., 2011). In addition to that lipopeptide and other compounds were active in the biocontrol of plant pathogens, such as bacilysin a dipeptide as described in B. amyloliquefaciens FZB42 and subtilin, a lantibiotic in B. subtilis (Koumoutsi et al., 2004). The AMPs genes identified in the isolated strains were Bacillomycin with a product size of 370 bp, Surfactin (201 bp), Iturin A (423 bp) and subtilin (375 bp) respectively (Chen et al., 2009; Mora et al., 2011). AMPs are considered as cyclic lipopeptides such as fengycin, iturin, bacillomycin, and surfactin. These compounds have been implicated in the biocontrol of several plant diseases. The presence of bacterial communities in Rhizosphere resulted in increased anti-fungal properties. This observation forms an optimistic basis for alternative approaches to stimulate efficient natural biocontrol.
Acknowledgments
The authors are grateful to the Department of Plant Pathology, University of Agricultural Sciences, Bengaluru and Vishweswaryya canal farm Mandya, India for providing research facilities.