Development qRT-PCR Protocol to Predict Strawberry Fusarium Wilt Occurrence
Article information
Abstract
Strawberry Fusarium wilt disease, caused by Fusarium oxysporum f. sp. fragariae, is the most devastating disease in strawberry production. The pathogen produces chlamydospores which tolerate against harsh environment, fungicide and survive for decades in soil. Development of detection and quantification techniques are regarded significantly in many soilborne pathogens to prevent damage from diseases. In this study, we improved specific-quantitative primers for F. oxysporum f. sp. fragariae to reveal correlation between the pathogen density and the disease severity. Standard curve r2 value of the specific-quantitative primers for qRT-PCR and meting curve were over 0.99 and 80.5°C, respectively. Over pathogen 105 cfu/g of soil was required to cause the disease in both lab and field conditions. With the minimum density to develop the wilt disease, the pathogen affected near 60% in nursery plantation. A biological control microbe agent and soil solarization reduced the pathogen population 2-fold and 1.5-fold in soil, respectively. The developed F. oxysporum f. sp. fragariae specific qRT-PCR protocol may contribute to evaluating soil healthiness and appropriate decision making to control the disease.
Introduction
Strawberry (Fragaria × ananassa) is one of the world’s most commercially important fruit crops. Many countries are cultivating strawberry at large scale and Korea has 6th scale cultivation areas worldwide and exported amount of strawberry about 2,152 tons annually. Diseases occurrence are one of the major limiting factor that severely impact the crop agronomic performance and lead to economic losses. Moreover, most strawberry cultivars are highly susceptible to several destructive and economically important pathogens such as Fusarium oxysporum f. sp. fragariae, Phytophthora fragariae, Colletotrichum acutatum, Verticillium dahliae, Botrytis cinerea, and Xanthomonas fragariae (Maas, 1998). Fusarium wilt of strawberry, named as Fusarium yellows leave caused by the vascular fungus, Fusarium oxysporum Schlechtend.: Fr. f. sp. fragariae Winks and Williams (1965) was first discovered and described in southeastern Queensland, Australia. The disease and the pathogen are also known in Japan and Korea (Nagaraian et al., 2004). Chlamydospores one of spore formation by the pathogen has very thick cell wall that could be survive in unfavorable environment and prevent to fungicide penetration (Tezuka and Makino, 1991; Winks and Williams, 1965). F. oxysporum f. sp. fragariae (FOF) is pathogenic only to strawberry, but not appeared in tomato, watermelon, cabbage, and passion fruit which are well-known hosts for same genus pathogen (Kodama, 1974). As Fusarium wilt in other crops, symptom development of in strawberry prefer to high temperatures that lead to wilt infected leaves and die rapidly. In addition, slimy white or pink masses of micro and macro conidia may produce on the infected plant tissues.
Typically, strawberry Fusarium wilt disease has been controlled by cultivation methods, such as covering fields with black vinyl or silver polyethylene films (Ashworth and Gaona, 1982; Kodama and Fukuit, 1982). Soil fumigation with chloropicrin, dichloropropenes and dichloropropane are known to reduce the pathogen density in soil. Soil solarization is one of process for soil sterilization that use transparent plastic film for rising temperature, which is harmful to soil-borne pathogens in soil via solar radiation and high temperature. Many weeds, pathogens such as fungi, bacteria and nematodes were extinct by high soil temperatures (Katan, 1981; Pinkerton et al., 2000; Raio et al., 1997). Furthermore, several alternative methods against strawberry Fusarium wilt have been developed recently. For example, sodium dichloroisocyanurate (NaDCC) and biological control agent, which effectively suppressed the pathogen and the disease occurrence (Cha et al., 2016; Kim et al., 2015).
Rapid, accurate diagnosis of disease and identification of causal pathogen are important process for effectively control of the diseases (Sankaran et al., 2010). Propriety and specific diagnosis of the pathogen are essential elements to manage the problems at the appropriate time (McCartney et al., 2003). Either isolation of the pathogen using selective media or biochemical, chemical, and immunological analyses are used as conventional methods to detect and identify the pathogen. These methods are enabled to recognize the existence of pathogens, but they require considerable time and serious labor concentration with various lab techniques (Lievens et al., 2005). Polymerase chain reaction (PCR) based assays have been demonstrated to detect the pathogens more specific and spend less time-consuming than the conventional techniques (Sankaran et al., 2010). The stretch of the best for management of diseases depend not only on the existence of pathogens but also on the pathogen inoculum density. Therefore, quantification of the pathogen density has been concerned as an important aspect of a part of disease control management (Bonants et al., 2004). Conventional quantification methods were considered in low efficacy and non-specific relatively, while quantification based on PCR techniques and quantification real-time PCR (qRT-PCR) provides a precise result of the pathogen diagnosis. Unlike end-point PCRs, It is possible to observe the specific time of the first reaction via specific primers. Because, qRT-PCRs carry out reaction that amplify products during each PCR cycle. Template quantification was estimated during the exponential phase of the reaction is highly specific (Capote et al., 2012; Schena et al., 2013).
In this study, we developed the strawberry Fusarium wilt pathogen specific qRT-PCR system to increase accuracy of FOF quantification in both lab and field condition. The system demonstrated that correlation between the pathogen density and the disease incidence/severity. Therefore, this system will contribute to evaluate the disease occurrence prediction in commercial field and assessment of fungicide or soil fumigation efficacy.
Materials and Methods
Primer design and condition of qRT-PCR
Han-Skippy gene region was used to design a primer set, the gene have 239 base pair (bp) inter-retrotransposon region and showed only F. oxysporum species was reported by Suga et al. (2013) (Supplementary Fig. 1). The gene sequences were analyzed by IDT SciTools OligoAnalyzer 3.1 software (Integrated DNA Technologies Inc., Coralville, USA) with length of amplicon was designed less than 200-bp and 3′ end of primers had unique nucleotide sequences. Amplification proceed of qRT-PCR added to 25 ul of SYBR-Green RealTime PCR Master Mix (Toyobo, Osaka, Japan) and 2 ul of each primer respectively. DNA templates of the pathogen which was extracted by CTAB method (Porebski et al., 2003), FOF strain F9 isolates (Cha et al., 2016) were used with an approximate concentration of 5 ug/μl and serial dilution from 650 ng/μl to 0.065 ng/μl 10-fold for standard curve determined on a Nano Drop ND2000C spectrophotometer (Thermo scientific, Waltham, MA, USA) and added distilled water up to 50 μl in 0.2 ml 8-tube PCR Strips and Caps (Bio-Rad, Hercules, CA, USA). The PCR cycling condition set at 95°C for 1 min and 40 cycles at 95°C for 15 s, 51°C for 15 s, 72°C for 45 s and melting curve was generated with 60 cycles from 65°C to 95°C increasing at each cycle by 0.5°C via CFX384 Real-Time System (Bio-Rad).
DNA extraction form soil samples
Fast DNA SPIN kit for soil (MP, Santa Ana, CA, USA) was used, 0.5 g–0.7 g soil sample put into Lysing Matrix E tube, added 978 μl SPB buffer and 122 μl MT buffer. Homogenization was performed via mini-bead beater-1 (BIOSPEC, Bartlesvile, OK, USA) for 10 min in 6.0 speed. Homogenized sample centrifuged at 13,000 rpm for 10 min before supernatants were transferred into a new 1.5-ml tube and added 250 μl PPS buffer then mixed by hand 10 times. The tube was incubated for 10 min at room temperature. After incubator, the sample was centrifuged at 13,000 rpm for 5 min to pellet precipitate. Supernatant was transferred into a new 15-ml falcon tube and added 1 ml Binding Matrix solution, mixed the solution and incubated at room temperature at 3 min. The supernatant discarded and 700 μl of the remainder solution transferred into a SPIN Filter tube and centrifuged at 13,000 rpm for 5 min and discarded the flow-through of the SPIN filter tube for two times. Added 500 μl prepared SEWS-M solution into the SPIN Filter tube and gently resuspended the pellet by pipetting. The tube was centrifuged at 13,000 rpm for 5 min, discarded the flow-through and centrifuged again with the same condition for remove the residual ethanol. The SPIN Filter transferred into a clean 2.0-ml catch tube and dried for 5 min at room temperature. Added 100 μl of DES buffer to the SPIN Filter tube and gently resuspend the pellet by pipetting then incubated at 55°C for 5 min. Volume of the DNA sample was measured, added 2 to 2.5 volumes of cold 100% ethanol, mixed well and placed at −20°C for 20 min, then centrifuged at 13,000 rpm for 15 min. The supernatant was carefully discard then add 1 ml 70% ethanol and pipetting the solution. The tube was centrifuged at 13,000 rpm for 1 min, abandoned the mixture carefully after dried at room temperature at least 20 min. The pellet was resuspended in 20 μl TE buffer then keep at −80°C until use.
qRT-PCR with standard soil samples
Accuracy of the primer set was examined with FOF (strain F9) DNA extracted from soil media and strawberry cultivation raw soil. Both types of soil were sterilized two times at 121°C for 15 min. The pathogen spore density was measured by Haemacytometer (SUPERIOR, Lauda-Konigshofen, Germany) under a microscope and calculated spore density to mix into in the soil media and raw soil, different density of the pathogen spores was amended (102, 104 and 106) with three replications. Chromosomal DNA per ng based on average weight of nucleotides, which is 650 Dalton per nucleotide. Median total length of F. oxysporum was 51.7572 Mb (van Dam et al., 2016) which indicated that F. oxysporum has 4474 unit per ng of DNA. Soil DNAs were extracted from the pathogen inoculated soils by Fast DNA SPIN kit (MP). The template DNA adjusted 5 ng/μl, added to 25 μl of SYBR-Green Realtime PCR Master Mix (Toyobo), 2 μl of the primer pair and up to 50 μl distilled water. The PCR cycling condition was same above-mentioned.
Correlation between the pathogen density in soil and disease occurrence
FOF F9 strain was grown on potato dextrose agar (PDA: potato dextrose 24 g, agar 20 g per l) at 27°C for 7 days. The mature fungal spores were collected by scarping with sterilized water then transfer into a 1.5 ml tube. The spores were adjusted different densities (107, 105, 103 and 101 cfu) via hemocytometer. Soil was added to plastic pots (22 × 60 × 18 cm), and 50-days-old strawberry (cv. Sulhyang) were planted in each pot. The treatments were arranged in a complete randomized block design and replicated five times. Plants were incubated in a growth chamber (25°C; 16 h light/8 h dark cycle), and the severity of disease were observed every 7 days up to 35 days after planting. The severity of the disease was evaluated using a 0–5 scale, every week: 0, healthy; 1, 1 to 3 leaves rolled and yellowed; 2, 3 to 4 leaves rolled and deformed; 3, chlorosis and early plant wilting; 4, necrosis and entire plant wilting; and 5, dead or nearly so. The results were statistically analyzed by Tukey’s HSD test (with significance set at p = 0.05) using Sigma Plot ver. 11.0 (Systat software INC., San Jose, CA, USA). Quantitative detection based on qRT-PCR was conducted to calculate the population in three times (0, 3 and 5 weeks).
Evaluation of eco-friendly Fusarium wilt disease control agents
Strawberry and the pathogen preparation were same as described above. Four different eco-friendly the disease control agents such as Phoscraft, Curamin (BVB SUBSTRATES, De Lier, The Netherlands), NaDCC (Kim et al., 2015), and Streptomyces griseus S4-7 (Cha et al., 2016) were evaluated their efficiency in the disease prevent. The agents were treated as Phoscraft (1000 times dilution), Curamin (2000 times dilution), NaDCC (100 ppm), 106 cfu of S4-7 and the pathogen (F9 strain) was inoculated at 105 cfu in each treatment. After 5 weeks of the treatment, the samples randomly collected from 3 pots of each treatment. The soil sampling for qRT-PCR and severity of the disease were same way to establish variation of the population in soil and effect of disease pathogenicity.
Efficiency of S4-7 on Fusarium wilt disease in nursery plantation and soil sterilization on the pathogen density in soil
To evaluate efficiency of S. griseus S4-7 on the pathogen suppression in field condition, strawberries (cv. Sulhyang) in two greenhouses which place in JinJu, Gyeongnam province were employed. Each greenhouse was divided into two sections: treatment and control areas replicated 3 times for each section, one of replication consist of 15 mother plants and their offspring plants were only treated in treatment area with 106 cfu of S4-7 in Jun 9th and Jul 6th, 2016 and soil sampling also carried out at same days. Disease incidence of the Fusarium wilt was investigated 5 times. Population of the pathogen in soil was measured via qRT-PCR with the developed primer set. Soil sterilization as solar heat method was testified in main cultivation area of the two greenhouses above mentioned. Soil samplings were conducted at before sterilization and after sterilization with 3 replications.
Results
Sensitivity and detection limit of the qRT-PCR primer
Detection of Fusarium wilt pathogen with end-point PCR, which amplified a transposable element called Han-Skippy was reported in a previous study (Suga et al., 2013). We developed more accurate and specific qRT-PCR system to detect the strawberry Fusarium wilt pathogen. Size of qRT-PCR amplicon was designed as 72 bp length (Table 1). The primer pair meet standard requirement such as number of hairpin structure, self and hetero dimer structure via IDT SciTools OligoAnalyzer 3.1 (Integrated DNA Technologies Inc.). Standard curve with the pathogen genomic DNA was generated with different concentration of gDNA from 650 ng/μl to 0.065 ng/μl as 10-fold serial dilution. Threshold cycle (Ct) value of each concentration of DNA showed 3.3 differentiation in Ct. R square value of the standard curve was 0.991 and melting curve indicated that generated amplicon with the primer showed clear product (Tm: 80.5°C) (Supplementary Fig. 2). The primer pair specially reacted with only FOF strain F9 but not others F. oxysporum (data not shown, please see discussion). To check, detection limit in both soil media and raw soil, the F9 spores were inoculated in both soils at 106, 104 and 102 cfu/g of soil concentration. As result, the qRT-PCR with 106 and 104 cfu/g of soil samples detected the pathogen level as 106, 105 cfu/g of soil. However, the qRT-PCR failed to distinguish between 102 and 103 pathogen densities in soils (Fig. 1). Taken together, detection limit of the pathogen in soil with the qRT-PCR primer is between 103 and 104 cfu/g in soil condition.
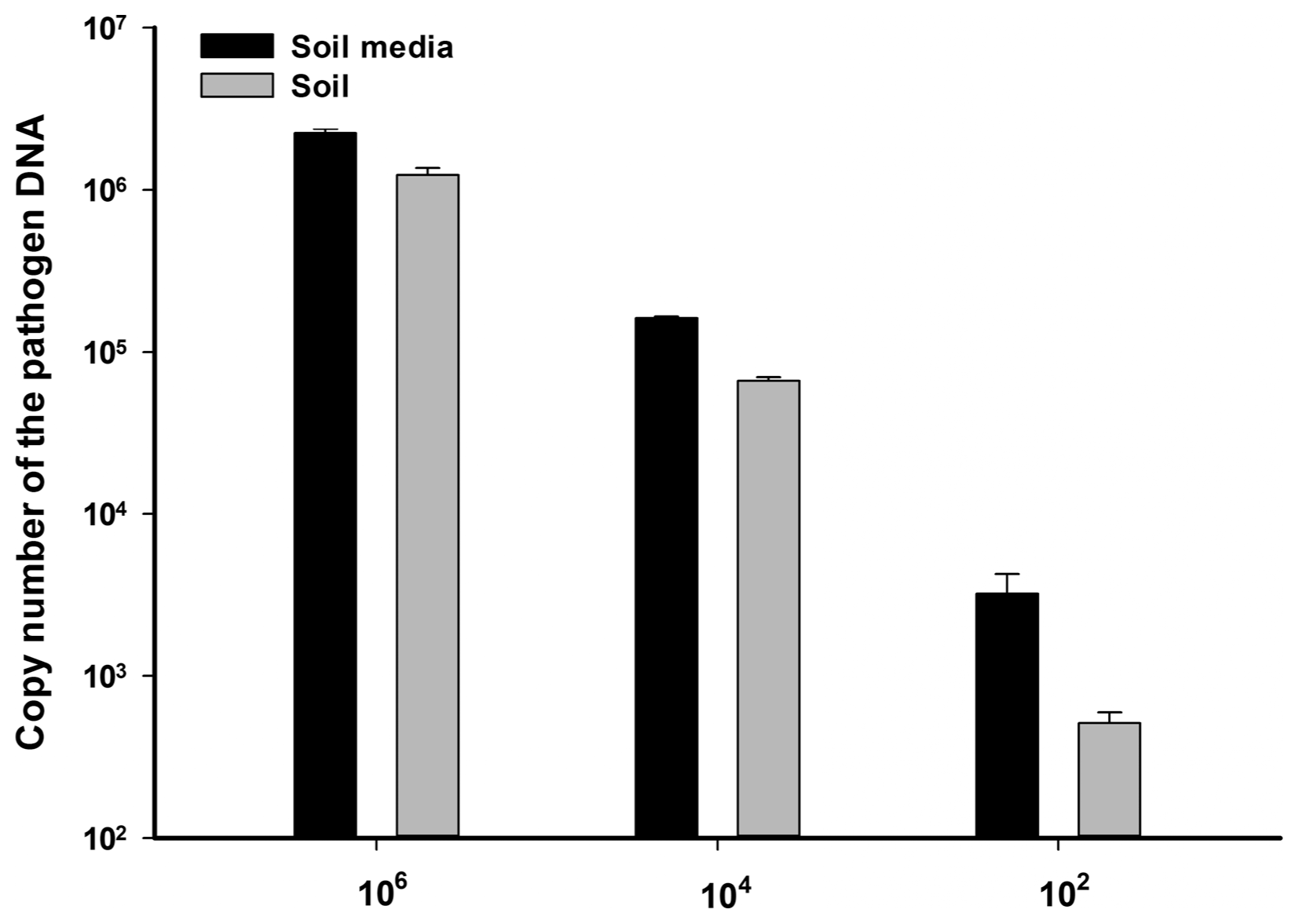
Detection limit assay of the pathogen in soil level. Different concentration (macroconidia 106, 104 and 102 cfu/gram of soil) was inoculated in two soil types (soil media and raw soil). Each soil was autoclaved two times, the pathogen concentration confirmed both hemocytometer and calculation of pathogen number based on whole genomic DNA weight of Fusarium oxysporum f. sp. fragariae.
Correlation between the pathogen density and the disease occurrence
Correlation between the disease occurrence and the pathogen density in soil was investigated with the qRT-PCR. When the pathogen existed over 105 cfu/g of soil, the Fusarium wilt disease clearly occurred. The pathogen densities at 105 and 107 cfu/g of soil showed high score of the disease severity about 4 and 5 in each treatment respectively (Fig. 2A). Regardless soil sampling times, the qRT-PCR detected the pathogen densities as 106 cfu when the pathogen spore was inoculated at 107 cfu/g of soil. However, the pathogen untreated control, 101 and 103 cfu/g of soil treated samples were detected as density of 104 level during the sampling times. This result matched with the detection limit of the pathogen both soil media and raw soil of the qRT-PCR (Fig. 1). Both 105 and 107 of the pathogen maintained the density above 105 cfu/g of soil during experimental period (Fig. 2B).

Correlation of the Fusarium wilt disease occurrence and the pathogen density in soil. Total 4 different densities of the pathogen were treated in pot which have been planted strawberry plants. (A) Disease index was evaluated 5 weeks after planting and severity of the wilt were evaluated on a 0–5 scale; 0 = healthy, 1 = 1–3 leaves rolled and yellowed, 2 = 3 leaves rolled and deformed, 3 = chlorosis and early plant wilting, 4 = necrosis and entire plant wilting, and 5 = dead. (B) Soil samples were amplified by qRT-PCR and calculate population of the pathogen. Different alphabet on the bar that mean is statistically different in Tukey HSD with p = 0.05.
Efficiency of eco-friendly agents against the Fusarium wilt disease
Currently, no chemical fungicide is registered to prevent strawberry Fusarium wilt disease. However, some eco-friendly agents are used by farmers, such as Phoscraft, Curamin, and NaDCC. A microbial agent, S. griseus S4-7, was reported exceptional antifungal activity against the strawberry Fusarium wilt pathogen (Cha et al., 2016). Five weeks after the treatments, Phoscraft, Curamin and NaDCC and positive control (F9, 105 cfu/g of soil) showed severe the disease occurrence, the disease severity scores were 4. However, S4-7 treated strawberry plants were showed the disease severity less than score 2 (Fig. 3A). The pathogen densities in each treatment of the eco-friendly agent were detected via the qRT-PCR. In untreated control, the qRT-PCR showed background level of reaction and in only pathogen treatment (105 cfu) showed 1.5 × 105 (Fig. 3B). Phoscraft, Curamin and NaDCC treatments failed to reduce the pathogen density in soil. However, S4-7 successfully reduced the FOF population in soil condition. The qRT-PCR result in S4-7 treatment showed less than 104 cfu of the pathogen 5 weeks after the treatment (Fig. 3B).
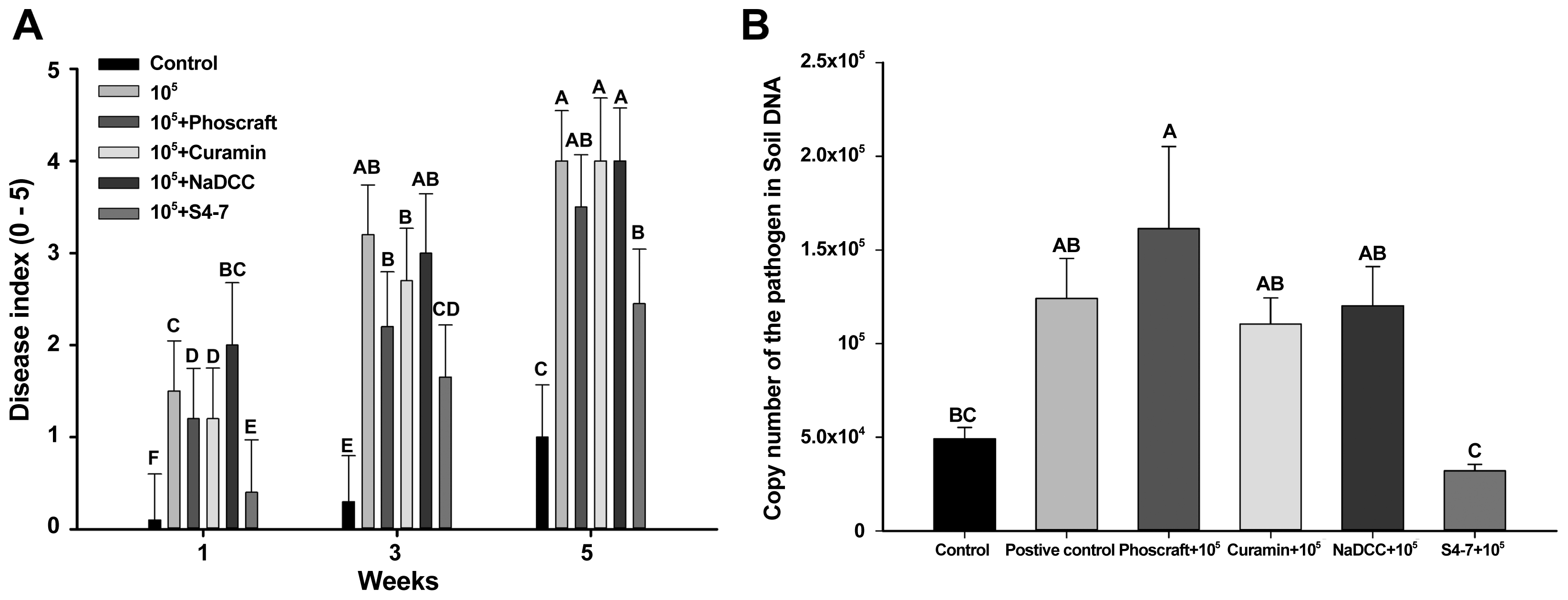
Evaluation of effectiveness of eco-friendly fungicides on the wilt disease. The pathogen was inoculated 105 cfu/gram of soil, the agents were treated only one time when the strawberry planted. (A) Disease index was performed 5 weeks after inoculation and severity of wilt were evaluated on a 0–5 scale; 0 = healthy, 1 = 1–3 leaves rolled, and yellowed, 2 = 3 leaves rolled and deformed, 3 = chlorosis and early plant wilting, 4 = necrosis and entire plant wilting, and 5 = dead. (B) At 5 weeks, soil DNA were extracted and amplified with the qRT-PCR primer set to calculate population of the pathogen. Different alphabet on the bar indicated statistically different with Tukey HSD (p = 0.05).
During nursery period of strawberry, Fusarium wilt disease occurrence showed less than 20% until early of July in year of 2016. However, the disease incidence was rapidly increased up to 60% between July 21 to August 4, 2016 in untreated control field (Fig. 4A). The pathogen population in untreated soil responsibly increased from background level to 105 cfu/g of soil (Fig. 4B), which is important pathogen density level as described above (Fig. 2). Interesting, there is two weeks interval between the pathogen density reached at the level of 105 cfu (July 6) and the disease incidence increased (July 21). In S4-7 treated strawberry showed minimum level of the disease occurrence (Fig. 4A) and the pathogen density in soil was decreased (Fig. 4B).
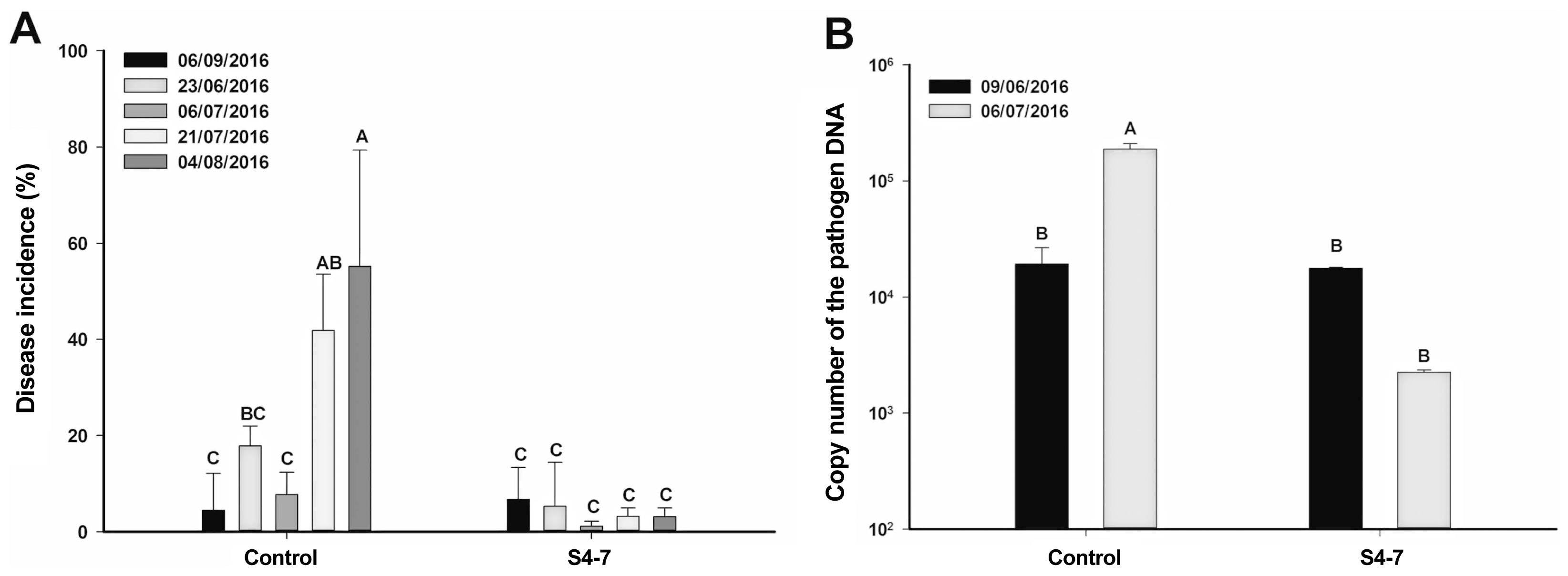
Biocontrol effect of S4-7 in Fusarium wilt disease in the strawberry nursery plantation. S4-7 (105 cfu/gram of soil) was treated twice, Jun 9th and Jul 6th, and soil samplings were carried out. (A) Disease incidence of the wilt disease was investigated 5 times from June to August. (B) The pathogen densities in soils.
Discussion
Several soil-borne pathogens cause serious economic losses worldwide. Therefore, rapid and accurate detection of pathogen is an important factor for optimal and proper control of these diseases. PCR primers and techniques have been developed for detection of Phytophthora nicotianae and P. cactorum (Li et al., 2011), Verticillium dahliae (Bilodeau et al., 2012) in strawberry. However, detection methods for the most serious disease, which is Fusarium wilt caused by F. oxysporum f. sp. fragariae, has not been developed yet. Additionally, we do not have reliable information about correlation between the disease severity and the pathogen population in soil of the disease. Therefore, in this study, specific primer pair based on specific nucleotide sequence region in FOF were developed that can detected in a various type of samples such as genomic DNA, soil media DNA and soil DNA with the different inoculum densities. Furthermore, correlation between population of the pathogen or spore numbers and the disease severity was revealed based on the qRT-PCR system.
The Fusarium detection primers were produced that numerous combination of primers based on several transposable elements such as Hop, Hor-net1, Foxy, Fofra and Skippy (Anaya and Roncero, 1995; Chalvet et al., 2003; Hua-Van et al., 2000). The specific genetic markers were investigated for development to detect the pathogen in formae speciales level via random amplified polymorphic DNA (RAPD) and Inter-retroelement amplified polymorphism PCR (IRAP-PCR) were reported (Lievens et al., 2008; Suga et al., 2013). However, the previous reports were not measured quantification or density of the pathogen in soil. Additionally, those techniques required a great deal of time and labor.
qRT-PCR diagnoses have been developed for quantification of other Fusarium spp. (Filion et al., 2003), F. oxysporum (Jimenez-Fernandez et al., 2010) and formae speciales of F. oxysporum in plant or soil samples (Lievens et al., 2006; Pasquali et al., 2004; Zambounis et al., 2007). Suga et al. (2013) reported a specific primer set for PCR which can detect formae speciales level of F. oxysporum. The primer set was designed based on sequences of two transposable elements Han and Skippy, which existed only F. oxysporum strains or races. The transposable elements were reported from F. oxysporum II5, F. oxysporum PHW808 F. oxysporum f. sp. melonis and F. oxysporum f. sp. lycopersici (Suga et al., 2013). We also developed the qRT-PCR primer set based on amplified sequences of the transposable elements via Genbank database. The standard curves and the melting curves with the qRT-PCR primers were demonstrated the primer set had specificity, reliable detection limit and similar detection pattern between pure DNA and soil extracted DNA.
Epidemiology of plant pathogens and levels of disease severity are closely associated by variation in virulence of the pathogen. The pathogen density and virulence are typically influenced to be positively in aspect of absorbing more nutrient from the host (Barrett et al., 2009). More the pathogen grows and more produce transmissible propagules following the time (Frank, 1996). Strawberry Fusarium wilt disease system showed that interaction between the pathogen density and rate of the diseases incidence and severity raise rapidly at 105 cfu pathogens were existence in soil. In nursery plantation, outbreak of Fusarium wilt was not happened when the population was detected under 105 cfu before July 6, 2016. On the other hand, initiation of the disease epidemic started once the pathogen was detected over 105 cfu population. Below copy number of 103 of the pathogen, the disease was not occurred, and the primer set also failed to detect, due to the detection limit of the qRT-PCR with soil samples. The detection limit may be caused by several problematics such as presence of polymerase inhibitors in soil which cause by reduction of the qRT-PCR sensitivity. This lead to hardness of initial amplification qRT-PCR cycling (Scarlett et al., 2013). Nevertheless, establishment of relation of the density with induced the disease outbreak was accomplished in this study and also the primers could be used to quantify a single fungal specie in soils. Although various chemical fungicides have been tried to control strawberry Fusarium wilt disease, no reliable fungicide has been registered yet. Therefore, in this study, efficiency of eco-friendly fungicides included Phoscraft, Curamin, NaDCC, and S4-7 were evaluated in terms of the disease control and the pathogen population with the qRT-PCR primer set. A microbial agent, S4-7 was reported by Cha et al. (2016). The agent was isolated and characterized from a suppressive soil against Fusarium wilt and was demonstrated exceptional the disease control ability in both lab and field conditions. Phoscraft, Curamin and NaDCC failed to control the Fusarium wilt disease and the qRT-PCT results revealed the pathogen densities in each treatment showed above 105 cfu. However, S4-7 treatment showed the lowest disease index. Not only the wilt disease suppressed by S4-7, but also the pathogen population in soil was decreased. Those finding also reproduced in strawberry nursey fields. Application of the diagnosis protocol was not only confirmation of control the disease but also contributed reviewing of agronomical control procedure to observe variation of the pathogen density through calculation of the biomass in the soil.
Supplementary Information
Acknowledgments
This study was supported by the Technology Development Program for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea (Project No. 315004-5).