Bursaphelenchus xylophilus, commonly known as pine wood nematode (PWN) is a causal agent of pine wilt disease (PWD) that causes extensive damage to forest ecosystem. The nematodes uses Monochamus beetles as vectors to infect mainly Pinus species and causes xylem dysfunction in their host, resulting in fatal wilting symptom (Mota and Vieira, 2008). In general, controls of PWD rely on aerial application of insecticides to prevent vector transmission. In addition, injection of chemicals, such as broad-spectrum of nematicides including avermectin and abamectin into the tree trunk is widely put in practice (Lee et al., 2003; Takai et al., 2003). Use of synthetic pesticides and chemicals raises great environmental concerns towards non-target organisms and human health (Jayaraj et al., 2016). In line with this, the nematicidal activities of bacteria have been regarded as alternative sources of ecofriendly biological controls of PWN (Eljounaidi et al., 2016; Zheng et al., 2016a).
Raoultella ornithinolytica strain MG (R. ornithinolytica MG hereafter) is a gram negative bacterium recently isolated from mountain-cultivated ginseng (MG) plants in South Korea (Khan et al., 2017). We found that ethyl acetate extract (EtOAc) of R. ornithinolytica MG culture has nematicidal activity against PWNs. To identify and understand potential genome-encoded factors that are associated with the nematicidal activity of this bacterium, here we set out to determine the draft genome of R. ornithinolytica MG. Furthermore, we predicted genes involved in production of compounds including secondary metabolites, which are potentially contributing to nematicidal activity.
The R. ornithinolytica MG strain was identified based on morphological attributes and 16S rDNA sequencing in our previous study (Khan et al., 2017). R. ornithinolytica MG strain was deposited in the Korean Collection for Type Cultures (deposition number: KCTC13338BP). The genomic DNA of R. ornithinolytica MG was extracted from an overnight bacterial culture grown at 37°C at 200 rpm in nutrient broth [peptone 0.5% w/v (Becton, Dickinson and Company, Franklin Lakes, NJ, USA), yeast extract 0.5% w/v (Duchefa Biochemie, Haarlem, The Netherlands), agar 0.5% w/v (Becton, Dickinson and Company), NaCl 0.5% w/v; pH 6.8]. Standard phenol-chloroform method was used for genomic DNA extraction (He, 2011; Maniatis et al., 1982).
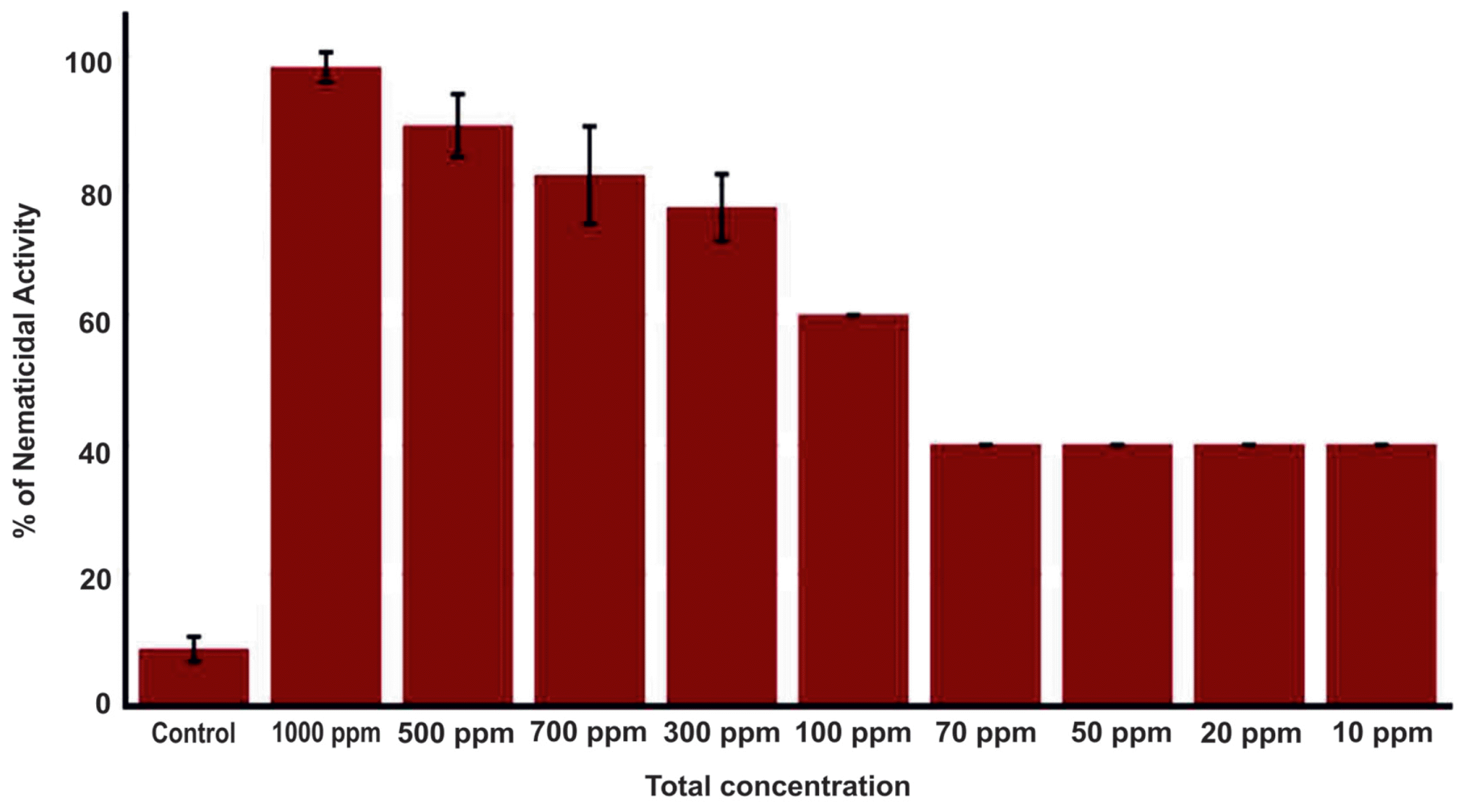
The 500 μl of R. ornithinolytica MG culture grown for 12 h was transferred to the 500 ml of Tryptic Soy Broth (TSB) (Becton, Dickinson and Company) in 1000 ml Erlenmeyer flask and incubated for 5 days at 30°C with 150 rpm. After 5 days, an equal volume of EtOAc (500 ml) (Duksan Reagents, Ansan, Korea) was added to the broth culture and mixed well by shaking vigorously, followed by sonication for 30 min. The EtOAc mixture was incubated for overnight on orbital shaker at 130 rpm and allowed to stand for 2 h. The top clear phase was transferred to a round flask and condensed in a rotary evaporator at 40°C (Paul et al., 2013). The crude EtOAc extract was dissolved in a solution of 3% Dimethyl sulfoxide (DMSO) (Junsei, Tokyo, Japan) and then dried completely. Approximately 100 B. xylophilus nematodes (mixture of L2, L3 and L4/adult stages) were taken in 90 μl of autoclaved distilled water in each well and the nematicidal activity of total metabolites of EtOAc extracts were tested by adding 10 μl at various concentrations (1000 ppm to 100 ppm) with six technical replications per concentration in a 96-well plate on different days, and 3% DMSO was used as control. The plates were then incubated for three days at 20°C and the numbers of alive/dead nematodes were counted at 12, 24 and 48 h under a microscope and the percentage of nematode mortality was calculated (Choi et al., 2006; Guo et al., 2017).
Genome sequencing was performed with a PacBioRS II (Pacific Biosciences, Menlo Park, California, USA) at Theragen Etex Co., Ltd (Suwon, Korea). Raw sequences were assembled using the hierarchical genome assembly process (HGAP3) de novo assembler (Chin et al., 2013), and the resulting genome was annotated by NCBI prokaryotic genome automatic annotation pipeline (PGAAP) (Tatusova et al., 2016). The draft genome sequence of R. ornithinolytica strain MG was deposited in GenBank under the accession number CP017802.
Secondary metabolite gene clusters in the R. ornithinolytica MG genome were predicted using anti-SMASH search (Weber et al., 2015). Comparative analysis of R. ornithinolytica strains were performed and visualized with CLC Genomics Workbench (v9.0.1) (QIAGEN, Aarhus, Denmark, http://www.clcbio.com).
Ethyl acetate extract of the bacterial culture was tested for its potential nematicidal activity by measuring proportion of nematodes that are killed by addition of varying concentration of extract. Compared to the control (3% DMSO), the extract showed about 40% nematicidal activity at concentrations ranging from 10 to 70 ppm (Fig. 1). Nematicidal activity started to increase at higher concentration (over 100 ppm) and peaked at 1000 ppm nearly annihilating B. xylophilus population under treatment. These results clearly suggest the potential of R. ornithinolytica MG as a source of compounds having activity against B. xylophilus.
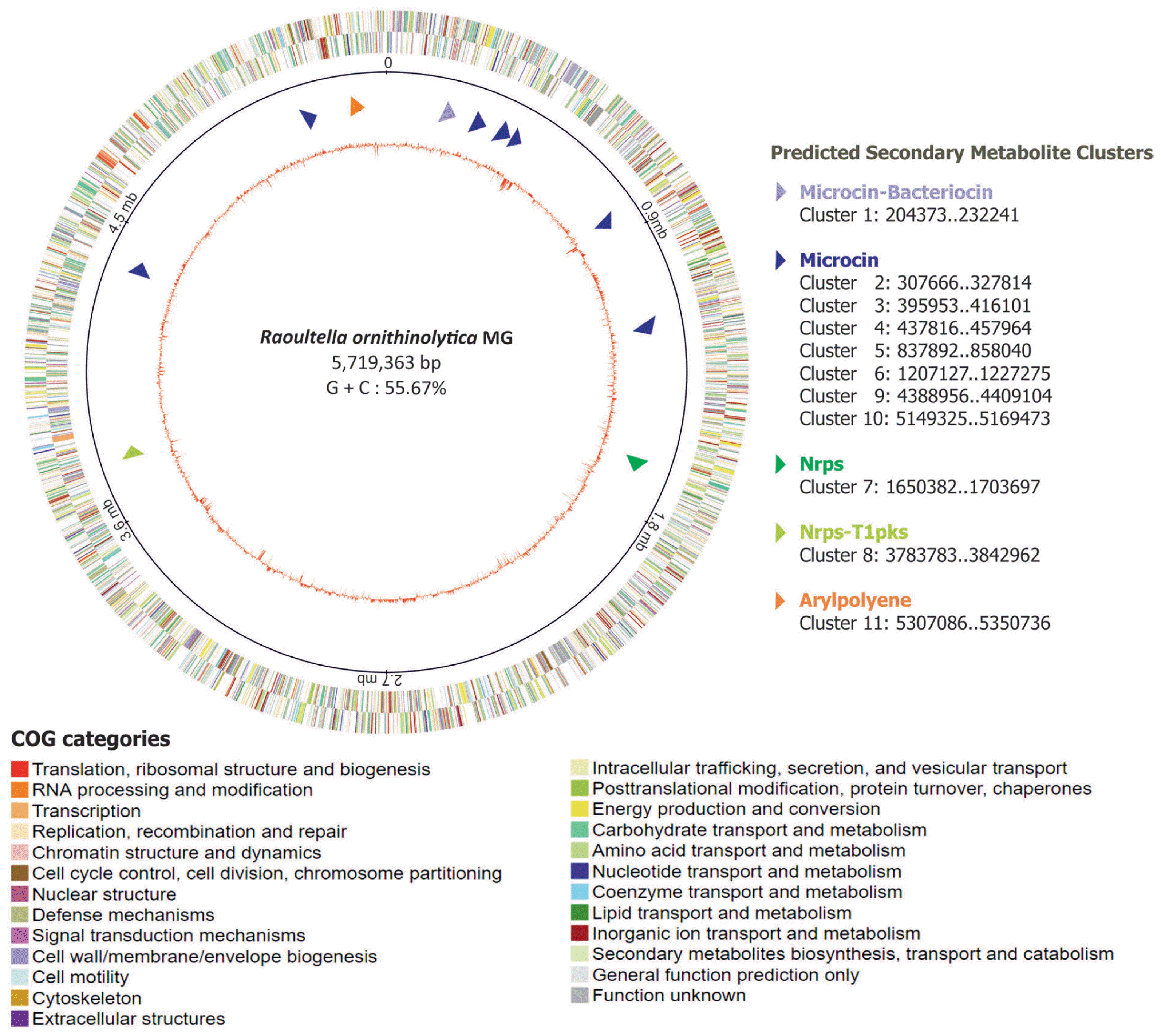
Given such activity of R. ornithinolytica MG against B. xylophilus, we carried out sequencing of bacterial draft genome to provide genomic basis for pinpointing genetic components that endow the bacteria with nematicidal activity. Subsequently, a total of 1,066,832,850 bp were sequenced in 114,707 reads, which were assembled using the hierarchical genome assembly process (HGAP3) de novo assembler, resulting in average coverage depth of 182x. This final assembly of the entire R. ornithinolytica MG was determined to be 5,719,363 bp with G + C content of 55.67% with 3 contigs. Among the contigs, the largest one was 5,499,520 bp long, accounting for about 96% of entire genome and the remaining contigs were 218,906 bp and 937 bp long, respectively.
A total of 5,322 genes in the draft genome of R. ornithinolytica MG was annotated by NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAAP). These includes 5,132 coding genes, 128 RNA genes and 62 pseudogenes (Table 1). Among 5,132 coding genes, 4,445 coding genes (86.61%) were assigned putative functions based on functional annotation. The subsystem features were annotated by RAST server (Aziz et al., 2008) and visualized by using CiVi web based visualization tool (Overmars et al., 2015) (Fig. 2).
In comparison, the complete genome of R. ornithinolytica strain B6 (Shin et al., 2013) from public database (NCBI Accession: CP004142) is composed of a circular chromosome of 5,398,151 bp with 55.9% GC content. A total of 4,909 coding genes, 79 tRNAs, and 25 rRNAs were annotated, and 4,070 coding genes (82.90%) were predicted to have putative functions. This implies that the R. ornithinolytica strain MG possesses larger genome encoding more number of genes than strain B6.
In order to mine the nematicidal factors in R. ornithinolytica MG, we took two approaches. In the first approach, we attempted to predict and catalogue gene clusters that are involved in secondary metabolite biosynthesis, potentially contributing to nematicidal activity against B. xylophilus. The anti-SMASH search for secondary metabolite biosynthesis gene clusters on the genome of R. ornithinolytica MG showed that there are 11 biosynthetic gene clusters related to the production of different types of secondary metabolites including Microcin (7 clusters), Microcin-Bacterocin (1 cluster), Nrps (1 cluster), Nrps-T1PKS (1cluster), and Arylpolyene (1 cluster) (Table 2, Supplementary Fig. 1, 2). The presence of these gene clusters was also observed in the genomes of R. ornithinolytica strains B6 (2,3-Butanediol-producing bacterium isolated from oil-contaminated soil) (NCBI Accession: CP004142) (Shin et al., 2013), R. ornithinolytica strain S12 (Lignin-degrading bacterium isolated from forest soil) (NCBI Accession: CP010557) (Bao et al., 2015) and A14 (Isolated from animal feces) (Leung et al., 2016) (NCBI Accession: CP008886.1) (Table 2). Interestingly, only three gene clusters such as Nrps (1 cluster), Arylpolyene (1 cluster) and Bacteriocin (1 cluster) were found on the genome of R. ornithinolytica strain S12, while the remaining two isolates contained the 11 clusters. In general, Microcin and Bacteriocin mimics siderophore so as to target and eliminate the enteric pathogens. The Arylpolyene gene clusters are similar to carotenoids. The Nrps cluster genes are known for their biological activities and pharmacological properties (Agrawal et al., 2016).
In the second approach, we examined presence/absence of 42 known nematicidal factors (Zheng et al., 2016b) in the R. ornithinolytica MG genome using BLASTP search (Altschul et al., 1990) (Supplementary Table 1). This search revealed presence of 9 potential nematicidal factors encoded in the genome. These include homologs of amidophosphoribosyltransferase (Xia et al., 2011), Calcium-transporting ATPase (Fan et al., 2007), Chitinase (including basic and endochitinases) and Serrawettin W2 (Fragment) (Zheng et al., 2016b) (Table 3).
To rule out the possibility that nematicidal activity of R. ornithinolytica MG is mediated by avermectin, we searched the genome for orthologous genes in avermectin (anti-nematode agent) biosynthetic gene (18 genes) (Kim et al., 2016). This showed that 5 key genes including AveR (Transcriptional regulator), AveC (Modification: spiroketal moiety C22-23 dehydration), AveE (Modification: Furan ring reduction), AveBVI and AveBVII (Modification: Addition of sugar moiety) are missing (Ikeda et al., 1999) (Supplementary Fig. 2), suggesting that the observed nematicidal activity of the bacterium cannot be attributed to avermectin.
Although genome sequences of a few R. ornithinolytica strains including the strain B6 were deposited in NCBI, the genome sequence of R. ornithinolytica MG is the first to be reported with nematicidal activity against B. xylophilus. Considering the nematicidal activity, the availability of R. ornithinolytica MG draft genome sequence would provide genetics and genomic basis in understanding and improving its application to controlling PWD.



 PDF Links
PDF Links PubReader
PubReader Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print






